Abstract

Background
Somatic mutations identified in patients with myelodysplastic syndromes (MDS) are associated with disease features and carry prognostic information independent of the International Prognostic Scoring System (IPSS) and the revised IPSS (IPSS-R). Risk models that include mutation information have been proposed, but not widely adopted. In practice, there is no consensus on how to best combine clinical information with tumor sequencing data to predict prognosis. To accomplish this, we must define the relevant genes to consider and accurately measure their prognostic impact. Here we examine the relationship between mutations in MDS-associated genes and clinically relevant measures, including overall survival, in a large, multi-center analysis of MDS patient cohorts collected around the globe.
Methods
Data on 3392 MDS patients gathered by members of the International Working Group for Prognosis in MDS-Molecular Committee were combined under the aegis of the MDS Foundation. Patients gave informed consent for collection of their data and tumor samples at their respective institutions in accordance with the Declaration of Helsinki. Samples were examined for somatic mutations primarily by next generation sequencing. Categorical variables were compared using a chi-squared test, while continuous variables were compared using a Wilcoxon rank-sum test. Overall survival (OS) was calculated from the date of the sequenced sample to the date of death and was censored at transplant or the last known follow-up time. P-values are two-sided and considered significant at the <0.001 level to adjust for multiple comparisons.
Results
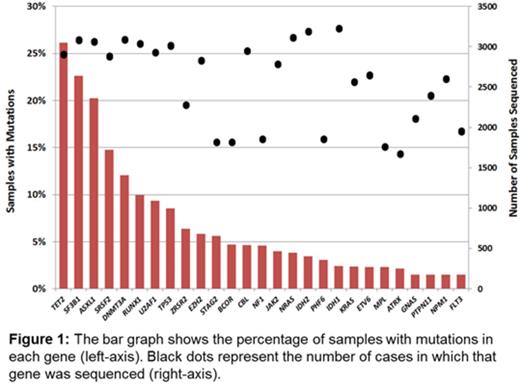
Survival data were available for 3200 patients with a median follow up of 3.7 years and included 1671 deaths. Median survival of the cohort was 2.88 years. The 27 genes sequenced in at least half of the cohort and mutated in > 1.5% of samples were included for analysis (Figure 1). Mutations in 12 genes were strongly associated with shorter OS in univariate analyses (p<0.001 for each gene): ASXL1, CBL, EZH2, IDH2, NF1, NRAS, PTPN11, RUNX1, SRSF2, STAG2, TP53, and U2AF1. Only mutations of SF3B1 were associated with a longer OS at this significance threshold. The large size of the cohort allowed for more precise estimates of survival in less frequently mutated genes. For example, mutations of IDH2 (present in 3.4% of cases, n=103) were associated with shorter OS (hazard ratio [HR] 1.61, 95% confidence interval [CI] 1.26-2.05; p=0.0001) whereas IDH1 mutations (present in 2.4% of cases, n=77) were only marginal (HR 1.29, CI: 0.97-1.72; p=0.082), demonstrating the distinct impact of mutations in these highly related genes.
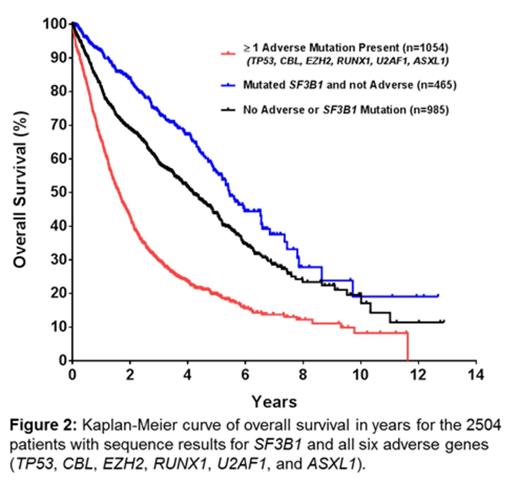
IPSS-R risk groups could be determined for 2173 patients and were strongly associated with OS. Adjusting the hazard ratio of death for IPSS-R risk groups identified several mutated genes with independent prognostic significance: TP53 (HR 2.37, CI 1.94-2.90), CBL (HR 1.57, CI 1.22-2.03), EZH2 (HR 1.55, CI 1.22-2.03), and RUNX1 (HR 1.50, CI 1.24-1.83). Mutations of U2AF1 (HR 1.29, CI 1.06-1.58) and ASXL1 (HR 1.21, CI 1.04-1.41) retained a more modest association with shorter OS. Adjustment for IPSS-R risk groups also moderated the favorable risk associated with mutations of SF3B1 (HR 0.83, CI 0.70-0.99). Patients without mutations in any of the 6 adverse genes above represented 58% of the fully sequenced cohort and had a longer median survival than patients with adverse mutations (4.8 years vs. 1.6 years respectively, p < 0.0001; Figure 2) even after correction for IPSS-R risk groups (adjusted HR 0.59, CI 0.51-0.67).
Multivariable analysis of this dataset will examine the combined contribution of mutated genes to prognosis. A mutation score based on survival risk will be proposed and internally validated. The impact of somatic mutation in patients traditionally considered lower risk will be explored.
Conclusions
This large study definitively validates the prognostic value of mutations in several MDS-associated genes while clarifying the significance of other, less frequently mutated ones. Mutations in several genes retain their prognostic significance after adjustment for IPSS-R risk groups, indicating that these select abnormalities could refine the prediction of prognosis when incorporated into a clinical scoring system such as the IPSS-RM. The results of this analysis will serve as the template with which to build an integrated molecular risk model for MDS.
Bejar:Alexion: Other: ad hoc advisory board; Celgene: Consultancy, Honoraria; Genoptix Medical Laboratory: Consultancy, Honoraria, Patents & Royalties: MDS prognostic gene signature. Haferlach:MLL Munich Leukemia Laboratory: Employment, Equity Ownership. Sekeres:Celgene Corporation: Membership on an entity's Board of Directors or advisory committees; TetraLogic: Membership on an entity's Board of Directors or advisory committees; Amgen: Membership on an entity's Board of Directors or advisory committees. Fenaux:Celgene Corporation: Honoraria, Research Funding; Amgen: Honoraria, Research Funding; Janssen: Honoraria, Research Funding; Novartis: Honoraria, Research Funding. Kern:MLL Munich Leukemia Laboratory: Employment, Equity Ownership. Shih:Novartis: Research Funding. Komrokji:Celgene: Consultancy, Research Funding; Incyte: Consultancy; Novartis: Research Funding, Speakers Bureau; Pharmacylics: Speakers Bureau. List:Celgene Corporation: Honoraria, Research Funding. Santini:celgene, Janssen, Novartis, Onconova: Honoraria, Research Funding. Campbell:14M genomics: Other: Co-founder and consultant. Ebert:Celgene: Consultancy; Genoptix: Consultancy, Patents & Royalties; H3 Biomedicine: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.

This icon denotes a clinically relevant abstract

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal