To the editor:
Recent large-scale sequencing studies report recurrent somatic mutations in the blood of elderly individuals in genes previously linked to clonal expansion of hematopoietic stem cells.1-4 Particularly for DNMT3A and TET2, a steep age-associated increase in the prevalence of somatic mutations is observed from middle age onward.2-4 In addition, prospective analyses performed in predominantly middle-aged individuals show an increased risk for all-cause mortality for carriers of such mutations as compared with noncarriers.3,4 Jointly, these data suggest a rapidly increasing vulnerability among the elderly for adverse health effects associated with clonal expansion of hematopoietic stem cells. However, prospective data on elderly somatic mutations carriers are scarce. We therefore investigated the association between all-cause mortality and carriership of somatic mutations in genes linked to clonal expansion of hematopoietic stem cells in a large elderly subsample (N = 864, 80 years and older) derived from 2 large-scale community-dwelling Dutch cohort studies.5,6
For the present study, we investigated whole-blood–derived genomes of 646 individuals of 80 years and older from the Rotterdam Study5 (RS; mean age at inclusion, 84.6 years; range, 80.0-105.8 years; supplemental Appendix 2, available on the Blood Web site) and 218 individuals of 89 years and older from the Leiden Longevity Study6 (LLS; mean age at inclusion, 94.0 years; range, 88.9-103.4 years; supplemental Appendix 2). Jointly, this elderly subsample consists of 597 participants aged 80 to 89 years and 267 participants aged over 90 years, which is twice the number of participants for the respective age categories as compared with any other study previously conducted on this topic.2-4 Selected elderly participants of the RS and LLS were followed for all-cause mortality for a median 8.7 and 9.2 years, respectively, which was sufficiently long to identify the age at death of 81.3% and 93.6% of the respective study subsamples. Methods of DNA sequencing and analysis are described in supplemental Appendix 3. The ethical committees of the involved institutes approved both studies, and written informed consent was obtained from all study participants.
Using this unique cohort of sequenced oldest old, we first set out to confirm the recurrent acquisition of somatic mutations in genes linked to clonal hematopoiesis in the blood of highly aged individuals. For this, we curated a list of 15 genes (supplemental Appendix 4) reported to harbor recurrent somatic mutations in the blood of normal individuals in any of the large-scale sequencing studies conducted to date.2-4 Thus, identified genes were analyzed for putative somatic mutations according to the gene-specific inclusion criteria set by Jaiswal et al (supplemental Appendix 4).4
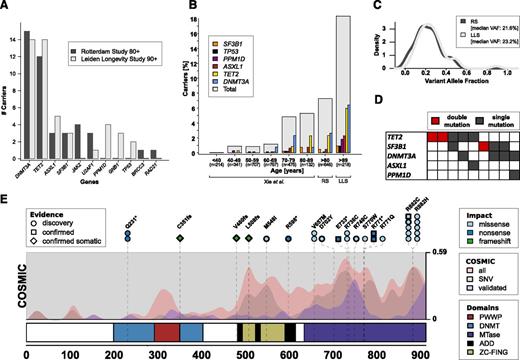
The mutational analysis identified 39 (6.0%) and 40 (18.3%) unique carriers of, respectively, 42 and 46 mutations for the RS and LLS elderly subsamples, respectively, predominantly in DNMT3A and TET2 (Figure 1A; supplemental Tables 1-2). The observed prevalence of somatic mutations in genes linked to clonal hematopoiesis in the RS and LLS elderly subsamples is consistent with the age-associated increase observed by Xie et al (Figure 1B).2 Our observation thus confirms the age-associated increase of detectable somatic mutations in genes previously linked to hematopoietic malignancies reported by Xie et al and extends this observation up to the highest ages.2
Characterization of identified variants in blood of the RS and LLS elderly subsample. (A) Mutations in genes previously linked to hematopoietic malignancies. Barplot of the number of individuals carrying a mutation, split by genes and study. Note that only 11 of the 15 investigated genes had a mutation (see supplemental Appendix 4 for a complete list). (B) Prevalence of carriers of somatic mutations stratified by age category using data of Xie et al2 and the observations in the RS and LLS elderly subsample. (C) Distribution of VAFs of the identified mutations. (D) Comutation plot of carriers with 2 independent mutations. (E) Overview of mutations in DNMT3A identified in the RS and LLS elderly subsample. Variants are annotated at the top with color coding to indicate the impact and a shape to indicate the types of follow-up experiments. Circles indicate mutations detected in our sequencing data; squares indicate mutations also validated by Sanger sequencing; diamonds indicate mutations also validated by Sanger sequencing and absent in an IBD2 matched sib, that is, confirming somatic variations. Mutations identified in multiple carriers are indicated with stacked annotations and those having bold borders were identified in the LLS. Missense variants are only included whenever they are present on a curated list of recurrently reported variants in the Catalogue Of Somatic Mutations In Cancer (COSMIC)7 assembled by Jaiswal et al4 . Domains: ADD, histone-binding domains; DNMT, DNA methyltransferase interaction domain; MTase, methyltransferase domain; PWWP, conserved DNA binding domain; ZC-FING, zinc finger domains. COSMIC, densities of somatic variants identified in hematopoietic or lymphoid tissue collected by the Catalogue Of Somatic Mutations In Cancer (COSMIC)7 database: all small variants (red), missense single nucelotide variants (SNVs) (gray), small variants confirmed to be of somatic origin (blue).
Characterization of identified variants in blood of the RS and LLS elderly subsample. (A) Mutations in genes previously linked to hematopoietic malignancies. Barplot of the number of individuals carrying a mutation, split by genes and study. Note that only 11 of the 15 investigated genes had a mutation (see supplemental Appendix 4 for a complete list). (B) Prevalence of carriers of somatic mutations stratified by age category using data of Xie et al2 and the observations in the RS and LLS elderly subsample. (C) Distribution of VAFs of the identified mutations. (D) Comutation plot of carriers with 2 independent mutations. (E) Overview of mutations in DNMT3A identified in the RS and LLS elderly subsample. Variants are annotated at the top with color coding to indicate the impact and a shape to indicate the types of follow-up experiments. Circles indicate mutations detected in our sequencing data; squares indicate mutations also validated by Sanger sequencing; diamonds indicate mutations also validated by Sanger sequencing and absent in an IBD2 matched sib, that is, confirming somatic variations. Mutations identified in multiple carriers are indicated with stacked annotations and those having bold borders were identified in the LLS. Missense variants are only included whenever they are present on a curated list of recurrently reported variants in the Catalogue Of Somatic Mutations In Cancer (COSMIC)7 assembled by Jaiswal et al4 . Domains: ADD, histone-binding domains; DNMT, DNA methyltransferase interaction domain; MTase, methyltransferase domain; PWWP, conserved DNA binding domain; ZC-FING, zinc finger domains. COSMIC, densities of somatic variants identified in hematopoietic or lymphoid tissue collected by the Catalogue Of Somatic Mutations In Cancer (COSMIC)7 database: all small variants (red), missense single nucelotide variants (SNVs) (gray), small variants confirmed to be of somatic origin (blue).
The fraction of reads annotated to the alternative alleles (variant allele fractions [VAFs]) is generally much lower for the identified mutations than the 50% expected for germline heterozygous variants (RS: median, 21.6%; interquartile range, 14.1%-29.6%) (LLS: median, 23.2%; interquartile range, 16.5%-31.7%) (Figure 1C). This finding indicates that the identified mutations were only present in a part of the sequenced blood cells, and thus corroborates the hypothesized clonal outgrowth of hematopoietic stem cells.
Compared with the 3 previous studies predominantly including middle-aged participants, a high prevalence of mutation carriers with 2 mutations was observed in the RS and LLS elderly subsample (Figure 1D; 9 in 864 [1.04%] vs 6 in 2636 [0.28%],2 18 in 12 380 [0.15%],3 and 49 in 17 182 [0.28%]4 ).
The spatial correlation between the identified variants within DNMT3A and TET2 with respect to the primary protein sequence and previous reports in Catalogue Of Somatic Mutations In Cancer (COSMIC)7 further corroborates our findings (Figure 1E; supplemental Figure 1). Additional Sanger sequencing experiments in the LLS elderly subsample (supplemental Appendix 5) confirmed the presence for 18 of 19 tested mutations in DNMT3A (Figure 1E diamonds and squares) and TET2 (supplemental Figure 1). Moreover, Sanger sequencing in siblings of 6 mutant carriers, who inherited the identical genetic alleles from their parents at these loci as the mutant carriers (supplemental Appendix 6), did not show the identified somatic mutations (6 of 6; Figure 1E diamonds), thus confirming that these variants in DNMT3A and TET2 were indeed acquired during life.
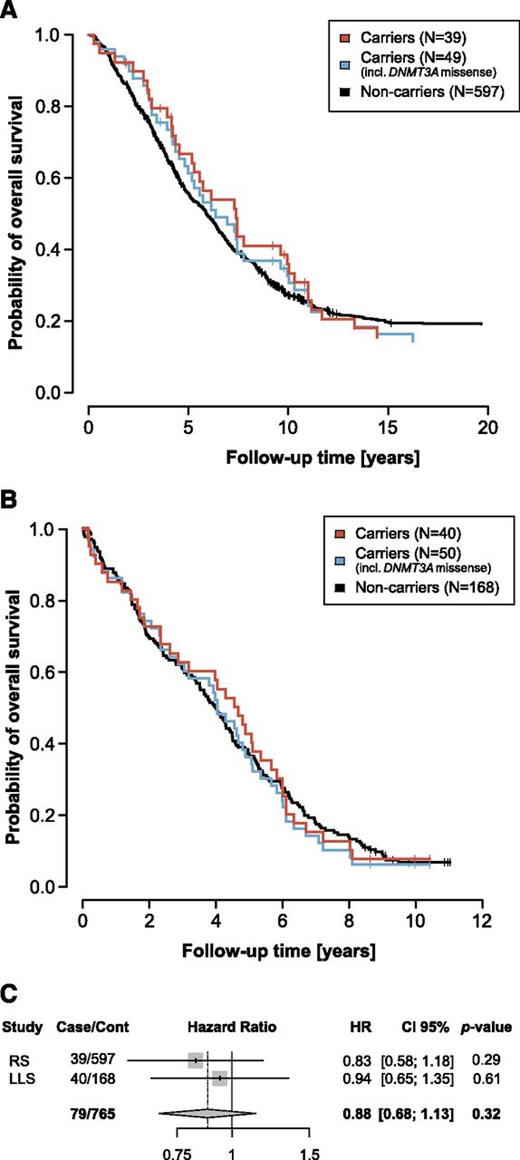
Having identified carriers of somatic mutations in genes linked to clonal hematopoiesis in the RS and LLS elderly subsample, we next assessed the impact on survival of carrying such mutations. When analyzing the impact of carriership of the identified somatic mutations in the 15 genes previously linked to clonal expansion of hematopoietic stem cells, no difference in survival was observed between carriers and noncarriers for the RS (hazard ratio [HR] = 0.83 [0.58-1.17], P = .29; Figure 2A) nor the LLS (HR = 0.94 [0.65-1.35], P = .61; Figure 2B; supplemental Appendix 6) elderly subsample. Also, a fixed-effect meta-analysis showed no indications of compromised survival (HR = 0.88 [0.68-1.13], P = .32; Figure 2C; supplemental Appendix 6).
Kaplan-Meier survival curves of the RS and LLS elderly subsample. (A) Kaplan-Meier curves for the 39 mutation carriers and 596 noncarriers in the RS elderly subsample. (B) Kaplan-Meier curves for the 40 mutation carriers and 168 noncarriers in the LLS elderly subsample. Because Jaiswal et al4 and Genovese et al3 do not agree on the status of DNMT3A missense mutations, we excluded DNMT3A missense mutation carriers from noncarriers in both the RS and LLS elderly subsample (supplemental Tables 6-7). (C) Forest plot combining the Cox proportional hazards analyses in the RS and LLS elderly subsample.
Kaplan-Meier survival curves of the RS and LLS elderly subsample. (A) Kaplan-Meier curves for the 39 mutation carriers and 596 noncarriers in the RS elderly subsample. (B) Kaplan-Meier curves for the 40 mutation carriers and 168 noncarriers in the LLS elderly subsample. Because Jaiswal et al4 and Genovese et al3 do not agree on the status of DNMT3A missense mutations, we excluded DNMT3A missense mutation carriers from noncarriers in both the RS and LLS elderly subsample (supplemental Tables 6-7). (C) Forest plot combining the Cox proportional hazards analyses in the RS and LLS elderly subsample.
Using DNA sequencing data in an elderly subsample derived from 2 large-scale community-dwelling Dutch cohort studies,5,6 we confirm that somatic mutations in genes previously linked to hematopoietic malignancies are common in the oldest old, especially in DNMT3A and TET2. Yet, in 2 independent studies, jointly comprising the largest sample in this age range to date, we found no indications that the potentially premalignant mutations compromise the 8- to 10-year survival of highly aged carriers.
In contrast to our findings, 2 recent large-scale sequencing studies in peripheral blood performed in 12 3003 and 17 1824 normal mostly middle-aged individuals found a significant increased risk for all-cause mortality among carriers of premalignant somatic mutations, predominantly in DNMT3A and TET2. The difference in mortality risk between middle-aged and highly aged people may lie in the fact that the oldest old suffer from many other comorbidities affecting their mortality rate. Causes of death or coincident morbidities at the time of death may support this hypothesis, however, such data were not available for our studies. Last, there is intrinsic selection bias when investigating the oldest old: the success of these highly aged individuals in coping with at least some of the adverse effects during aging could limit the ability to detect adverse health effects associated with age-related clonal expansion.
A possible limitation of our study might relate to the lower sequencing depth, allowing for a less sensitive detection of variants characterized by a low allele fraction. However, when Jaiswal et al stratified their mortality analyses on the median VAF, they observed that the observed increased risk on mortality could largely be attributed to carriers of variants with the largest VAF.4 Also, the relatively modest size could limit our study. However, a power analysis (supplemental Appendix 6) indicated that we should be able to detect a significant increased risk of all-cause mortality among mutation carriers compared with noncarriers.
We conclude that, unlike previous reports in predominantly middle-aged individuals,3,4 somatic mutations in genes linked to clonal expansion of hematopoietic stem cells do not compromise the 8- to 10-year survival in the oldest old.
Authorship
The online version of this article contains a data supplement.
Acknowledgments: The authors thank the study participants of the Leiden Longevity Study and the Rotterdam Study, the staff, and participating general practitioners and pharmacists of both studies. The generation and management of the exome sequencing data for the Rotterdam Study was executed by the Human Genotyping Facility of the Genetic Laboratory of the Department of Internal Medicine, Erasmus Medical Center (MC), The Netherlands. The authors thank Mr Pascal Arp, Ms Mila Jhamai, Mr Marijn Verkerk for their help in creating the Rotterdam Study Exome Sequencing database. In addition, the authors thank Prof Veelken and the reviewers for their useful comments on the manuscript.
This work was supported by the Medical Delta (COMO), Pfizer Inc (USA), and European Union’s Seventh Framework Programme (FP7/2007-2011) under grant agreement number 259679. The Leiden Longevity Study was financially supported by the Innovation-Oriented Research Program on Genomics (SenterNovem IGE05007), and the next-gen sequencing by The Netherlands Consortium for Healthy Ageing (NCHA; grant 050-060-810), all in the framework of The Netherlands Genomics Initiative (NGI), Netherlands Organisation for Scientific Research (NWO) and by Biobanking and Biomolecular Research Infrastructure Netherlands (BBMRI-NL; www.bbmri.nl), a research infrastructure financed by the Dutch government (NWO 184.021.007). The Rotterdam Study was supported by Erasmus MC and Erasmus University, Rotterdam, Netherlands Organization for Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly, the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII), and the Municipality of Rotterdam. The exome sequencing data set was funded by the NGI/NWO sponsored NCHA (project no. 050-060-810), by the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, and by a Complementation Project of the BBMRI-NL (project no. CP2010-41).
Contribution: E.B.v.d.A., S.J.P., M.J.T.R., M.B., and P.E.S. conceived the analyses and wrote the paper; E.B.v.d.A., S.J.P., J.D., M.H.M., S.P., J.v.R., R.K., and J.B.J.v.M. performed data processing and the statistical analyses; H.E.D.S., N.L., and W.J.d.D. performed validation experiments; A.G.U., A.H., A.J.M.d.C., D.R.C., G.-J.B.v.O., and P.E.S. conceived the employed studies and contributed reagents and materials for the experiments; and J.J.H.-D. provided statistical counseling.
Conflict-of-interest disclosure: S.J.P. was previously employed by the for-profit health care company Pfizer Inc, for which he also has a stock ownership and research funding to disclose. S.J.P. is currently employed by the for-profit health care company 23andME. S.P. is currently employed by the for-profit health care company Pfizer Inc, for which she also has a stock ownership and research funding to disclose. In addition, S.P. has an intellectual property interest to disclose. The remaining authors declare no competing financial interests.
David R. Cox died on January 22, 2013.
A complete list of the members of the Genome of The Netherlands Consortium appears in supplemental Appendix 1.
Correspondence: Erik B. van den Akker, Leiden University Medical Center, Building 2 (Research), Room T5-46, Einthovenweg 20, 2333 ZC, Leiden, The Netherlands; e-mail: e.b.van_den_akker@lumc.nl.