Key Points
In myeloma patients, trisomy 3 improved time to progression and trisomies 3 and/or 5 improved overall survival.
In contrast, trisomy 21 significantly worsened overall survival.
Abstract
The prognosis of multiple myeloma is mainly dependent upon chromosomal changes. The 2 major abnormalities driving poor outcome are del(17p) and t(4;14). However, the outcome of these high-risk patients is not absolutely uniform, with some patients presenting long survival. We hypothesized that these better outcomes might be related to concomitant “good-risk” chromosomal changes exploring hyperdiploidy. We analyzed a large series of 965 myeloma patients, including 168 patients with t(4;14) and 126 patients with del(17p), using high-throughput single-nucleotide polymorphism arrays after plasma cell sorting. As expected, trisomic chromosomes were highly associated. Using the LASSO model, we found that only chromosome 3, when trisomic, was associated with a longer progression-free survival and that 3 trisomies modulated overall survival (OS) in myeloma patients: trisomies 3 and 5 significantly improved OS, whereas trisomy 21 worsened OS. In patients with t(4;14), trisomies 3 and/or 5 seemed to overcome the poor prognosis. For the first time, using a specific modeling approach, we show that not all trisomies display the same prognostic impact. This finding could be important for routine assessment of prognosis in myeloma, and some high-risk patients with a traditional evaluation could in fact be standard-risk patients.
Introduction
In multiple myeloma (MM), prognosis is mainly dependent on the chromosomal abnormalities present in tumor plasma cells.1-3 So far, no “good” prognosis abnormalities have been described. Some abnormalities, such as hyperdiploidy or the translocation t(11;14), are associated with standard- as opposed to high-risk features, which include the translocations t(4;14), t(14;16), or t(14;20); the deletion 17p (del(17p)); and the gains of the 1q arm (1q gains).1-5 The worst chromosomal abnormality is del(17p), especially when present in the majority of plasma cells, which is observed in 7% to 8% of patients. For instance, in the Intergroupe Francophone du Myélome (IFM) experience in transplant-eligible patients, the median overall survival (OS) was 22 months.3 The second important chromosomal change is t(4;14), observed in 10% to 15% of patients. In the same patient population, the median OS was 41 months.3 The MAF translocations, that is, t(14;16) and t(14;20), are not routinely assessed because of their low incidence (3% and 1%, respectively); in a previous study, we showed that t(14;16) and 1q gains were not independent prognostic factors.6 Thus, in IFM routine practice, only del(17p) and t(4;14) are evaluated at diagnosis and define the true high-risk patients.
Three studies have addressed the issue of prognostic heterogeneity in high-risk patients, with opposite conclusions.7-9 The first study concluded that concomitant trisomies did not modify the outcome of high-risk patients,7 whereas a second one from the same group showed that concomitant trisomies completely overcame the poor prognosis of high-risk abnormalities.8 A more recent study from the Medical Research Council (MRC) suggested that trisomies did not modify the outcome of high-risk patients.9 These 3 studies used either fluorescence in situ hybridization (FISH) techniques, evaluating only the presence of trisomies 3, 7, 9, and 15, or DNA content evaluation by flow cytometry. One of the main limitations of the first 2 studies was the relatively limited number of high-risk patients enrolled (20 and 115 patients, respectively). In the third study, the number of high-risk patients was higher, but the definition of high risk was different and included 1q gains.
In order to clearly answer this important question, we conducted a very large study based on extensive genomic analysis of 965 patients analyzed by single-nucleotide polymorphism (SNP) array. The aims of this study were to describe the trisomies involved in myeloma, to study how they are associated with each other, to test their relationships with progression-free survival and OS, and to investigate the possibility of their influence on the effect of t(4;14) and del(17p).
Patients and methods
We analyzed by SNP array (Affymetrix, Santa Clara, CA), using either the SNP6.0 array or the Cytoscan array, depending on the time of analysis, a cohort of 965 patients diagnosed in one of the IFM centers. All patients provided signed consent for these genetic analyses in accordance with the Declaration of Helsinki. Bone marrow samples were obtained at the time of diagnosis, before treatment initiation, and were shipped overnight to a central laboratory. Upon receipt, mononuclear bone marrow cells were sorted using the RoboSep system, according to the manufacturer’s recommendations (StemCell Technologies, Vancouver, BC, Canada). Only samples with >80% of plasma cells after sorting were kept for analysis by SNP array. All patients were treated routinely, and selection was based on the availability of enough frozen plasma cells and sufficient data on clinical characteristics, treatment, and follow-up. A total of 56.5% of patients were male, and the median age was 60.5 years. Regarding treatment, 191 patients received a VAD (vincristine, Adriamycin, and dexamethasone) induction followed by high-dose melphalan and autologous stem cell transplant (ASCT) (20%), 521 patients received a VD (Velcade and dexamethasone) induction followed by high-dose melphalan and ASCT (54%), and 26% received a nonintensive treatment because of older age (>65 years). At the time of analysis, 60.1% of the patients were still alive. High-risk patients were defined by the presence of del(17p) in more than 60% of plasma cells assessed by interphase FISH of sorted plasma cells, as previously reported, and/or by the presence of t(4;14), which was also evaluated by FISH using specific probes from Abbott (Paris, France). For hyperdiploidy, we assessed on SNP arrays trisomies of the chromosomes 3, 5, 7, 9, 11, 15, 17, 18, 19, and 21. Because it has been shown in childhood acute lymphoblastic leukemia10 and myeloma11 that the number of trisomies is important for outcome (hyperdiploidy >50 was associated with a better outcome than hyperdiploidy 47 to 50), we also counted the number of chromosomes on the SNP arrays for each patient and defined hypodiploidy (<46 chromosomes), pseudodiploidy (46 chromosomes), mild hyperdiploidy (47 to 50 chromosomes), and large hyperdiploidy (>50 chromosomes)
Statistical analysis
The Pearson χ2 test was used to compare associations between trisomies, between high-risk and non–high-risk myeloma, and between genders. Differences in age between patients with and without a specific trisomy were compared using the Student t test. Time to progression (TTP) was defined as the time interval between diagnosis and progression. OS was defined as the time from myeloma diagnosis until death due to any cause. Patients were censored at the date of last contact if no event was reported. Survival curves, medians, and interquartile ranges (IQRs) were obtained using the Kaplan-Meier method, and comparisons between groups were made using log-rank test.
We first reported the association between each trisomy and TTP and OS using a univariate Cox model. For multivariate modeling, we included in the model t(4;14), del(17p), treatment (no ASCT vs VAD + [high-dose therapy]/ASCT vs VD + [high-dose therapy]/ASCT), and all of the trisomies. As the correlation between trisomy variables was very high (multicollinearity), the traditional multivariate Cox model produces estimates that are both biased and unstable, leading to an arbitrary selection of important variables. Among the penalized regression methods developed to overcome this statistical problem is a L1 penalized Cox regression using the LASSO (least absolute shrinkage and selection operator)12 adjusted for treatment. The LASSO tends to select only a few variables among a set of variables highly correlated, enabling one to perform variable selection and thus allowing the selection of a sparser model. The regularization parameter that is used for penalization was estimated using 10-fold crossvalidation. The penalty term was only applied on the trisomy variables, whereas t(4;14), del(17p), and treatment were left unpenalized, as they have already shown their predictive value in previous studies. To produce a consistent selection of variables, this analysis was repeated on 500 replications obtained by bootstrapping the original data (BOLASSO). The variables selected after bootstrapping were the only ones that were nonzero (ie, that have a predictive value) in at least 80% of the replications. Once the variables were selected, we repeated the same analysis for testing first-order interactions between the selected trisomies and t(4;14), del(17p), and treatment groups, and penalties were only applied to interaction effects.
In the last step, we fit a Cox model including t(4;14), del(17p), treatment, and the selected trisomies, adjusting further for age, sex, and β2-microglobulin level. Ultimately, we separately tested the independent effect of the 2 proxy variables commonly used for exploring hyperdiploidy (ie, high hyperdiploidy and presence of at least one trisomy). The proportionality assumption was verified by evaluating Cox-Snell residuals and log-log plot. A 2-sided P value <.05 was considered significant. Statistical analyses were performed on the R package “penalized”13 and Stata statistical software, release 11.2 (Stata Corporation, College Station, TX).
Results
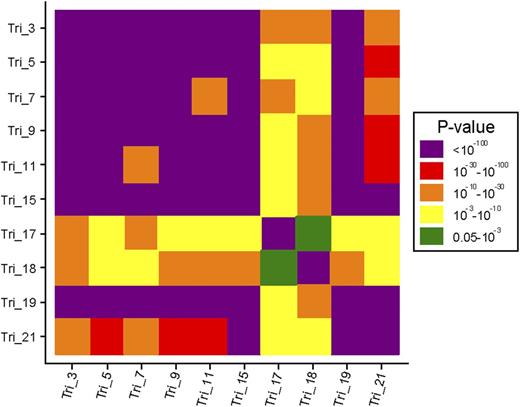
Translocation t(4;14) was observed in 168 patients and del(17p) in 126 patients; 26 patients presented both abnormalities. At least one trisomy was found in 61% of patients. The most commonly observed trisomy was chromosome 9 (48%), followed by chromosomes 15 (47%), 19 (46%), 5 (38%), 3 (36%), 11 (33%), 7 (26%), 21 (23%), 18 (11%), and 17 (5%). Only 13% of patients had a pseudodiploid karyotype, whereas hypodiploidy, mild hyperdiploidy, and large hyperdiploidy were observed in 35%, 14%, and 38% of patients, respectively. No gender difference in trisomy distribution was found, except for trisomy 3 (41% vs 29%, P < .001) and trisomy 19 (50% vs 41%, P = .007), which were more frequent in male than female patients. Trisomy 5 was the only age-related trisomy. The mean age was slightly higher in patients with a trisomy 5 than in patients without this trisomy (61.6 years vs 59.5 years, P = .002). The distribution of trisomies by risk groups is shown in Table 1, and associations between trisomies are reported in Figure 1.
Distribution of trisomies and chromosome number according to t(4;14) and del(17p) profiles
| . | Standard risk (n = 633) . | t(4;14) (n = 168) . | χ2P* . | del(17p) (n = 126) . | χ2P† . | χ2P‡ . |
|---|---|---|---|---|---|---|
| Trisomy 3 | 244 (38.5) | 46 (27.4) | .007 | 34 (27.0) | .014 | .867 |
| Trisomy 5 | 306 (48.3) | 15 (8.9) | <.001 | 26 (20.6) | <.001 | .002 |
| Trisomy 7 | 189 (29.9) | 18 (10.7) | <.001 | 23 (18.3) | .008 | .035 |
| Trisomy 9 | 360 (56.9) | 32 (19.0) | <.001 | 40 (31.7) | <.001 | .004 |
| Trisomy 11 | 260 (41.1) | 17 (10.1) | <.001 | 25 (19.8) | <.001 | .009 |
| Trisomy 15 | 336 (53.1) | 45 (26.8) | <.001 | 40 (31.7) | <.001 | .225 |
| Trisomy 17 | 40 (6.3) | 8 (4.8) | .450 | 1 (0.8) | .012 | .061 |
| Trisomy 18 | 83 (13.1) | 7 (4.2) | .001 | 13 (10.3) | .389 | .025 |
| Trisomy 19 | 335 (52.9) | 43 (25.6) | <.001 | 36 (28.6) | <.001 | .396 |
| Trisomy 21 | 177 (28.0) | 17 (10.1) | <.001 | 18 (14.4) | .001 | .190 |
| At least one trisomy | 422 (66.8) | 69 (41.1) | <.001 | 59 (47.2) | <.001 | .123 |
| Number of chromosomes | ||||||
| 46 | 94 (14.8) | 17 (10.1) | 11 (8.7) | |||
| <46 | 157 (24.8) | 102 (60.7) | 71 (56.3) | |||
| 47-50 | 82 (13.0) | 26 (15.5) | 20 (15.9) | |||
| ≥51 | 300 (47.4) | 23 (13.7) | <.001 | 24 (19.0) | <.001 | .468 |
| . | Standard risk (n = 633) . | t(4;14) (n = 168) . | χ2P* . | del(17p) (n = 126) . | χ2P† . | χ2P‡ . |
|---|---|---|---|---|---|---|
| Trisomy 3 | 244 (38.5) | 46 (27.4) | .007 | 34 (27.0) | .014 | .867 |
| Trisomy 5 | 306 (48.3) | 15 (8.9) | <.001 | 26 (20.6) | <.001 | .002 |
| Trisomy 7 | 189 (29.9) | 18 (10.7) | <.001 | 23 (18.3) | .008 | .035 |
| Trisomy 9 | 360 (56.9) | 32 (19.0) | <.001 | 40 (31.7) | <.001 | .004 |
| Trisomy 11 | 260 (41.1) | 17 (10.1) | <.001 | 25 (19.8) | <.001 | .009 |
| Trisomy 15 | 336 (53.1) | 45 (26.8) | <.001 | 40 (31.7) | <.001 | .225 |
| Trisomy 17 | 40 (6.3) | 8 (4.8) | .450 | 1 (0.8) | .012 | .061 |
| Trisomy 18 | 83 (13.1) | 7 (4.2) | .001 | 13 (10.3) | .389 | .025 |
| Trisomy 19 | 335 (52.9) | 43 (25.6) | <.001 | 36 (28.6) | <.001 | .396 |
| Trisomy 21 | 177 (28.0) | 17 (10.1) | <.001 | 18 (14.4) | .001 | .190 |
| At least one trisomy | 422 (66.8) | 69 (41.1) | <.001 | 59 (47.2) | <.001 | .123 |
| Number of chromosomes | ||||||
| 46 | 94 (14.8) | 17 (10.1) | 11 (8.7) | |||
| <46 | 157 (24.8) | 102 (60.7) | 71 (56.3) | |||
| 47-50 | 82 (13.0) | 26 (15.5) | 20 (15.9) | |||
| ≥51 | 300 (47.4) | 23 (13.7) | <.001 | 24 (19.0) | <.001 | .468 |
Comparison between t(4;14) and standard-risk patients.
Comparison between del(17p) and standard-risk patients.
Comparisons between t(4;14) and del(17p) patients; patients with both abnormalities (n = 26) were excluded from this comparison, and 64 patients with missing data on t(4;14) or del(17p) were excluded from this analysis.
Graphical representation of the association between trisomies.P values were derived from a χ2 test. Tri, trisomy.
Graphical representation of the association between trisomies.P values were derived from a χ2 test. Tri, trisomy.
With a median follow-up time of 4.5 years, the median TTP was 2.6 years (IQR 1.5-4.5) in the standard-risk group, 1.4 years (IQR 0.9-2.5) among t(4;14) patients, and 1.2 years (IQR 0.6-2.4) among del(17p) patients. Median TTP according to chromosome number was 1.8 years (IQR 0.9-3.4), 2.5 years (IQR 1.5-3.9), 2.0 years (IQR 1.1-4.2), and 2.4 years (IQR 1.5-4.2) for patients with hypodiploidy, pseudodiploidy, mild hyperdiploidy, and large hyperdiploidy, respectively (P = .0005). For OS, the median survival was 8.7 years (IQR 4.7, not reached) in the standard-risk group, 3.5 years (IQR 1.6-6.8) among t(4;14) patients, and 2.7 years (IQR 1.1-4.6) among del(17p) patients. The median OS according to chromosome number was 5.0 years (IQR 2.2-9.1), 5.9 years (IQR 3.6-10.0), 5.6 years (IQR 3.0-8.2), and 9.1 years (IQR 4.2, not reached) for patients with hypodiploidy, pseudodiploidy, mild hyperdiploidy, and large hyperdiploidy, respectively (P < .0001). All the trisomies had a protective effect on survival in univariate analysis, except for trisomies 17, 18, and 21, which did not modify TTP or OS prognosis (Table 2).
Unadjusted effect of trisomies and hyperdiploidy on TTP and OS
| . | TTP . | OS . | ||
|---|---|---|---|---|
| HR (95% CI) . | P . | HR (95% CI) . | P . | |
| Trisomy 3 | 0.8 (0.7-0.9) | <.001 | 0.6 (0.5-0.8) | <.001 |
| Trisomy 5 | 0.8 (0.7-0.9) | .003 | 0.5 (0.4-0.7) | <.001 |
| Trisomy 7 | 0.8 (0.7-1.0) | .038 | 0.8 (0.6-1.0) | .030 |
| Trisomy 9 | 0.8 (0.7-1.0) | .018 | 0.6 (0.5-0.8) | <.001 |
| Trisomy 11 | 0.8 (0.7-0.9) | .001 | 0.6 (0.4-0.7) | <.001 |
| Trisomy 15 | 0.8 (0.7-0.9) | .003 | 0.6 (0.5-0.8) | <.001 |
| Trisomy 17 | 1.0 (0.7-1.3) | .752 | 1.0 (0.6-1.5) | .815 |
| Trisomy 18 | 1.0 (0.8-1.2) | .690 | 1.1 (0.8-1.5) | .683 |
| Trisomy 19 | 0.8 (0.7-1.0) | .008 | 0.6 (0.5-0.8) | <.001 |
| Trisomy 21 | 0.9 (0.8-1.1) | .282 | 1.0 (0.8-1.2) | .748 |
| At least one trisomy | 0.9 (0.8-1.0) | .075 | 0.7 (0.6-0.9) | .004 |
| Number of chromosomes | ||||
| 46 | 1 | .014 | 1 | |
| <46 | 1.3 (1.1-1.7) | .150 | 1.5 (1.1-2.1) | .014 |
| 47-50 | 1.2 (0.9-1.6) | 1.4 (0.9-2.0) | .113 | |
| ≥51 | 1.0 (0.8-1.2) | .710 | 0.7 (0.5-1.0) | .086 |
| . | TTP . | OS . | ||
|---|---|---|---|---|
| HR (95% CI) . | P . | HR (95% CI) . | P . | |
| Trisomy 3 | 0.8 (0.7-0.9) | <.001 | 0.6 (0.5-0.8) | <.001 |
| Trisomy 5 | 0.8 (0.7-0.9) | .003 | 0.5 (0.4-0.7) | <.001 |
| Trisomy 7 | 0.8 (0.7-1.0) | .038 | 0.8 (0.6-1.0) | .030 |
| Trisomy 9 | 0.8 (0.7-1.0) | .018 | 0.6 (0.5-0.8) | <.001 |
| Trisomy 11 | 0.8 (0.7-0.9) | .001 | 0.6 (0.4-0.7) | <.001 |
| Trisomy 15 | 0.8 (0.7-0.9) | .003 | 0.6 (0.5-0.8) | <.001 |
| Trisomy 17 | 1.0 (0.7-1.3) | .752 | 1.0 (0.6-1.5) | .815 |
| Trisomy 18 | 1.0 (0.8-1.2) | .690 | 1.1 (0.8-1.5) | .683 |
| Trisomy 19 | 0.8 (0.7-1.0) | .008 | 0.6 (0.5-0.8) | <.001 |
| Trisomy 21 | 0.9 (0.8-1.1) | .282 | 1.0 (0.8-1.2) | .748 |
| At least one trisomy | 0.9 (0.8-1.0) | .075 | 0.7 (0.6-0.9) | .004 |
| Number of chromosomes | ||||
| 46 | 1 | .014 | 1 | |
| <46 | 1.3 (1.1-1.7) | .150 | 1.5 (1.1-2.1) | .014 |
| 47-50 | 1.2 (0.9-1.6) | 1.4 (0.9-2.0) | .113 | |
| ≥51 | 1.0 (0.8-1.2) | .710 | 0.7 (0.5-1.0) | .086 |
Using the BOLASSO procedure, only trisomy 3 was selected as being useful for predicting TTP (ie, the regression coefficients were nonzero in at least 400 of the 500 samples), and all other trisomies were selected in <50% of replications. After adjusting for age, sex, β2-microglobulin level, and treatments, patients with a trisomy of chromosome 3 had a significantly improved TTP compared with patients without trisomy 3 (adjusted hazard ratio [aHR], 0.8; 95% confidence interval [CI], 0.6-0.9; P = .002; Figure 2). No interactions were found between trisomy 3 and treatment group or high-risk profiles.
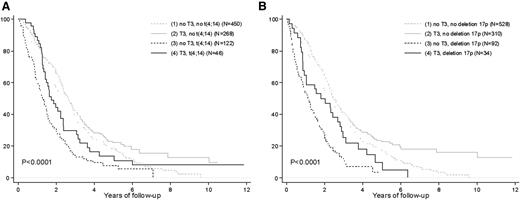
Trisomy 3 improves the PFS of high-risk patients. Kaplan-Meier estimates of TTP according to (A) t(4;14) and trisomy 3 and (B) del(17p) and trisomy 3.
Trisomy 3 improves the PFS of high-risk patients. Kaplan-Meier estimates of TTP according to (A) t(4;14) and trisomy 3 and (B) del(17p) and trisomy 3.
For predicting OS, trisomies 3, 5, and 21 were selected. Trisomies of chromosomes 17, 9, 19, 15, 7, 11, and 18 were nonzero in 41%, 51%, 52%, 67%, 69%, 75%, and 76% of replications, respectively. After adjusting for age, sex, β2-microglobulin level, and treatments, patients with a trisomy of chromosome 3 (aHR, 0.7; 95% CI, 0.5-0.9; P = .01) or 5 (aHR, 0.7; 95% CI, 0.5-0.9; P = .02) had a significantly improved OS compared with patients who lacked the given trisomy, whereas patients with a trisomy of chromosome 21 (aHR, 1.4; 95% CI, 1.0-2.0; P = .04) had worse outcome than patients lacking this trisomy (Table 3). Testing interactions between those trisomies and high-risk profile and treatment groups, none were reported to be consistently significant meaning that the protective effect reported for trisomy of chromosome 5 or 3 (Figure 3), or the risk effect reported for trisomy of chromosome 21 (Figure 4) was the same whatever the initial high-risk profile of patients and whatever the group of treatment. After inclusion into the model of these specific trisomies, neither large hyperdiploidy, as measured by a number of chromosomes >50 (aHR, 0.9; 95% CI, 0.5-1.7; P = .813), nor at least one trisomy (aHR, 1.3; 95% CI, 0.9-1.7; P = .154) had a protective effect on OS. Compared with the mild hyperdiploidy group, large hyperdiploidy patients more frequently had trisomy 3 (38.7% vs 76.1%; P < 10−3), trisomy 5 (32.8% vs 84.5%; P < 10−3), or a co-occurrence of both protective trisomies (9.5% vs 64.0%; P < 10−3). Similarly trisomies 3, 5, and 3 and 5 were reported in 58.6%, 62.7%, and 42.4% of patients having at least one trisomy, respectively.
Multivariate Cox model adjusted for age, sex, and β2-microglobulin level (N = 767)
| . | No. of patients . | No of events . | aHR . | 95% CI . | P . |
|---|---|---|---|---|---|
| Treatment | |||||
| No HDM | 182 | 71 | 1 | — | — |
| VAD-HDM | 137 | 100 | 0.78 | (0.51-1.18) | .242 |
| VD-HDM | 448 | 130 | 0.59 | (0.40-0.87) | .007 |
| Translocation (4;14) | |||||
| No | 623 | 210 | 1 | — | — |
| Yes | 144 | 91 | 2.20 | (1.69-2.86) | <.001 |
| Deletion 17p | |||||
| No | 676 | 232 | 1 | — | — |
| Yes | 91 | 69 | 3.23 | (2.45-4.26) | <.001 |
| Trisomy 3 | |||||
| No | 494 | 223 | 1 | — | — |
| Yes | 273 | 78 | 0.66 | (0.48-0.91) | .011 |
| Trisomy 5 | |||||
| No | 469 | 221 | 1 | — | — |
| Yes | 298 | 80 | 0.66 | (0.47-0.93) | .017 |
| Trisomy 21 | |||||
| No | 584 | 235 | 1 | — | — |
| Yes | 183 | 66 | 1.41 | (1.02-1.96) | .040 |
| . | No. of patients . | No of events . | aHR . | 95% CI . | P . |
|---|---|---|---|---|---|
| Treatment | |||||
| No HDM | 182 | 71 | 1 | — | — |
| VAD-HDM | 137 | 100 | 0.78 | (0.51-1.18) | .242 |
| VD-HDM | 448 | 130 | 0.59 | (0.40-0.87) | .007 |
| Translocation (4;14) | |||||
| No | 623 | 210 | 1 | — | — |
| Yes | 144 | 91 | 2.20 | (1.69-2.86) | <.001 |
| Deletion 17p | |||||
| No | 676 | 232 | 1 | — | — |
| Yes | 91 | 69 | 3.23 | (2.45-4.26) | <.001 |
| Trisomy 3 | |||||
| No | 494 | 223 | 1 | — | — |
| Yes | 273 | 78 | 0.66 | (0.48-0.91) | .011 |
| Trisomy 5 | |||||
| No | 469 | 221 | 1 | — | — |
| Yes | 298 | 80 | 0.66 | (0.47-0.93) | .017 |
| Trisomy 21 | |||||
| No | 584 | 235 | 1 | — | — |
| Yes | 183 | 66 | 1.41 | (1.02-1.96) | .040 |
HDM, high-dose melphalan.
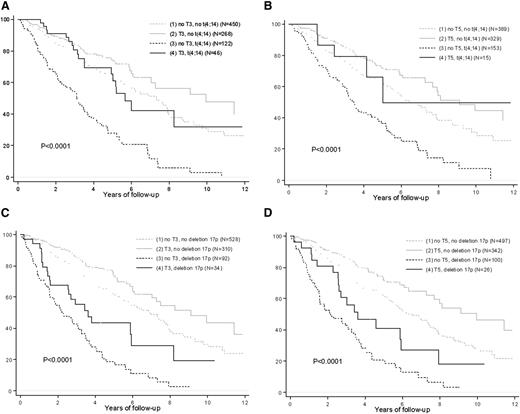
Trisomies 3 and 5 improve the OS of high-risk patients. Kaplan-Meier estimates of OS according to (A) t(4;14) and trisomy 3, (B) t(4;14) and trisomy 5, (C) del(17p) and trisomy 3, and (D) del(17p) and trisomy 5.
Trisomies 3 and 5 improve the OS of high-risk patients. Kaplan-Meier estimates of OS according to (A) t(4;14) and trisomy 3, (B) t(4;14) and trisomy 5, (C) del(17p) and trisomy 3, and (D) del(17p) and trisomy 5.
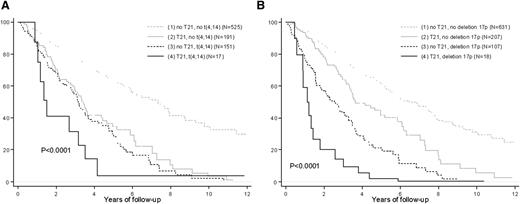
Trisomy 21 worsens the OS of high-risk patients. Kaplan-Meier estimates of OS according to (A) t(4;14) and trisomy 21 and (B) del(17p) and trisomy 21. The curves were adjusted on trisomies 3 and 5.
Trisomy 21 worsens the OS of high-risk patients. Kaplan-Meier estimates of OS according to (A) t(4;14) and trisomy 21 and (B) del(17p) and trisomy 21. The curves were adjusted on trisomies 3 and 5.
Supplementary analyses were conducted for exploring the effects of the trisomies 3, 5, and 21 on deletion 1p32 (supplemental Figures 1 and 2 available on the Blood Web site) and 1q gain (supplemental Figures 3 and 4), and the results were similar to the ones presented in this report, as were the survival curves including only transplanted patients (supplemental Figures 5-7).
Discussion
Along with the International Staging System,14 chromosomal abnormalities observed in the tumor plasma cells represent the most important prognostic factor in MM. Among all the chromosomal changes described in MM, del(17p) and t(4;14) are the 2 major abnormalities that impair the survival of patients. We previously reported that del(17p) has an important prognostic value only if present in the major clone. This finding was recently confirmed by an independent study.15 Even though the median OS of patients displaying these high-risk features is short in all the series reported in the literature, some of these patients present longer survivals. This is especially true for patients with t(4;14). The most consistent explanation could be that other chromosomal changes may have a protective effect, even in these high-risk patients. A recent study from the Mayo Clinic group8 suggested that the presence of trisomies in some of these high-risk patients may improve outcome. This study, based on the FISH technique, analyzed only trisomies 3, 7, 9, and 15 and grouped t(4;14) and del(17p) patients in the same category, impairing the study of heterogeneity of survival among t(4;14) patients. Independently of the nature of the trisomy, they showed that the overall prognosis was dependent only on the presence of trisomy, totally overcoming the prognostic value of the high-risk group. This finding is of major importance for the management of these patients, indicating that trisomy assessment is mandatory for all patients with del(17p) or t(4;14). Another recent study from the MRC9 reported opposite findings, claiming that trisomies do not modify the outcome of high-risk patients. These apparently contradictory results might be explained by several study differences, especially in regard to the definition of high risk. In the MRC study, high risk was defined as the presence of t(4;14), del(17p), MAF translocations, or 1q gains. We previously showed that 1q gains cannot be considered as a high-risk feature, as they are associated with a median OS >5 years.5
Recently, more sophisticated global genomic technologies (eg, comparative genomic hybridization/SNP array, next-generation sequencing) have been used to assess chromosomal changes in MM.16-18 They have vastly confirmed the cytogenetic/FISH data, with a much higher level of definition. In a preliminary report, we showed that concomitant genetic changes worsened the outcome of high-risk patients.19 In order to address the issue of the role of trisomies in modulating the outcome of high-risk patients, we performed a large, SNP-array–based study of 965 patients with MM. To specifically address the issue of trisomies in high-risk patients, we enriched the series with such patients, explaining the apparently higher incidences of del(17p) and t(4;14) in this cohort. This overrepresentation of high-risk patients could have biased the HR estimates, but this bias has been studied in epidemiology and found to be very small.20 In this series (the largest never published on high-risk patients), we showed that trisomy 3 and/or 5 overcame the poor prognosis of t(4;14) and improved that of del(17p), whereas trisomy 21 worsened the prognostic value of both t(4;14) and del(17p). Regarding TTP, only trisomy 3 showed a statistically significant association with a better outcome.
In most of the studies exploring hyperdiploidy or trisomies in myeloma patients, proxy variables as categorization of the number of chromosomes or at least one trisomy are used to summarize the risk of a set of specific trisomies that is too large to model and to prevent high collinearity. Partitioning hyperdiploidy according to the number of chromosomes (47 to 50 and 51 to 58), we have clearly shown that only patients with large hyperdiploidy also have a high frequency of protective trisomies. This is easily explained by the high association between trisomies. Consequently, only high hyperdiploidy can be reported as a protective marker for OS in myeloma patients, because the higher the number of chromosomes, the more likely some protective trisomies will be present. This result can explain why Lim et al only reported a protective effect in the high hyperdiploidy group. Moreover, we also reported that only 3 trisomies were consistently reported as having a prognostic effect in our patients; for 2 of them, the effect was protective, and for trisomy of chromosome 21, the effect was worsening survival. This result demonstrated that a proxy of hyperdiploidy based on the presence of at least one trisomy, as reported by Kumar et al, is not in itself pertinent, because not all trisomies are relevant and the effects on survival probability of trisomies can go in the opposite direction.
The concordance of our results with those of Kumar et al (even though they did not study the same trisomies) is again explained by the high association between trisomies. In our data set, 82% of patients with trisomy 7, 75% of patients with trisomy 9, and 76% of patients with trisomy 15 also had trisomy 5. So, studying those trisomies, they have also measured the effect of trisomy 5 while this trisomy was not selected in their study. Therefore, high hyperdiploidy or at least one trisomy can only be regarded as a risk indicator and not as a risk factor. Furthermore, when making future treatment decisions based on a patient’s cytogenetic profile, we need to focus on specific risk factors and not on oversimplified markers in order to better understand the pathogenesis of myeloma and correctly stratify patients into prognostic groups. Our work, by enlightening the greater importance of the presence of trisomies 3, 5, and 21 among all the trisomies described in myeloma patients, takes place in this framework.
Our group previously demonstrated the protective effect of chromosome 5 in a series of 192 patients.16 Trisomy of chromosome 3 has never been specifically studied in myeloma. How these trisomies impact OS remains an unresolved question. These trisomies lead to a copy-number gain of hundreds of genes, preventing any target gene search. The worsening effect of trisomy 21 has never been reported previously in myeloma. This can be explained by the strategy of modeling most often used in multivariable analysis. During the first step, only variables statistically significant at a prespecified threshold (commonly 20%) in nonadjusted analyses are included into the model. Therefore, when this strategy is used, it is probable that trisomy 21 receives little attention from statisticians and clinicians, as the nonadjusted risk reported for trisomy 21 is almost null (HR, 0.96; 95% CI, 0.8-1.2; P = .748). Again, it is cumbersome to select a specific target gene responsible for this prognostic effect.
Our study is strengthened by the use of a statistical approach allowing the selection of predictive variables among highly correlated predictors, and it is reinforced by bootstrapping procedures that avoid unstable selection of variables. This resampling method incorporates a small change in the data that subsequently can lead to some changes in the estimates of effect. Therefore, using bootstrapping, we controlled the stability of our results. Even though the choice of a cutoff for the proportion of bootstrap sample for which a variable is retained in order to include it in the final model is arbitrary, the choice of a higher threshold (90%) will only retain trisomy 3 for OS prognosis, and no trisomy will have been selected by choosing a stringent threshold (100%). The large sample population, long follow-up, and the SNP-array methods are also strong points that reinforced our findings. Nevertheless, some limitations still need to be mentioned. Our study was adjusted for conventional high-risk cytogenetic factors affecting the risk of death, but residual confounding due to other chromosomal abnormalities (ie, gain (1q), del(1p)) not considered for the selection of trisomies may still exist. Indeed, despite the large sample studied, some groups of patients were underrepresented when adding other genomic changes in our analysis. Among patients with coexistent del(17p) and del(1p32), only 8 patients had trisomy 3 and 5 patients had trisomy 5 or 21. The group sizes were even smaller for patients with t(4;14) and del(1p32). Moreover, this study was mainly exploratory, and because we were the first to select specific trisomies to understand why myeloma patients with large hyperdiploidy have better OS, our results need to be validated in other populations.
In conclusion, this large study found that specific trisomies could impact TTP and OS in high-risk patients. For the first time, we have shown that not all trisomies have the same prognostic effect. These results, if confirmed in other populations, could have direct consequences on prognostic assessment modalities. For patients with high-risk features, SNP array should be performed, because the presence of trisomies 3 and 5 seems to abrogate the poor outcome value of t(4;14).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was partially supported by the Cancer Pharmacology of Toulouse and Region (CAPTOR) program and a National Institutes of Health, National Cancer Institute PO1 grant (CA100707-12).
Authorship
Contribution: M.-L.C., J.C., and H.A.-L. designed the research, performed the experiments, analyzed the data, and wrote the paper; V.L.-C. and E.Y. performed the statistical analyses and wrote the paper; and all other coauthors provided samples and clinical data and approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hervé Avet-Loiseau, Unit for Genomics in Myeloma, IUC-Oncopole, 1 Ave Irène Joliot-Curie, 31059 Toulouse, France; e-mail: avet-loiseau.h@chu-toulouse.fr.
References
Author notes
M.-L.C. and J.C. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal