Abstract
Metabolic remodeling of cancer is controlled by metabolic enzymes having oncogenic or tumor suppressor functions, along with oncogenes and tumor suppressors, which cooperate with the tissue environment to define specific metabolic profiles (Yuneva et al. Cell met 2012). Dysregulated metabolic pathways contribute to the pathogenesis of Acute Myeloid Leukemia (AML), as demonstrated for IDH1/2 mutations, which force the production of the oncometabolite 2-Hydroxyglutarate (Ward et al. Cancer Cell 2010) and can be selectively targeted (Wang et al. Science 2013). However, the genetic determinants of leukemia metabolic plasticity are largely unexplored.
To identify metabolism-related pathogenic mechanisms in AML, we screened 886 AML cases for targeted genomic alterations and performed Whole Exome Sequencing of 143 leukemia samples (100 bp paired-end, HiSeq2000, Illumina), focusing our analysis on 37 AML cases (34 at diagnosis and 3 at relapse). Mutations were called by MuTect and GATK. Moreover, transcriptional analysis was performed on bone marrow cells from 59 AML cases (≥80% blasts) and 7 healthy controls (HTA2.0, Affymetrix).
By mapping the mutated genes into functional categories, we identified a previously undescribed class of mutations targeting metabolism-related genes, that we define metabolic acute leukemia genes (MALGs). MALG was the most represented category after signaling pathways (76/915 genomic alterations) and 29/37 patients carried at least one MALG mutation. MALG mutations mostly targeted biosynthesis and catabolism of lipids and of CoA (ACP2, PANK2), bioenergetic pathways, metabolism of amino acids and nucleotides (NUDT18, IMPDH2). Notably, IMPDH2 is a target of MYC, a known regulator of cancer cell metabolism, and balances the nucleotide pool required for DNA replication (Liu et al, Plos one 2008). IMPDH2 was not only mutated but also upregulated at mRNA level in AML compared with controls (p=0.0001), suggesting an oncogenic function of the gene in AML, which is under investigation. Moreover, MYC transcriptional network was affected by additional mutations targeting genes regulating MYC activity (HUWE1, ZBTB17, TRRAP) and degradation (HEPACAM). Mutations in amino acid metabolism affected the synthesis/degradation of serine (PHGDH), glycine (SHMT2), proline (PRODH), tryptophan(CYP1B1) and glutamate (OPLAH), with a glutamate-related metabolic signature being also enriched in AML. These results may be highly relevant to AML therapy, since they may identify patients suitable to glutaminase inhibitor treatment, which is under development by pharmaceutical companies.
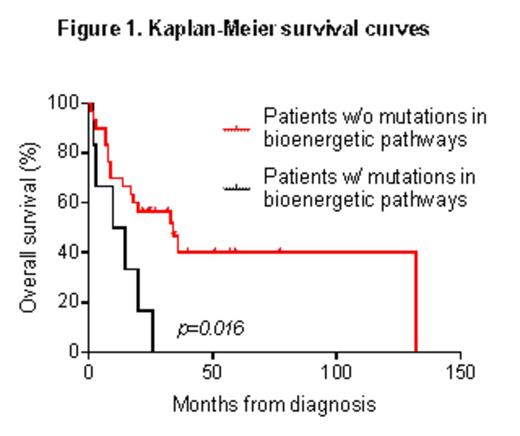
An additional subset of patients displayed mutations in glucose-dependent bioenergetic pathways: glycolysis (GPI), oxidative phosphorylation (ND1, ND4, ND5, CYTB) and pentose phosphate pathway (H6PD, PGLS). These mutations were mutually exclusive with KRAS/NRAS alterations, which were detected in 8/37 samples. Indeed, oncogenic KRAS stimulates glucose uptake and channeling of glucose intermediates into pentose phosphate pathway (Ying et al. Cell 2012). Mutations in the bioenergetic pathways occurred across different cytogenetic groups and were associated with a poor outcome in terms of overall survival (p=0.016 Fig.1) in our AML cohort.
Along with mutations in KRAS- and MYC-oncogenic pathways, which are known to control metabolic processes, we identified a novel functional category of mutated genes involved in metabolism (MALG) in AML. Our results may suggest different types of metabolic remodeling across leukemia subgroups. Mutations targeting a common downstream metabolic pathway are mutually exclusive in our cohort, as shown by KRAS and genes involved in glucose-dependent bioenergetic processes. Glucose metabolism predicts clinical outcome and chemotherapy response in AML (Chen et al. Blood 2014). Our data further suggest that the mutational screening of glucose-related MALGs may define a new subgroup of patients, which could not be identified by cytogenetic analysis. These findings may have implication for AML treatment, since metabolic alterations and genomic determinants of metabolic remodeling are promising targets for tailored therapies, as recently shown for glutaminase and IDH1/2 inhibitors.
Acknowledgments: EHA Research Fellowship award, FP7 NGS-PTL project, ELN, AIL, AIRC, progetto Regione-Università 2010-12
Soverini:Novartis, Briston-Myers Squibb, ARIAD: Consultancy. Cavo:JANSSEN, CELGENE, AMGEN: Consultancy. Martinelli:Novartis: Consultancy, Speakers Bureau; Pfizer: Consultancy; MSD: Consultancy; Ariad: Consultancy; BMS: Consultancy, Speakers Bureau; ROCHE: Consultancy; AMGEN: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal