Key Points
In AML with normal cytogenetics, age, response to induction, and FLT3-ITD allow for an estimate of outcome after allogeneic HSCT in CR1.
Neither variation of classical transplant techniques nor development of chronic GVHD outweighs the negative impact of FLT3-ITD.
Abstract
To analyze the influence of distinct combinations of molecular aberrations on outcome after allogeneic hematopoietic stem cell transplantation (HSCT) for cytogenetically normal acute myeloid leukemia (CN-AML), a retrospective registry analysis was performed on 702 adults undergoing HSCT in first complete remission (CR). Patients were grouped according to presence or absence of NPM1 mutations (NPM1mut) and FLT3 internal tandem duplications (FLT3-ITD). Double-negative patients were evaluated for mutations of the CCAAT/enhancer binding protein α gene (CEBPα). The influence of genotypes on relapse, non-relapse mortality, leukemia-free survival (LFS) and overall survival (OS), and a prognostic classification combining NPM1/FLT3-ITD profile and classical risk factors were calculated. Two-year OS from HSCT was 81 ± 5% in NPM1mut/FLT3wt, 75 ± 3% in NPM1wt/FLT3wt, 66 ± 3% in NPM1mut/FLT3-ITD, and 54 ± 7% in NPM1wt/FLT3-ITD (P = .003). Analysis of CEBPα among patients with NPM1wt/FLT3wt revealed excellent results both in patients with CEBPαmut and with a triple negative genotype (2-year OS: 100%/77 ± 3%). In a Cox-model of predefined variables, age, FLT3-ITD and >1 course of chemotherapy to reach CR were risk factors associated with inferior outcome, regardless of NPM1 mutational status, variations of transplant protocols, or development of graft-versus-host disease. In a prognostic risk classification, 2-year OS/LFS rates were 88 ± 3%/79 ± 4% without any, 77 ± 2%/73 ± 3% with one, and 53 ± 4%/50 ± 4 with ≥2 risk factors (P = .003/.002).
Introduction
Allogeneic hematopoietic stem cell transplantation (alloHSCT) offers a strong antileukemic effect in acute myeloid leukemia (AML), although the benefit in terms of overall survival (OS) is compromised by nonrelapse mortality (NRM).1 In first complete remission (CR1), the indication for alloHSCT is frequently based on genetic risk factors. In general, transplantation is recommended for patients with unfavorable cytogenetics, and discouraged for patients with favorable cytogenetic aberrations, whereas data are less clear in the intermediate cytogenetic subgroup.2,3
In patients with intermediate cytogenetics, and particularly cytogenetically normal AML (CN-AML), molecular aberrations play a decisive role in prognosis.2,4-7 Therefore, international guidelines recommend testing for the two most frequent molecular markers (ie, the mutation of the nucleophosmin1 gene [NPM1mut], and internal-tandem duplication of the fms-related tyrosine kinase 3 gene [FLT3-ITD]), as well as the mutations of the CCAAT/enhancer-binding protein α gene (CEBPα), as part of routine diagnostics in newly diagnosed AML.2 Among other factors, the indication for alloHSCT in CN-AML achieving CR is frequently based on the molecular profile, in particular on the presence of an FLT3-ITD, although data on the role of alloHSCT even in this particular subgroup remain controversial,5,8-11 and the negative prognostic value of this aberration is maintained in the allogeneic setting.12 Recent data suggest that the mutual interaction of co-occurring molecular aberrations, rather than one single aberration, might be decisive for clinical outcome. In particular, the prognostic significance of FLT3-ITD is thought to be modified by NPM1mut.13-15 Nevertheless, it is not clear so far from clinical data, whether patient subgroups characterized by different combinations of molecular markers do have different outcomes after alloHSCT, because the numbers of transplanted patients in reported series are relatively small.5,11,16 With this background, the Acute Leukemia Working Party (ALWP) of European Group of Blood and Bone Marrow Transplantation (EBMT) performed a retrospective, registry-based analysis to provide data on risk factors and OS after alloHSCT in CR1 in large, molecularly defined subgroups of patients with CN-AML.
Patients and methods
Inclusion criteria and data collection
EBMT is a voluntary organization including >500 transplantation centers that are required to file annual follow-up reports on all consecutive HSCT, based on patients’ written informed consent in accordance with the Declaration of Helsinki. After approval by the ALWP board, adult patients with de novo AML were selected from this database according to the following criteria: (1) first alloHSCT in CR1 (excluding CR with incomplete recovery, CRi) between 2006 and 2012; (2) HLA-identical related or at least 7 of 8 antigen (HLA A, B, DR, DQ)-matched unrelated donor (RD/MUD); (3) normal karyotype; and (4) available information on the presence or absence of NPM1mut and FLT3-ITD at the time of diagnosis. Cytogenetics and molecular genetics were performed by the referring institutions according to local standards.
Data extracted from the database and completed by transplant centers upon additional request included age, gender, donor relationship and HLA compatibility, conditioning regimen, graft source, graft-versus-host disease (GVHD) prophylaxis, disease response, incidence of GVHD and relapse after HSCT, survival status, date and cause of death, and last follow-up. To ensure quality of the data, physicians reviewed submitted data and made personal contact with reporting centers to clarify doubtful information.
Definitions and statistics
Remission and relapse,2 conditioning intensity,17 and GVHD18 were defined and classified as described.
The probabilities of acute (aGVHD) and chronic (cGVHD), NRM, and relapse were calculated by using the cumulative incidence estimator to accommodate competing risks. For NRM, relapse was the competing risk, and for relapse, the competing risk was NRM. For aGVHD and cGVHD, death without the event was the competing risk. The Gray test was used for comparisons. OS and leukemia-free survival (LFS) were calculated from date of HSCT using Kaplan-Meier estimates. For all prognostic analyses, continuous variables were categorized and the median was used as a cutoff point. cGVHD was included as a time-dependent variable. A Cox proportional hazards model was used for multivariate regression. Variables differing in terms of distribution between the groups and factors conceptually important were included in the model. Results are expressed as hazard ratio (HR) with 95% confidence interval (CI).
All tests were two-sided. The type I error rate was fixed at .05 for determination of factors associated with time-to-event outcomes. SPSS 19.0 and R 3.0.1 software packages were used.
Results
Information on molecular markers was available in 702 patients. Data on 15 patients reported earlier12 were updated for the present analysis, whereas results in 687 patients had not been analyzed previously. The median age was 51 years, 80% received peripheral blood stem cell grafts. Fifty-five percent each had related, 45% had unrelated (8/8 match, n = 49; 10/10 match, n = 225; 9/10, n = 49) donors. Conditioning was myeloablative (MAC) in 47%, and reduced (RIC) or nonmyeloablative (NMA) in 53%. Based on the presence of NPM1mut and FLT3-ITD at diagnosis, patients were grouped into 4 different genotypes: NPM1wt/Flt3wt (n = 290, 41%), NPM1mut/FLT3wt (n = 68, 10%), NPM1wt/FLT3-ITD (n = 75, 11%), and NPM1mut/FLT3-ITD (n = 269, 38%). Molecular subgroups were well balanced with respect to the majority of characteristics. However, imbalances were observed concerning the interval from diagnosis to CR (9 days longer in the NPM1wt groups) and to alloHSCT (10 days longer in FLT3wt groups), the number of induction courses to reach CR1 (higher in the NPM1wt/FLT3-ITD group), the year of transplantation (1 year earlier in the FLT3wt groups), and the intensity of the conditioning (more MAC in the FLT3-ITD groups; see Table 1 for detailed patient characteristics).
Characteristics of NPM1/FLT3-ITD molecular subgroups among 702 patients undergoing allogeneic HSCT in first CR for AML with normal cytogenetics, as of molecular subgroups
| . | Total (n = 702) . | NPM1wt/FLT3wt (n = 290) . | NPM1mut/FLT3wt (n = 68) . | NPM1wt/ FLT3-ITD (n = 75) . | NPM1mut/ FLT3-ITD (n = 269) . | P . |
|---|---|---|---|---|---|---|
| Age (y), median (range) | 51 (18-71) | 52 (21-70) | 52 (24-67) | 47 (19-70) | 51 (18-71) | .05 |
| Interval diagnosis to CR1 (days), median (range) | 43 (10-210) | 47 | 38 | 48 | 39 | <.001 |
| Interval CR1 to HSCT (days), median (range) | 106 (11-643) | 110 | 107 | 98 | 104 | .5 |
| Interval diag to HSCT (days), median (range) | 155 (55-714) | 162 | 160 | 153 | 149 | .02 |
| Year of HSCT, median | 2010 | 2009 | 2009 | 2010 | 2010 | <.001 |
| Patient sex, n (%) | ||||||
| Male | 357 (51%) | 162 (56%) | 30 (44%) | 41 (55%) | 124 (46%) | .07 |
| Female | 345 (49%) | 128 (44%) | 38 (56%) | 34 (45%) | 145 (54%) | |
| Donor sex , n (%) | ||||||
| Male | 419 (60%) | 182 (63%) | 42 (63%) | 42 (56%) | 153 (57%) | .49 |
| Female | 280 (40%) | 108 (37%) | 25 (37%) | 33 (44%) | 114 (43%) | |
| Female donor for male patient, n (%) | ||||||
| No | 577 (83%) | 235 (81%) | 58 (87%) | 58 (77%) | 226 (85%) | .33 |
| Yes | 122 (18%) | 55 (19%) | 9 (13%) | 17 (23%) | 41 (15%) | |
| Donor type, n (%) | ||||||
| HLA id sibling | 383 (55%) | 163 (56%) | 38 (56%) | 38 (51%) | 144 (54%) | .82* |
| UD | 319 (45%) | 127 (44%) | 30 (44%) | 37 (49%) | 125 (47%) | |
| 10/10 AG match | 225 | 93 | 19 | 25 | 88 | .51† |
| 9/10 AG match | 49 | 21 | 7 | 6 | 15 | |
| 8/8 AG match | 45 | 13 | 4 | 6 | 22 | |
| Conditioning, n (%) | ||||||
| Myeloablative | 330 (47%) | 116 (40%) | 31 (46%) | 43 (57%) | 140 (52%) | .007 |
| BuCy | 162 | 67 | 15 | 16 | 64 | |
| BuFlu | 41 | 11 | 1 | 6 | 23 | |
| CyTBI | 88 | 27 | 7 | 14 | 40 | |
| Other | 39 | 11 | 8 | 7 | 13 | |
| Reduced, n (%) | 370 (53%) | 174 (60%) | 37 (54%) | 32 (43%) | 127 (48%) | |
| BuFlu | 192 | 90 | 16 | 13 | 73 | |
| FluMel | 55 | 18 | 8 | 5 | 24 | |
| Other | 123 | 66 | 13 | 14 | 30 | |
| CMV serostatus (donor/patient), n (%) | ||||||
| neg/neg | 193 (28%) | 80 (28%) | 19 (29%) | 25 (33%) | 68 (25%) | .86 (.06 for neg/neg vs other) |
| pos/neg | 71 (10%) | 33 (11%) | 5 (8%) | 8 (11%) | 24 (9%) | |
| neg/pos | 170 (24%) | 67 (23%) | 14 (21%) | 16 (21%) | 72 (27%) | |
| pos/pos | 268 (38%) | 110 (38%) | 28 (42%) | 26 (35%) | 103 (39%) | |
| Stem cell source, n (%) | ||||||
| BM | 159 (23%) | 62 (21%) | 18 (27%) | 15 (20%) | 64 (24%) | .72 |
| PB | 543 (77%) | 228 (79%) | 50 (74%) | 60 (80%) | 205 (76%) | |
| Number of induction courses to reach CR1, n (%) | ||||||
| 1 | 486 (75%) | 200 (75%) | 47 (73%) | 35 (52%) | 204 (81%) | <.001 |
| ≥2 | 163 (25%) | 66 (25%) | 17 (27%) | 33 (49%) | 47 (19%) | |
| Total number of chemotherapy courses before HSCT, n (%) | ||||||
| 1 | 83 (29%) | 38 (29%) | 11 (29%) | 8 (28%) | 26 (32%) | .27 |
| 2 | 112 (40%) | 56 (42%) | 18 (47%) | 12 (41%) | 26 (32%) | |
| 3 | 62 (22%) | 33 (25%) | 4 (11%) | 5 (17%) | 20 (24%) | |
| >3 | 25 (9%) | 6 (5%) | 5 (13%) | 4 (14%) | 10 (12%) | |
| Use of ATG, n (%) | ||||||
| No | 387 (55%) | 156 (54%) | 36 (53%) | 43 (58%) | 152 (57%) | .85 |
| Yes | 312 (45%) | 133 (46%) | 32 (47%) | 31 (42%) | 116 (43%) | |
| HLA id sibling, n (%) | ||||||
| No ATG | 268 (71%) | 111 (69%) | 24 (63%) | 26 (70%) | 107 (75%) | .46 |
| ATG | 112 (29%) | 51 (31%) | 14 (37%) | 11 (30%) | 36 (25%) | |
| URD, n (%) | ||||||
| No ATG | 119 (37%) | 45 (35%) | 12 (40%) | 17 (46%) | 45 (36%) | .67 |
| ATG | 200 (63%) | 82 (65%) | 18 (60%) | 20 (54%) | 80 (64%) | |
| GVHD prophylaxis, n (%) | ||||||
| CyA-based | 635 (91%) | 266 (92%) | 61 (90%) | 67 (92%) | 241 (90%) | .43 |
| CyA+MTX | 322 | 123 | 33 | 33 | 133 | |
| CyA+MMF | 187 | 92 | 13 | 26 | 56 | |
| CyA+other | 126 | 51 | 15 | 8 | 52 | |
| Tacrolimus-based | 37 (5%) | 16 (6%) | 2 (3%) | 4 (5%) | 15 (6%) | |
| Tacrolimus+MMF | 21 | 14 | 2 | 1 | 4 | |
| Tacrolimus+other | 16 | 2 | — | 3 | 9 | |
| Other | 30 | 8 | 5 | 4 | 13 | |
| 3% | 2% | 7% | 3% | 4% | ||
| Acute GVHD after HSCT, n (%) | ||||||
| AGVH <II | 498 (74%) | 199 (72%) | 56 (85%) | 52 (70%) | 191 (73%) | .16 |
| AGVH ≥II | 180 (27%) | 78 (28%) | 10 (15%) | 22 (30%) | 70 (27%) |
| . | Total (n = 702) . | NPM1wt/FLT3wt (n = 290) . | NPM1mut/FLT3wt (n = 68) . | NPM1wt/ FLT3-ITD (n = 75) . | NPM1mut/ FLT3-ITD (n = 269) . | P . |
|---|---|---|---|---|---|---|
| Age (y), median (range) | 51 (18-71) | 52 (21-70) | 52 (24-67) | 47 (19-70) | 51 (18-71) | .05 |
| Interval diagnosis to CR1 (days), median (range) | 43 (10-210) | 47 | 38 | 48 | 39 | <.001 |
| Interval CR1 to HSCT (days), median (range) | 106 (11-643) | 110 | 107 | 98 | 104 | .5 |
| Interval diag to HSCT (days), median (range) | 155 (55-714) | 162 | 160 | 153 | 149 | .02 |
| Year of HSCT, median | 2010 | 2009 | 2009 | 2010 | 2010 | <.001 |
| Patient sex, n (%) | ||||||
| Male | 357 (51%) | 162 (56%) | 30 (44%) | 41 (55%) | 124 (46%) | .07 |
| Female | 345 (49%) | 128 (44%) | 38 (56%) | 34 (45%) | 145 (54%) | |
| Donor sex , n (%) | ||||||
| Male | 419 (60%) | 182 (63%) | 42 (63%) | 42 (56%) | 153 (57%) | .49 |
| Female | 280 (40%) | 108 (37%) | 25 (37%) | 33 (44%) | 114 (43%) | |
| Female donor for male patient, n (%) | ||||||
| No | 577 (83%) | 235 (81%) | 58 (87%) | 58 (77%) | 226 (85%) | .33 |
| Yes | 122 (18%) | 55 (19%) | 9 (13%) | 17 (23%) | 41 (15%) | |
| Donor type, n (%) | ||||||
| HLA id sibling | 383 (55%) | 163 (56%) | 38 (56%) | 38 (51%) | 144 (54%) | .82* |
| UD | 319 (45%) | 127 (44%) | 30 (44%) | 37 (49%) | 125 (47%) | |
| 10/10 AG match | 225 | 93 | 19 | 25 | 88 | .51† |
| 9/10 AG match | 49 | 21 | 7 | 6 | 15 | |
| 8/8 AG match | 45 | 13 | 4 | 6 | 22 | |
| Conditioning, n (%) | ||||||
| Myeloablative | 330 (47%) | 116 (40%) | 31 (46%) | 43 (57%) | 140 (52%) | .007 |
| BuCy | 162 | 67 | 15 | 16 | 64 | |
| BuFlu | 41 | 11 | 1 | 6 | 23 | |
| CyTBI | 88 | 27 | 7 | 14 | 40 | |
| Other | 39 | 11 | 8 | 7 | 13 | |
| Reduced, n (%) | 370 (53%) | 174 (60%) | 37 (54%) | 32 (43%) | 127 (48%) | |
| BuFlu | 192 | 90 | 16 | 13 | 73 | |
| FluMel | 55 | 18 | 8 | 5 | 24 | |
| Other | 123 | 66 | 13 | 14 | 30 | |
| CMV serostatus (donor/patient), n (%) | ||||||
| neg/neg | 193 (28%) | 80 (28%) | 19 (29%) | 25 (33%) | 68 (25%) | .86 (.06 for neg/neg vs other) |
| pos/neg | 71 (10%) | 33 (11%) | 5 (8%) | 8 (11%) | 24 (9%) | |
| neg/pos | 170 (24%) | 67 (23%) | 14 (21%) | 16 (21%) | 72 (27%) | |
| pos/pos | 268 (38%) | 110 (38%) | 28 (42%) | 26 (35%) | 103 (39%) | |
| Stem cell source, n (%) | ||||||
| BM | 159 (23%) | 62 (21%) | 18 (27%) | 15 (20%) | 64 (24%) | .72 |
| PB | 543 (77%) | 228 (79%) | 50 (74%) | 60 (80%) | 205 (76%) | |
| Number of induction courses to reach CR1, n (%) | ||||||
| 1 | 486 (75%) | 200 (75%) | 47 (73%) | 35 (52%) | 204 (81%) | <.001 |
| ≥2 | 163 (25%) | 66 (25%) | 17 (27%) | 33 (49%) | 47 (19%) | |
| Total number of chemotherapy courses before HSCT, n (%) | ||||||
| 1 | 83 (29%) | 38 (29%) | 11 (29%) | 8 (28%) | 26 (32%) | .27 |
| 2 | 112 (40%) | 56 (42%) | 18 (47%) | 12 (41%) | 26 (32%) | |
| 3 | 62 (22%) | 33 (25%) | 4 (11%) | 5 (17%) | 20 (24%) | |
| >3 | 25 (9%) | 6 (5%) | 5 (13%) | 4 (14%) | 10 (12%) | |
| Use of ATG, n (%) | ||||||
| No | 387 (55%) | 156 (54%) | 36 (53%) | 43 (58%) | 152 (57%) | .85 |
| Yes | 312 (45%) | 133 (46%) | 32 (47%) | 31 (42%) | 116 (43%) | |
| HLA id sibling, n (%) | ||||||
| No ATG | 268 (71%) | 111 (69%) | 24 (63%) | 26 (70%) | 107 (75%) | .46 |
| ATG | 112 (29%) | 51 (31%) | 14 (37%) | 11 (30%) | 36 (25%) | |
| URD, n (%) | ||||||
| No ATG | 119 (37%) | 45 (35%) | 12 (40%) | 17 (46%) | 45 (36%) | .67 |
| ATG | 200 (63%) | 82 (65%) | 18 (60%) | 20 (54%) | 80 (64%) | |
| GVHD prophylaxis, n (%) | ||||||
| CyA-based | 635 (91%) | 266 (92%) | 61 (90%) | 67 (92%) | 241 (90%) | .43 |
| CyA+MTX | 322 | 123 | 33 | 33 | 133 | |
| CyA+MMF | 187 | 92 | 13 | 26 | 56 | |
| CyA+other | 126 | 51 | 15 | 8 | 52 | |
| Tacrolimus-based | 37 (5%) | 16 (6%) | 2 (3%) | 4 (5%) | 15 (6%) | |
| Tacrolimus+MMF | 21 | 14 | 2 | 1 | 4 | |
| Tacrolimus+other | 16 | 2 | — | 3 | 9 | |
| Other | 30 | 8 | 5 | 4 | 13 | |
| 3% | 2% | 7% | 3% | 4% | ||
| Acute GVHD after HSCT, n (%) | ||||||
| AGVH <II | 498 (74%) | 199 (72%) | 56 (85%) | 52 (70%) | 191 (73%) | .16 |
| AGVH ≥II | 180 (27%) | 78 (28%) | 10 (15%) | 22 (30%) | 70 (27%) |
ATG, antithymocyte globulin; BuCy, busulfan/cyclophosphamide; BuFlu, busulfan/fludarabin; CR1, first complete remission; CyA, cyclosporin A; CyTBI, cyclophosphamide/total body irradiation; FluMel, fludarabin/melphalan; MMF, mycophenolate mofetil; MTX, methotrexate; UD, unrelated donor.
HLA identical sibling vs unrelated donors.
Different subgroups of unrelated donors (10/0 vs 9/10 vs 8/8 AG match).
Relapse and NRM after alloHSCT
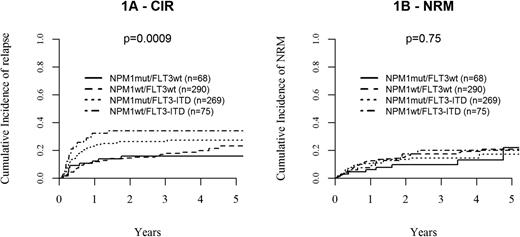
Concerning cumulative incidence of relapse (CIR), the molecular subgroups differed significantly according to FLT3 mutational status, with patients with FLT3-ITD showing a higher CIR (26 ± 3% and 34 ± 6% at 2 years in patients with and without concomitant NPM1mut, respectively) compared with patients lacking FLT3-ITD (2-year CIR: 16 ± 3% and 14 ± 2% in patients with and without NPM1mut, respectively; global P value between FLT3-ITD and FLT3wt: .0009). In contrast, the presence or absence of NPM1mut did not significantly influence CIR in both FLT3-ITD and FLT3wt. Molecular subgroups did not show any influence on NRM (global P value: .75; Figure 1 and supplemental Table 1).
Relapse and non-relapse mortality after alloHSCT according to molecular subgroups. Cumulative incidence plots illustrating the combined influence of NMP1 and FLT3 mutational status on (A) relapse (CIR) and (B) non-relapse mortality (NRM).
Relapse and non-relapse mortality after alloHSCT according to molecular subgroups. Cumulative incidence plots illustrating the combined influence of NMP1 and FLT3 mutational status on (A) relapse (CIR) and (B) non-relapse mortality (NRM).
In the multivariate model, FLT3-ITD (HR 2.23, 95%CI 1.44-3.46, P = .0003) and the number of courses of induction chemotherapy to reach CR1 (HR 1.50, 95% CI 1.02-2.22, P = .04) showed significant influence on CIR, whereas variations of the transplant procedure such as donor choice, (sibling vs unrelated), intensity of the conditioning, total body irradiation (TBI) and use of ATG had no influence. Younger age (HR: 3.42, 95% CI 1.98-5.91, P < .0001), RIC (HR 0.57, 95% CI 0.34-0,97, P = .04), and a shorter interval between achievement of CR and date of alloHSCT (HR 0.55, 95% CI 0.34-0.90, P = .02), but not molecular subtype, intensity of the conditioning, or donor type (including 1 AG-mismatched unrelated donors) were protective against NRM (Table 2).
Multivariate analysis of risk factors for 2-y outcome after allogeneic HSCT
| . | P . | HR . | 95% CI . | |
|---|---|---|---|---|
| Lower . | Upper . | |||
| CIR | ||||
| FLT3-ITD vs FLT3wt | .0003 | 2.23 | 1.44 | 3.46 |
| NPM1mut vs NPM1wt | .47 | 0.85 | 0.56 | 1.31 |
| RIC vs MAC | .032 | 1.25 | 0.81 | 1.93 |
| age > median | .21 | 1.31 | 0.86 | 1.99 |
| UD vs HLA id | .18 | 0.77 | 0.52 | 1.13 |
| Interval CR1 to HSCT > median | .94 | 0.99 | 0.67 | 1.45 |
| Year of HSCT > median | .32 | 0.81 | 0.54 | 1.22 |
| Number of induction courses for CR1 > 1 | .04 | 1.50 | 1.02 | 2.22 |
| NRM | ||||
| FLT3-ITD vs FLT3wt | .37 | 1.28 | 0.74 | 2.19 |
| NPM1mut vs NPM1wt | .50 | 0.83 | 0.49 | 1.42 |
| RIC vs MAC | .04 | 0.57 | 0.34 | 0.97 |
| age > median | .00001 | 3.42 | 1.98 | 5.91 |
| UD vs HLA id | .10 | 1.47 | 0.93 | 2.31 |
| Interval CR1 to HSCT > median | .02 | 1.80 | 1.11 | 2.91 |
| Year of HSCT > median | .41 | 0.81 | 0.48 | 1.35 |
| Number of induction courses for CR1 > 1 | .23 | 1.36 | 0.82 | 2.24 |
| OS | ||||
| FLT3-ITD vs FLT3wt | .001 | 1.85 | 1.29 | 2.66 |
| NPM1mut vs NPM1wt | .24 | 0.81 | 0.57 | 1.15 |
| RIC vs MAC | .53 | 0.89 | 0.62 | 1.28 |
| age > median | .0000005 | 2.54 | 1.77 | 3.66 |
| UD vs HLA id | .41 | 1.14 | 0.84 | 1.56 |
| Interval CR1 to HSCT > median | .08 | 1.33 | 0.96 | 1.83 |
| Year of HSCT > median | .41 | 0.86 | 0.60 | 1.23 |
| Number of induction courses for CR1 > 1 | .04 | 1.37 | 0.99 | 1.91 |
| LFS | ||||
| FLT3-ITD vs FLT3wt | .001 | 1.77 | 1.27 | 2.48 |
| NPM1mut vs NPM1wt | .32 | 0.84 | 0.61 | 1.18 |
| RIC vs MAC | .60 | 0.91 | 0.65 | 1.28 |
| age > median | .0002 | 1.90 | 1.36 | 2.66 |
| UD vs HLA id | 1.00 | 1.00 | 0.75 | 1.34 |
| Interval CR1 to HSCT > median | .14 | 1.25 | 0.93 | 1.67 |
| Year of HSCT > median | .23 | 0.82 | 0.60 | 1.13 |
| Number of induction courses for CR1 > 1 | .02 | 1.43 | 1.06 | 1.95 |
| . | P . | HR . | 95% CI . | |
|---|---|---|---|---|
| Lower . | Upper . | |||
| CIR | ||||
| FLT3-ITD vs FLT3wt | .0003 | 2.23 | 1.44 | 3.46 |
| NPM1mut vs NPM1wt | .47 | 0.85 | 0.56 | 1.31 |
| RIC vs MAC | .032 | 1.25 | 0.81 | 1.93 |
| age > median | .21 | 1.31 | 0.86 | 1.99 |
| UD vs HLA id | .18 | 0.77 | 0.52 | 1.13 |
| Interval CR1 to HSCT > median | .94 | 0.99 | 0.67 | 1.45 |
| Year of HSCT > median | .32 | 0.81 | 0.54 | 1.22 |
| Number of induction courses for CR1 > 1 | .04 | 1.50 | 1.02 | 2.22 |
| NRM | ||||
| FLT3-ITD vs FLT3wt | .37 | 1.28 | 0.74 | 2.19 |
| NPM1mut vs NPM1wt | .50 | 0.83 | 0.49 | 1.42 |
| RIC vs MAC | .04 | 0.57 | 0.34 | 0.97 |
| age > median | .00001 | 3.42 | 1.98 | 5.91 |
| UD vs HLA id | .10 | 1.47 | 0.93 | 2.31 |
| Interval CR1 to HSCT > median | .02 | 1.80 | 1.11 | 2.91 |
| Year of HSCT > median | .41 | 0.81 | 0.48 | 1.35 |
| Number of induction courses for CR1 > 1 | .23 | 1.36 | 0.82 | 2.24 |
| OS | ||||
| FLT3-ITD vs FLT3wt | .001 | 1.85 | 1.29 | 2.66 |
| NPM1mut vs NPM1wt | .24 | 0.81 | 0.57 | 1.15 |
| RIC vs MAC | .53 | 0.89 | 0.62 | 1.28 |
| age > median | .0000005 | 2.54 | 1.77 | 3.66 |
| UD vs HLA id | .41 | 1.14 | 0.84 | 1.56 |
| Interval CR1 to HSCT > median | .08 | 1.33 | 0.96 | 1.83 |
| Year of HSCT > median | .41 | 0.86 | 0.60 | 1.23 |
| Number of induction courses for CR1 > 1 | .04 | 1.37 | 0.99 | 1.91 |
| LFS | ||||
| FLT3-ITD vs FLT3wt | .001 | 1.77 | 1.27 | 2.48 |
| NPM1mut vs NPM1wt | .32 | 0.84 | 0.61 | 1.18 |
| RIC vs MAC | .60 | 0.91 | 0.65 | 1.28 |
| age > median | .0002 | 1.90 | 1.36 | 2.66 |
| UD vs HLA id | 1.00 | 1.00 | 0.75 | 1.34 |
| Interval CR1 to HSCT > median | .14 | 1.25 | 0.93 | 1.67 |
| Year of HSCT > median | .23 | 0.82 | 0.60 | 1.13 |
| Number of induction courses for CR1 > 1 | .02 | 1.43 | 1.06 | 1.95 |
HLA, human leukocyte antigen; MAC, myeloablative conditioning; MUD, matched unrelated donor.
GVHD
Cumulative incidence of acute GVHD grades 2-4 and cGVHD was 29 ± 2% and 40 ± 2%, respectively, with no differences among molecular subgroups (global P = .23 for aGVHD, .27 for cGvHD; see supplemental Table 1 for details). No significant influence of cGvHD on CIR could be detected either in the entire cohort or within molecular subgroups (P = .30/.20 among FLT3wt+/−NPM1mut, .90/.96 among FLT3-ITD+/−NPM1mut, respectively) when including cGVHD into the model as a time-dependent variable.
OS and LFS after alloHSCT
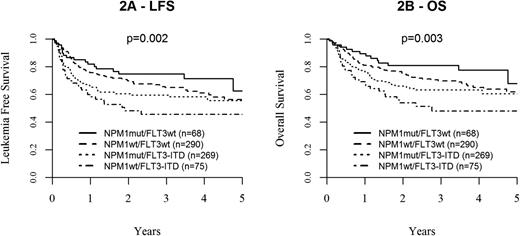
With a median follow-up of 26 months from transplantation among survivors, 2-year OS and LFS for the entire cohort were 70 ± 2% and 64 ± 2%, respectively. Molecular subgroups had a strong influence on outcome (global P = .003 for OS, P = .002 for LFS), with the best outcome observed in the NPM1mut/FLT3wt group (2-year OS: 81 ± 5%, LFS: 75 ± 5%). Notably, NPM1wt/FLT3wt patients showed similarly favorable results (2-year OS: 75 ± 3% and LFS: 70 ± 3%), whereas outcome was clearly inferior in patients harboring an FLT3-ITD (2-year OS: 66 ± 3%/LFS: 60 ± 7% in NPM1mut/FLT3-ITD and 54 ± 7%/48 ± 7% in NPM1wt/FLT3-ITD). Thus, in the presence of FLT3-ITD, NPM1mut showed a positive trend, but did not significantly alter outcome results (P = .15 for OS, P = .13 for LFS; Figure 2A, LFS, B, OS; supplemental Table 1).
Leukemia free survival and overall survival according to molecular subgroups. Kaplan-Meier plots showing the influence of distinct molecular subgroups based on NMP1 and FLT3 mutational status on (A) LFS and (B) OS.
Leukemia free survival and overall survival according to molecular subgroups. Kaplan-Meier plots showing the influence of distinct molecular subgroups based on NMP1 and FLT3 mutational status on (A) LFS and (B) OS.
Using a Cox model for multivariate analysis, the presence of an FLT3-ITD (HR 1.85, 95% CI 1.29-2.66, P = .001 for OS; HR 1.77, 95% CI 1.27-2.48, P = .001 for LFS) and age above the median (HR 2.54, 95% CI 1.77-3.66, P < .0001 for OS; HR 1.90, 95% CI 1.36-2.66, P = .0002 for LFS) were the main risk factors for outcome. Further, the number of induction courses to reach CR1 was of borderline significance (HR 1.37, 95% CI 0.99-1.91, P = .06 for OS; HR 1.43, 95% CI 1.06-1.95, P = .02 for LFS; Table 2). As with CIR, outcome was not influenced either by modifications of the transplant regimen (including donor type, donor match, and intensity of the conditioning) or development of GVHD.
Impact of mutated CEBPα among double-negative (NPM1wt/Flt3wt) patients
To further subdivide the NPM1wt/Flt3wt cohort, the role of the mutational status of CEBPα was analyzed in 151 informative patients. Thus, 2-year OS/LFS among triple-negative patients (n = 138, 91%) was 77 ± 3%/72 ± 3%, whereas 13 patients (9%) harboring a CEPBα mutation had an OS/LFS of 100%/92 ± 3%.
Prognostic risk classification
Based on the 3 independent risk factors (FLT3-ITD, age above the median, >1 induction course to reach CR1), a prognostic classification for outcome of CN-AML after alloHSCT was developed. Outcome parameters were significantly influenced by the score (none vs 1 vs 2 or 3 factors; P = .003 for OS, P = .002 for LFS, P = .0002 for CRI, P = .01 for NRM; Table 3 and Figure 3). The classification was then validated in an independent cohort of an earlier study from our group.12 Although the 2 cohorts differed significantly with respect to important variables (eg, intensity of the conditioning, year of transplant, length of follow-up), the prognostic value of the classification was confirmed (P < .0001 for LFS, OS and CIR, respectively).
Prognostic score for 2-year outcome after alloHSCT in CN-AML
| . | N . | LFS (% ± SE) . | OS (% ± SE) . | CIR (% ± SE) . | NRM (% ± SE) . |
|---|---|---|---|---|---|
| 0 factors | 104 | 79 ± 4 | 88 ± 3 | 14 ± 4 | 7 ± 3 |
| 1 factor | 322 | 73 ± 3 | 77 ± 2 | 15 ± 2 | 12 ± 2 |
| 2 or 3 factors | 223 | 50 ± 4 | 53 ± 4 | 31 ± 3 | 20 ± 2 |
| P | .002 | .003 | .0002 | .01 |
| . | N . | LFS (% ± SE) . | OS (% ± SE) . | CIR (% ± SE) . | NRM (% ± SE) . |
|---|---|---|---|---|---|
| 0 factors | 104 | 79 ± 4 | 88 ± 3 | 14 ± 4 | 7 ± 3 |
| 1 factor | 322 | 73 ± 3 | 77 ± 2 | 15 ± 2 | 12 ± 2 |
| 2 or 3 factors | 223 | 50 ± 4 | 53 ± 4 | 31 ± 3 | 20 ± 2 |
| P | .002 | .003 | .0002 | .01 |
SE, standard error.
Presence of Flt3-ITD, age > median, and >1 course of chemotherapy to reach CR1 were included as risk factors.
Prognostic classification of outcome after allogeneic HSCT for CN-AML in first complete remission, based on independent prognostic parameters (FLT3-ITD, age, and the number of induction courses to achieve CR). Estimate of (A) LFS, (B) OS, (C) CIR, and (D) NRM.
Prognostic classification of outcome after allogeneic HSCT for CN-AML in first complete remission, based on independent prognostic parameters (FLT3-ITD, age, and the number of induction courses to achieve CR). Estimate of (A) LFS, (B) OS, (C) CIR, and (D) NRM.
Discussion
In the largest study presented thus far on the role of molecular markers in adult CN-AML patients undergoing alloHSCT in CR1, significant differences among genetic subgroups were observed. Thus, in addition to patient age, FLT3-ITD, but not NPM1mut, was identified as a decisive factor for outcome. NRM and GVHD were not influenced by the molecular profile. By providing data on OS in high numbers of recently transplanted patients, including both related and unrelated donors, as well as reduced and myeloablative transplants, the results firmly establish which outcome can be expected after alloHSCT in different molecular subgroups of CN-AML. Given the fact that leukemia relapse as the decisive event for outcome after alloHSCT for AML was observed at a median of <6 months from alloHSCT both after RIC and MAC transplants,19-21 a follow-up longer than 2 years seemed to be reasonable. Further, the data allowed for a prognostic classification of patients undergoing alloHSCT for CN-AML.
Strict inclusion criteria and an extensive survey among participating centers, including repeated questionnaires and personal contacts, ensured high patient numbers and data quality. Nevertheless, the nature of a retrospective, registry-based study implicates limitations.
First, the EBMT registry only provides data on patients who in fact underwent alloHSCT. Therefore, we are not able to answer the question of whether alloHSCT should be offered to all patients diagnosed with CN-AML and one of the molecular subgroups defined here.
Second, we could not determine the mutant/wild-type allele ratio (AR) of FLT3-ITD nor the insertion site of FLT3-ITD in the majority of patients. Both variables have been described to play a major role for outcome after conventional therapy and alloHSCT, and also seemed to modify the role of other mutations, such as NPM1mut.16,22-24 However, heterogeneity, methodologic problems, and the relatively low sensitivity of most polymerase chain reaction assays, as well as a missing general agreement concerning a cutoff level for the FLT3-ITD/wild-type AR,11,16,23-27 have prompted the suggestion to generally classify all non-APL FLT3-ITD cases as poor risk.10,28 Further, no role of the mutant/wild-type AR on CIR after alloHSCT could be shown by the recent AML-SG study,11 and next-generation sequencing revealed the presence of different FLT3-ITD clones within the same patient both at diagnosis and during the course of the disease.29,30 Therefore, and in accordance with several well-accepted prognostic models,3,5,31 and recent classification systems7 for CN-AML, we decided to limit our analysis to the general presence or absence of FLT3-ITD.
Third, we do not have data on other recently identified mutations possibly modifying the prognostic role of both FLT3wt and FLT3-ITD patient subsets, such as TET2, DNMT3A, ASXL1, and IDH1/2. The prognostic role and mutual interaction of these mutations is a matter of ongoing research,32 and the role of different genotypes might vary according to the applied therapy, as shown for high-dose daunorubicin.7 Further, integration of several mutations into a clinically based prognostic scoring system is difficult, as demonstrated in a recent study by the German AML-SG, where high numbers of missing data on concurrent mutations precluded the inclusion of these variables in a multivariable model on outcome.11 Hence, for the time being, the data in molecular subgroups, which are based on the 2 most frequent molecular markers, as well as the proposed prognostic classification, might be a reasonable tool to estimate the outcome after alloHSCT in CR1 in a given patient with CN-AML.
The role of the general presence of FLT3-ITD for LFS and CIR, even after alloHSCT, has been shown previously for patients undergoing myeloablative conditioning for predominantly matched sibling transplants,12 although it had not been observed by others.9,33 In addition to confirming the negative influence of FLT3-ITD in a larger cohort including unrelated transplants and RIC, and extending it to an analysis on OS, we also looked for variations within the transplant procedure to identify strategies for improvement in this high-risk cohort. However, when adjusting for confounding factors, no unrelated donor, modified intensity of the conditioning, nor the use of ATG or inclusion of TBI into the preparative regimen could be shown to cause a significant difference among patients with FLT3-ITD. Similarly, development of cGVHD did not significantly protect against relapse. Hence, it seems unlikely that the negative prognostic value of FLT3-ITD might be abrogated by modification of the traditional components of the transplant procedure. This strongly argues in favor of the integration of innovative approaches into the transplant strategies. As an example, FLT3 inhibitors, which have been studied either as part of the induction treatment,34-36 as bridging to alloHSCT28 or as maintenance after alloSCT, should be further evaluated in randomized trials to improve outcome in this subgroup of patients.
As shown earlier,5,37 NPM1mut defined a subgroup with excellent prognosis among patients with FLT3wt. In the context of alloHSCT, this is ascribed to the presence of a particular strength of the allogeneic immune response.38 In contrast, the previously described protective role of NPM1mut in patients bearing FLT3-ITD13-15 could not be unequivocally confirmed by our data. Hence, this co-occurrence might either play no major role after alloHSCT, or the influence of NPM1mut might be limited to patients with a low FLT3-ITD/wild-type AR.16,24 However, in a recent AML-SG study, no impact of a concurrent NPM1 mutation could be demonstrated either.11 Integrated genetic profiling data further revealed a modification of the prognostic role of FLT3-ITD by other mutations not evaluated in our study (eg, TET2 or DNMT3A).7
Patients with a double-negative genotype (NPM1wt/FLT3wt) were further characterized by the presence or absence of CEBPα, the third molecular aberration generally recommended for testing in newly diagnosed AML.2 Accordingly, even triple-negative patients (n = 138) showed an excellent outcome after alloHSCT, although having been identified to bear an increased risk in earlier studies.5 This confirms data suggesting a potent graft-vs-leukemia effect in this particular subgroup.39 Longer follow-up might be required to confirm this observation, because this subgroup was the only one showing late relapses beyond 3 years from HSCT. In contrast, the excellent outcome of 13 patients with NPM1wt/FLT3wt and mutated CEBPα should not be overinterpreted, given low numbers and missing information, whether or not CEBPα mutation was bi-allelic.40
In conclusion, our data allow for a reliable prognostic estimate of outcome in different, well-defined molecular subgroups of patients with CN-AML after alloHSCT in CR1, with a remarkable impact of age and FLT3-ITD. Additional molecular features such as FLT3-ITD allelic burden or insertion site of FLT3-ITD,11,41 as well as the simultaneous search for co-occurring and potentially interacting molecular markers, might refine the accuracy of the estimate. The relevance of these additional characteristics should, however, be evaluated specifically in the setting of alloSCT, and in reliable numbers of patients. The study had not been designed to answer the question of whether patients with certain molecular subgroups should or should not undergo alloHSCT in CR1, nor can the findings be transferred to the entire patient population with newly diagnosed CN-AML. Nevertheless, the data might provide a basis for the decision between transplant and nontransplant consolidation strategies by giving a clear idea of the outcome to be expected after alloHSCT in a certain patient. In FLT3-ITD CN-AML, modifications of traditional transplant techniques did not improve outcome. Hence, studies evaluating the inclusion of innovative components, such as FLT3-inhibitors, are warranted.
Preliminary results of this study were presented at the annual meetings of the American Society of Hematology, New Orleans, LA, December 7-10, 2013, and the European Group of Blood and Marrow Transplantation, Milan, Italy, March 30 to April 2, 2014.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Following EBMT publication rules, co-authorship was offered to centers contributing the highest number of patients. Nevertheless, the authors highly appreciate the contribution by many physicians and data managers throughout the EBMT, who made this analysis possible. The authors further wish to acknowledge the enormous help by the data managers in the Acute Leukemia Working Party office in Paris, and by Daniela Engel and Astrid Bader, Augsburg, Germany.
The work was supported by an unrestricted research grant by Novartis Inc.
Authorship
Contribution: C.S., M.L., J.E., A.N., and M.M. designed the study, performed the analysis, and interpreted and discussed the results; C.S. wrote the manuscript; M.L., J.E., A.N., and M.M. refined the manuscript; E.P. managed the data; M.L. performed the statistical analysis; G.S., E.D., L.V., A.H., J.H.B., N.M., J.C., P.C., J.M., P.J., D.B., S.L., and N.I. contributed the largest numbers of patients, critically reviewed the manuscript, and made substantial contribution to the interpretation of the data and the final text; and F.B., F.C., C.G., B.S., and S.G. contributed to the design of the study, critically reviewed the manuscript, and made substantial contributions to the interpretation of the data and the final text.
Conflict-of-interest disclosure: C.S. has received an unrestricted research grant from Novartis Inc. The remaining authors declare no competing financial interests.
A list of contributing centers is provided in the supplemental Appendix.
Correspondence: Christoph Schmid, Stem Cell Transplantation Unit, Klinikum Augsburg, Ludwig-Maximilians-University of Munich, Stenglinstr. 2, D-86156 Augsburg, Germany; e-mail: christoph.schmid@klinikum-augsburg.de.
References
Author notes
J.E., A.N., and M.M. contributed equally to this study.