To the editor:
Minimal residual disease (MRD) monitoring via antigen receptor quantitative polymerase chain reaction (qPCR) is an important predictor of outcome in childhood acute lymphoblastic leukemia (ALL), is rigorously standardized within the EuroMRD consortium and has a greater sensitivity than flow cytometry (FC), which has been used in other trials.1 However, qPCR is laborious, expensive, and time consuming because of the development of patient-specific assays. MRD detection based on next-generation sequencing (NGS) of antigen receptor gene rearrangements is a promising tool that permits sequencing of large numbers of rearranged V-(D)-J segments and thus provides the picture of not only residual leukemia but also the normal immune repertoire of respective cells.2-5
To date, no correlation study on larger number of samples has been performed to determine how the discrepancies between NGS and qPCR data would affect clinical data. Moreover, there are no data investigating the impact of normal B-cell compartment reconstitution after induction treatment in ALL on prognosis. We developed a simple, cost effective, and easily adoptable approach using 2-round PCR amplifying virtually all immunoglobulin heavy chain (IgH) rearrangements6 and NGS on Ion Torrent/Proton sequencers (supplemental Methods; available on the Blood Web site). We compared the NGS-MRD results with current techniques7 in patients treated by Berlin-Frankfurt-Munster (BFM)-based protocol and investigated the changes in the treatment stratification using the new method.
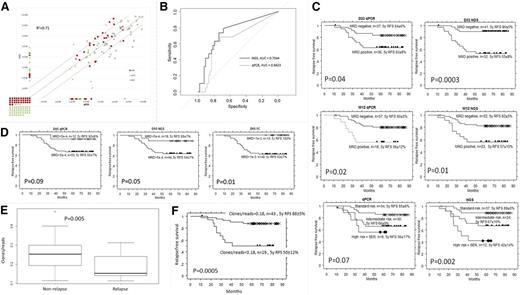
Altogether, we sequenced 210 samples from 76 patients (62× day 15, 73× day 33, 75× day 78) with a median coverage of 729 842 reads per sample. The overall correlation of NGS- and qPCR-MRD in all samples was satisfactory (R2 = 0.72; Figure 1A). A total of 32 samples (15%) were positive by NGS/negative by qPCR or vice versa, causing a shift in BFM risk stratification in 25 patients (33%), mainly between standard-risk (SR) and intermediate-risk (IR) groups in the low-positive patients. Five patients would be reassigned from the IR to the slow early responder (SER) group (4 of them relapsed), and 2 patients from SER to IR (1 of them relapsed).
The comparison of MRD as measured by qPCR and NGS, the prognostic significance of NGS, qPCR and FC at various treatment time points and the impact of IgH repertoire diversity on outcome. (A) The comparison of MRD as measured by qPCR and NGS. MRD at different time points (days 15, 33, and 78) by qPCR (x-axis) and NGS (y-axis). (B) Receiver operating characteristic curves and corresponding areas under the curve (AUC) statistics for day 33 NGS-MRD and day 33 qPCR-MRD prediction of relapse show better specificity of NGS over qPCR. (C) Prognostic significance of MRD at days 33 and 78 assessed simultaneously by qPCR and NGS. Kaplan-Meier survival plots show relapse-free survival (RFS) of patients with ALL based on MRD at day 33, MRD at day 78, and assigned via BFM risk group stratification into 3 risk groups using combined day 33 and day 78 MRD levels. At day 33, NGS defined a slightly larger (41 vs 37 patients, not significant) group of patients with negative MRD than qPCR, who had a similarly good prognosis as those defined by qPCR, even though 22% of patients in the NGS negative group were qPCR positive. The superiority of NGS over qPCR at day 33 was confirmed by deviance analysis of a multivariate Cox model, which showed a significant improvement of NGS prediction over qPCR (P = .003). As shown in the supplemental Methods, the total cohort was enriched for patients with relapse, which causes artificially low RFS in respective subgroups. (D) Prognostic significance of MRD at day 15 assessed by qPCR, NGS, and FC. Kaplan-Meier survival plots show RFS of patients with ALL based on MRD at day 15. (E) The impact of IgH repertoire diversity on outcome. Normalized IgH repertoire diversity expressed as number of clones/number of reads at day 78 for patients with and without relapse. The samples with NGS-MRD higher than 10−4 were excluded from the analysis. (F) RFS according to normalized IgH repertoire diversity at day 78. Kaplan-Meier survival RFS of patients with ALL based on repertoire diversity at day 78. Patients with clonal diversity (number of clones/number of reads) of less than 0.18 at day 78 had a 5-year RFS = 50 ± 12% compared with 88 ± 5% in other patients (P = .0005). Interestingly, of 9 patients who relapsed in the group with low diversity, 4 were stratified into the SR group according to current qPCR-based stratification (combined day 33/day 78). d15, day 15; d33, day 33; d78, day 78.
The comparison of MRD as measured by qPCR and NGS, the prognostic significance of NGS, qPCR and FC at various treatment time points and the impact of IgH repertoire diversity on outcome. (A) The comparison of MRD as measured by qPCR and NGS. MRD at different time points (days 15, 33, and 78) by qPCR (x-axis) and NGS (y-axis). (B) Receiver operating characteristic curves and corresponding areas under the curve (AUC) statistics for day 33 NGS-MRD and day 33 qPCR-MRD prediction of relapse show better specificity of NGS over qPCR. (C) Prognostic significance of MRD at days 33 and 78 assessed simultaneously by qPCR and NGS. Kaplan-Meier survival plots show relapse-free survival (RFS) of patients with ALL based on MRD at day 33, MRD at day 78, and assigned via BFM risk group stratification into 3 risk groups using combined day 33 and day 78 MRD levels. At day 33, NGS defined a slightly larger (41 vs 37 patients, not significant) group of patients with negative MRD than qPCR, who had a similarly good prognosis as those defined by qPCR, even though 22% of patients in the NGS negative group were qPCR positive. The superiority of NGS over qPCR at day 33 was confirmed by deviance analysis of a multivariate Cox model, which showed a significant improvement of NGS prediction over qPCR (P = .003). As shown in the supplemental Methods, the total cohort was enriched for patients with relapse, which causes artificially low RFS in respective subgroups. (D) Prognostic significance of MRD at day 15 assessed by qPCR, NGS, and FC. Kaplan-Meier survival plots show RFS of patients with ALL based on MRD at day 15. (E) The impact of IgH repertoire diversity on outcome. Normalized IgH repertoire diversity expressed as number of clones/number of reads at day 78 for patients with and without relapse. The samples with NGS-MRD higher than 10−4 were excluded from the analysis. (F) RFS according to normalized IgH repertoire diversity at day 78. Kaplan-Meier survival RFS of patients with ALL based on repertoire diversity at day 78. Patients with clonal diversity (number of clones/number of reads) of less than 0.18 at day 78 had a 5-year RFS = 50 ± 12% compared with 88 ± 5% in other patients (P = .0005). Interestingly, of 9 patients who relapsed in the group with low diversity, 4 were stratified into the SR group according to current qPCR-based stratification (combined day 33/day 78). d15, day 15; d33, day 33; d78, day 78.
NGS-MRD positivity at day 33 provided a more accurate prediction of relapse than qPCR-MRD positivity (Figure 1B-C) (5-year RFS NGS-negative/positive: 90 ± 5% vs 53 ± 9%, P = .0003; PCR-negative/positive: 84 ± 6% vs 63 ± 8%, P = .04). Deviance analysis of a multivariate Cox model showed significant improvement of NGS prediction over qPCR (P = .003). At day 78, the predictive value of NGS was almost identical to qPCR (Figure 1C). Combined day 33 and 78 information used for defining SR, IR, and high risk (+ SER) groups on BFM trials gave slightly better results than qPCR (Figure 1C). In consensus with published reports,8,9 both 8-color FC and qPCR defined a subgroup of patients with low MRD as early as day 15, who had a superior prognosis. The NGS approach identified a similarly large group of low-risk patients with excellent outcome (Figure 1D).
NGS data provide information not only about MRD but also about the rest of the B-lymphoid repertoire. We assessed clonal heterogeneity of the IgH repertoire in investigated samples, using the Vidjil algorithm10 (supplemental Methods). Interestingly, the patients with relapse had significantly lower IgH repertoire diversity at days 33 and 78. Even after removal of patients with NGS-MRD higher than 10−4, the patients with lower clonal diversity at day 78 had significantly worse 5-year RFS than other patients (P = .0005; Figure 1F).
In conclusion, the NGS risk group assignment is partly different from current approaches. This is mostly because of discrepancies in low-positive samples in SR/IR groups, but with no negative impact on final outcome. The NGS even provided a more precise prediction of relapse than qPCR at day 33 in our limited cohort. However, a prospective validation study comparing both methods will be needed to definitely accept NGS as the replacement for qPCR. A redefinition of stratification criteria for childhood ALL will be only the final step of a complex process that has just started. The main challenge in NGS-MRD methodology is standardization aiming at highly reproducible results between different centers, as it was achieved previously within the Euro-MRD group for qPCR. A European network, the EuroClonality-NGS Consortium, has been formed to standardize the workflow of analytics, preanalytics, and bioinformatics. Before this is accomplished, the methodology described in our study can serve as a broadly accessible and relatively inexpensive noncommercial solution for centers that do not perform antigen receptor qPCR but want to start using benefits of MRD monitoring in their clinical setting.
The online version of this article contains a data supplement.
Authorship
Acknowledgments: This work was supported by grants from the Internal Grant Agency of the Czech Ministry of Health (IGA MZCR NT14343), the Grant Agency of the Czech Republic (GACR, project of Centre of Excellence No. P302/12/G101) (J.T.), the Charles University Grant Agency (GAUK 394214) (M.K.), and the University Hospital Motol, Prague, Czech Republic (00064203) (K.M.).
Contribution: M.K. prepared the sequencing libraries, performed the sequencing runs, and performed the data analysis; K.M. prepared the sequencing libraries and performed the sequencing runs; V.B.-M. performed the sequencing runs; M.N. and E.M. performed the FC analysis; K.F. and J. Stuchly developed the bioinformatic tools for MRD tracking; M.G. and M.S. performed the repertoire analysis; C.P. and M.B. developed the protocol for library preparation and interpreted the data; M.F. developed the protocol for library preparation; J. Stary and J.T. planned the study and interpreted the data; and E.F. planned the study, interpreted the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jan Trka, Charles University, 2nd School of Medicine, V Úvalu 84, Prague, 15006, Czech Republic; e-mail: jan.trka@lfmotol.cuni.cz.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal