Key Points
Ezh2 loss in hematopoietic stem cells predisposes mice to develop heterogeneous hematologic malignancies.
Ezh1 is essential to maintain hematopoiesis in the setting of Ezh2 loss.
Abstract
Recent genome sequencing revealed inactivating mutations in EZH2, which encodes an enzymatic component of polycomb-repressive complex 2 (PRC2), in patients with myelodysplastic syndrome (MDS), myeloproliferative neoplasms (MPNs), and MDS/MPN overlap disorders. We herein demonstrated that the hematopoietic-specific deletion of Ezh2 in mice induced heterogeneous hematopoietic malignancies. Myelodysplasia was detected in mice following the deletion of Ezh2, and resulted in the development of MDS and MDS/MPN. Thrombocytosis was induced by Ezh2 loss and sustained in some mice with myelodysplasia. Although less frequent, Ezh2 loss also induced T-cell acute lymphoblastic leukemia and the clonal expansion of B-1a B cells. Gene expression profiling showed that PRC2 target genes were derepressed upon the deletion of Ezh2 in hematopoietic stem and progenitor cells, but were largely repressed during the development of MDS and MDS/MPN. Chromatin immunoprecipitation–sequence analysis of trimethylation of histone H3 at lysine 27 (H3K27me3) revealed a compensatory function of Ezh1, another enzymatic component of PRC2, in this process. The deletion of Ezh1 alone did not cause dysplasia or any hematologic malignancies in mice, but abolished the repopulating capacity of hematopoietic stem cells when combined with Ezh2 loss. These results clearly demonstrated an essential role of Ezh1 in the pathogenesis of hematopoietic malignancies induced by Ezh2 insufficiency, and highlighted the differential functions of Ezh1 and Ezh2 in hematopoiesis.
Introduction
Polycomb-group (PcG) proteins function in the maintenance of gene silencing through histone modifications. They form several distinct polycomb-repressive complexes (PRCs), including canonical and noncanonical PRC1 and PRC2.1,2 PRC2 consists of Eed, Suz12, and Ezh2 or its closely related homolog Ezh1. Ezh1 and Ezh2 are the catalytic components of PRC2 and catalyze the mono-, di-, and trimethylation of histone H3 at lysine 27 (H3K27me1/me2/me3). PcG proteins, including PRC2 components, have been characterized as general regulators of stem cells as well as tumorigenesis.1,3,4 Eed and Ezh1, the core component of PRC2, were previously found to be essential for the maintenance of hematopoietic stem cells (HSCs).5,6 Although Ezh2 is dispensable for the self-renewal capacity of adult bone marrow (BM) HSCs,7 it is required to promote MLL-AF9–driven acute myeloid leukemia (AML) in mice,8-10 and the forced expression of Ezh2 in HSCs was shown to cause myeloproliferative neoplasms (MPNs) in mice.11
Comprehensive genome sequence studies initially identified activating mutations in EZH2 in follicular and diffuse large B-cell lymphomas,12 thereby supporting the oncogenic function of PRC2 in cancer. However, inactivating mutations in EZH2 have also been identified in patients with myelodysplastic syndrome (MDS) (3%∼7%), MPNs (3%∼13%), and MDS/MPN overlap disorders (8%∼13%), which are all clonal myeloid disorders originating from HSCs.13-20 Given that EZH2, which is located at chromosome 7q36.1, is frequently involved in chromosomal abnormalities such as −7 and 7q−, in hematologic malignancies,21 these findings suggest a tumor suppressor role for EZH2 in these HSC disorders. Other components of PRC2, EED and SUZ12, are also targeted in somatic inactivating mutations, although the frequencies of their mutations were found to be markedly lower than those of EZH2 mutations.20 ASXL1, which plays an important role in the recruitment and/or stability of PRC2,22 has also been shown to carry inactivating mutations in patients with MDS, MPN, and MDS/MPN.20,23 Thus, the role of PRC2 in hematologic malignancies is complex.19
These findings prompted us to investigate the impact of inactivating EZH2 mutations on hematopoiesis and its interplay with coinciding mutations such as TET2 and RUNX1 in mice. In our previous studies, the concurrent depletion of Ezh2 and Tet2 in mice markedly accelerated the development of MDS and MDS/MPN and Ezh2 loss significantly promoted RUNX1S291fs-induced MDS.24,25 These findings support the tumor suppressor function of EZH2 in the context of myelodysplastic disorders. However, Ezh2 loss attenuated the predisposition of RUNX1S291fs-induced MDS cells to leukemic transformation,25 which is consistent with the finding that inactivating mutations of EZH2 are rare in de novo and secondary AML patients.12,20,26,27 In our previous studies, we also noted that the deletion of Ezh2 alone induced myelodysplastic disorders after a long latency; however, detailed analysis was not performed.24,25
In contrast to EZH2, no somatic mutations have been identified in EZH1 in hematologic malignancies.20 As described above, conditional ablation of Ezh1 in adult BM HSCs induces significant loss of adult HSCs, with concomitant impairment of their self-renewal capacity mainly due to derepression of Cdkn2a.5 It is known that Ezh1 complements Ezh2 in maintaining pluripotency of Ezh2−/− embryonic stem cells (ESCs), and depletion of Ezh1 abolishes residual methylation at H3K27 and derepresses H3K27me3 target genes, resulting in failure of the maintenance of ESC identity.28 Given that Ezh2-deficient HSCs maintain self-renewal capacity,7 Ezh1 may function as a critical backup enzyme also in the BM in the setting of Ezh2 loss.
In order to understand the pathological role of inactivating EZH2 mutations in myeloid malignancies, we herein analyzed mice with the hematopoietic cell-specific deletion of Ezh2 and determined how Ezh2 loss in HSCs predisposed mice to develop heterogeneous hematologic malignancies. We also examined the role of Ezh1 in the maintenance of hematopoiesis and in disease progression in the setting of Ezh2 loss.
Materials and methods
Mice
Ezh2fl/fl mice29 were crossed with Rosa26::Cre-ERT mice (TaconicArtemis GmbH) to achieve the conditional deletion of Ezh2. These mice were injected with 100 μL of tamoxifen dissolved in corn oil at a concentration of 10 mg/mL intraperitoneally for 5 consecutive days to induce Cre-ERT activity. Ezh1 constitutive knockout mice were obtained from Thomas Jenuwein’s laboratory (Max Planck Institute of Immunobiology and Epigenetics, Freiburg, Germany) and will be reported elsewhere. Part of its phenotypes has been reported previously.30,31 C57BL/6 mice congenic for the Ly5 locus (CD45.1) were purchased from Sankyo-Laboratory Service (Tsukuba, Japan). All animal experiments were performed in accordance with our institutional guidelines for the use of laboratory animals and approved by the Review Board for Animal Experiments of Chiba University (approval ID: 25-104).
Purification of hematopoietic cells
Mononuclear cells from BM were isolated on Ficoll-Paque PLUS (GE Healthcare) and incubated with a mixture of biotin-conjugated monoclonal antibodies (mAbs) against lineage markers including Gr-1, Mac-1, Ter-119, B220, interleukin-7Rα (IL-7Rα), CD4, and CD8α. The cells were further stained with allophycocyanin (APC)-Cy7–conjugated streptavidin and a combination of mAbs including phycoerythrin (PE)–Sca-1 and APC–c-Kit. The Pacific Blue–CD45.2 mAb was used as an additional marker for donor-derived cells.
BM transplantation
BM cells from CD45.2 mutant mice (test cells) were transplanted IV into 8- to 12-week-old CD45.1 recipients irradiated at a dose of 9.0 to 9.5 Gy with or without BM cells from 8- to 12-week-old CD45.1 congenic mice (competitor cells). The chimerism of donor-derived hematopoietic cells was monitored by flow cytometry. The proportion of donor cells was evaluated by dividing the number of CD45.2-single-positive cells by the total number of CD45-positive cells (CD45.1 + CD45.2).
Gene set enrichment analysis
We performed gene set enrichment analysis (GSEA) as described.32 We excluded genes not detected in microarray analyses and those with no reads in both samples to be compared in RNA-sequence analyses from GSEA.
Statistical analysis
The statistical significance of differences was measured by the unpaired 2-tailed Student t test. When the variance was judged as significantly different by f test and the number of samples was enough (n > 11), we used the Mann-Whitney nonparametric test. These tests were carried out using Graph Pad Prism, version 6.
Results
Deletion of Ezh2 caused myelodysplasia
To decipher the pathological role of inactivating EZH2 mutations in hematologic malignancies, we transplanted BM cells from Cre-ERT control (wild-type [WT]) and Cre-ERT;Ezh2fl/fl CD45.2 mice (8-12 weeks old) without competitor cells into lethally irradiated CD45.1 recipient mice and deleted Ezh2 by intraperitoneal injection of tamoxifen at 6 to 8 weeks posttransplantation (supplemental Figure 1A, available on the Blood Web site). We confirmed the efficient deletion of Ezh2 in hematopoietic cells from Ezh2Δ/Δ mice by genomic polymerase chain reaction (PCR) (supplemental Figure 2). We hereafter referred to the recipient mice reconstituted with WT and Ezh2Δ/Δ cells as WT and Ezh2Δ/Δ mice, respectively.
As we reported previously,7 chimerism of the lymphoid lineage, particularly of the B lymphoid lineage, was significantly reduced in the peripheral blood (PB) whereas that of the myeloid lineage was increased over time after the deletion of Ezh2 (supplemental Figure 1C). Correspondingly, the white blood cell count in the PB was significantly decreased in Ezh2Δ/Δ mice, whereas the platelet count significantly increased (supplemental Figure 1B). Anemia was also more severe in Ezh2Δ/Δ mice than in control mice (supplemental Figure 1B). However, BM analysis 3 months after the deletion of Ezh2 did not detect any significant changes in the BM cell counts and the numbers of CD34−LSK HSCs and LSK hematopoietic stem and progenitor cells (HSPCs) (supplemental Figure 1D). The proportion of Annexin V+ cells in CD71+Ter119+ erythroblasts was mildly increased in Ezh2Δ/Δ mice compared with WT mice, albeit not statistically significant, implicating enhanced inefficient hematopoiesis in the BM, a feature compatible with myelodysplastic disorders (supplemental Figure 1E). Morphologic dysplasia, which is characteristic of myelodysplastic disorders, became gradually evident in the PB of Ezh2Δ/Δ mice over time, including pseudo-Pelger-Huët cells, hypersegmented neutrophils, Howell-Jolly bodies, and giant platelets (supplemental Figure 1F). These findings indicate that deletion of Ezh2 caused myelodysplasia in mice.
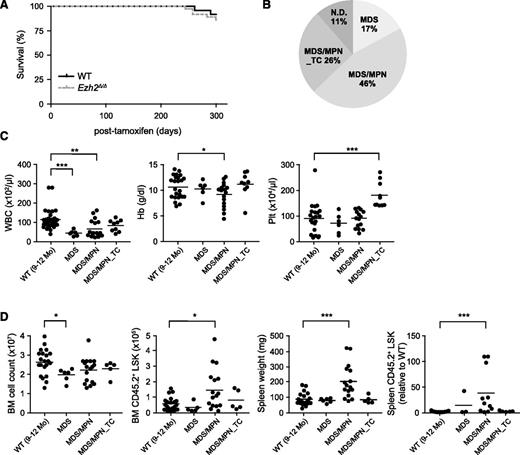
Ezh2Δ/Δ mice developed myelodysplastic disorders
Although no significant difference was observed in the survival of Ezh2Δ/Δ mice by 10 months after the deletion of Ezh2 (Figure 1A), they developed myelodysplastic disorders during a long observation period (Figure 1B; supplemental Table 1). We previously reported the development of MDS/MPN, but not MDS in Ezh2Δ/Δ mice.24 However, Ezh2Δ/Δ mice in the present study developed both MDS/MPN and MDS (Figure 1B). All Ezh2Δ/Δ mice showed morphologic dysplasia of hematopoietic cells as presented above (supplemental Figure 1F; supplemental Table 1). MDS/MPN mice exhibited myeloproliferative features, including splenomegaly with active extramedullary hematopoiesis, an increase in the number of LSK cells in the BM and/or the spleen, and chronic myelomonocytic leukemia (CMML)-like monocytosis in the PB. In contrast, MDS mice did not display any obvious myeloproliferative features, but were more likely to develop cytopenia (Figure 1C-D; supplemental Table 1). Some Ezh2Δ/Δ mice developed MDS/MPN that was characterized by a significant increase in platelet numbers and mild myeloproliferative features in the BM and spleen (Figure 1C-D; supplemental Table 1). We categorized these mice as MDS/MPN with thrombocytosis (MDS/MPN_TC). Thus, Ezh2Δ/Δ mice developed myelodysplastic disorders including MDS, MDS/MPN, and MDS/MPN_TC.
Ezh2Δ/Δ mice developed myelodysplastic disorders. (A) Kaplan-Meier survival curves of WT (n = 24) and Ezh2Δ/Δ (n = 32) mice. These survival curves were generated from 3 independent experiments. (B) A pie graph showing the types of hematologic malignancies observed in Ezh2Δ/Δ mice (n = 32) during a long observation period. (C) PB cell counts in Ezh2Δ/Δ mice 9 to 12 months after the deletion of Ezh2. WBC, Hb, and Plt counts in the PB from WT (n = 24) and Ezh2Δ/Δ MDS (n = 6), MDS/MPN (n = 17), and MDS/MPN_TC (n = 9) mice are plotted as dots and the mean values are indicated as bars. (D) Absolute numbers of total BM cells and LSK cells in a unilateral femur, and spleen weights and numbers of LSK cells in the spleen relative to WT are plotted as dots and mean values are indicated as bars (WT, n = 21; MDS, n = 6; MDS/MPN, n = 16; MDS/MPN_TC, n = 5). Statistical significance of difference was measured by unpaired 2-tailed Student t test or Mann-Whitney test when the variance was judged as significantly different by f test. *P < .05, **P < .01, ***P < .001. Hb, hemoglobin; ND, not determined; Plt, platelet; WBC, white blood cell.
Ezh2Δ/Δ mice developed myelodysplastic disorders. (A) Kaplan-Meier survival curves of WT (n = 24) and Ezh2Δ/Δ (n = 32) mice. These survival curves were generated from 3 independent experiments. (B) A pie graph showing the types of hematologic malignancies observed in Ezh2Δ/Δ mice (n = 32) during a long observation period. (C) PB cell counts in Ezh2Δ/Δ mice 9 to 12 months after the deletion of Ezh2. WBC, Hb, and Plt counts in the PB from WT (n = 24) and Ezh2Δ/Δ MDS (n = 6), MDS/MPN (n = 17), and MDS/MPN_TC (n = 9) mice are plotted as dots and the mean values are indicated as bars. (D) Absolute numbers of total BM cells and LSK cells in a unilateral femur, and spleen weights and numbers of LSK cells in the spleen relative to WT are plotted as dots and mean values are indicated as bars (WT, n = 21; MDS, n = 6; MDS/MPN, n = 16; MDS/MPN_TC, n = 5). Statistical significance of difference was measured by unpaired 2-tailed Student t test or Mann-Whitney test when the variance was judged as significantly different by f test. *P < .05, **P < .01, ***P < .001. Hb, hemoglobin; ND, not determined; Plt, platelet; WBC, white blood cell.
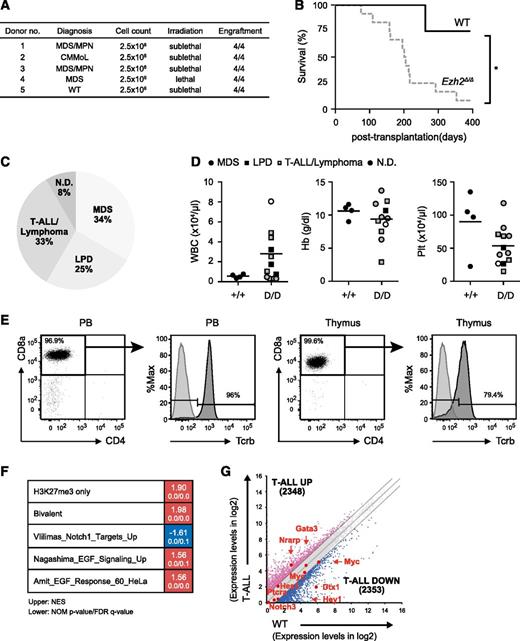
Serial transplantation of Ezh2Δ/Δ hematopoietic cells caused heterogeneous hematologic malignancies
To clarify whether the myelodysplastic disorders induced by Ezh2 loss were transplantable, we transplanted BM cells from MDS and MDS/MPN mice ∼10 months after the deletion of Ezh2 into secondary recipients (Figure 2A). In contrast to the primary transplantation, Ezh2Δ/Δ mice developed hematologic malignancies markedly earlier after secondary transplantation (Figure 2B; supplemental Table 2). Four recipient mice, which developed T-cell acute lymphoblastic leukemia (ALL) of a CD45.1+ host cell origin, were excluded from the present study (supplemental Table 2). Ezh2Δ/Δ mice developed more diverse diseases, including MDS, T-ALL/lymphoma, and lymphoproliferative disease (LPD) of B-1a B cells (Figure 2C-D; supplemental Table 2).
Serial transplantation of Ezh2Δ/Δ hematopoietic cells caused heterogeneous hematologic malignancies. (A) Summary of secondary transplantation. A total of 2.5 × 106 BM cells from MDS and MDS/MPN mice ∼10 months after the deletion of Ezh2 were transplanted into secondary recipients irradiated at a lethal (8.5 Gy) or sublethal (6.5 Gy) dose. The engraftment status is indicated. (B) A Kaplan-Meier survival curve of WT (n = 4) and Ezh2Δ/Δ (n = 12) mice in secondary transplantation. Four recipient mice, which developed CD45.1+ T-ALL of a host cell origin, were excluded. *P < .05. Statistical significance was measured by log-rank test. (C) A pie graph showing the types of hematologic malignancies observed in Ezh2Δ/Δ mice (n = 12) in the secondary transplantation. Four recipient mice, which developed CD45.1+ T-ALL of a host cell origin, were excluded. (D) PB cell counts of moribund mice and those surviving at 370 days after secondary transplantation. WBC, Hb, and Plt counts in the PB of Ezh2Δ/Δ MDS, LPD, and T-ALL/lymphoma mice are plotted as dots and the mean values are indicated as bars. (E) Flow cytometric profiles of donor-derived CD45.2+ T-ALL/lymphoma cells in the PB and thymus obtained from representative mice with T-ALL/lymphoma (11F8_004, supplemental Table 2) at 52 weeks posttransplantation. Unstained cells served as a control for Tcrβ staining. (F) Gene expression profiling of Ezh2Δ/Δ T-ALL cells. RNA sequence was performed using Ezh2Δ/Δ BM CD8+ T-ALL cells obtained from a Ezh2Δ/Δ mouse with T-ALL and WT BM CD8+ T cells. GSEA was performed using the RNA-sequence data and GSEA plots are shown. The NES, NOM P value, and FDR q value are indicated. (G) Scatter diagrams showing RNA-sequence data. The signal levels of RefSeq genes (RPKM + 1 in log2) in Ezh2Δ/Δ BM CD8+ T-ALL cells and WT BM CD8+ T cells are plotted. The light gray lines represent the boundaries for twofold increase and twofold decrease, respectively. The number of genes upregulated and downregulated more than twofold in Ezh2Δ/Δ T-ALL cells compared with WT cells are indicated. The representative direct target genes of Notch1 are shown as red dots. FDR, false discovery rate; NES, normalized enrichment score; NOM, nominal.
Serial transplantation of Ezh2Δ/Δ hematopoietic cells caused heterogeneous hematologic malignancies. (A) Summary of secondary transplantation. A total of 2.5 × 106 BM cells from MDS and MDS/MPN mice ∼10 months after the deletion of Ezh2 were transplanted into secondary recipients irradiated at a lethal (8.5 Gy) or sublethal (6.5 Gy) dose. The engraftment status is indicated. (B) A Kaplan-Meier survival curve of WT (n = 4) and Ezh2Δ/Δ (n = 12) mice in secondary transplantation. Four recipient mice, which developed CD45.1+ T-ALL of a host cell origin, were excluded. *P < .05. Statistical significance was measured by log-rank test. (C) A pie graph showing the types of hematologic malignancies observed in Ezh2Δ/Δ mice (n = 12) in the secondary transplantation. Four recipient mice, which developed CD45.1+ T-ALL of a host cell origin, were excluded. (D) PB cell counts of moribund mice and those surviving at 370 days after secondary transplantation. WBC, Hb, and Plt counts in the PB of Ezh2Δ/Δ MDS, LPD, and T-ALL/lymphoma mice are plotted as dots and the mean values are indicated as bars. (E) Flow cytometric profiles of donor-derived CD45.2+ T-ALL/lymphoma cells in the PB and thymus obtained from representative mice with T-ALL/lymphoma (11F8_004, supplemental Table 2) at 52 weeks posttransplantation. Unstained cells served as a control for Tcrβ staining. (F) Gene expression profiling of Ezh2Δ/Δ T-ALL cells. RNA sequence was performed using Ezh2Δ/Δ BM CD8+ T-ALL cells obtained from a Ezh2Δ/Δ mouse with T-ALL and WT BM CD8+ T cells. GSEA was performed using the RNA-sequence data and GSEA plots are shown. The NES, NOM P value, and FDR q value are indicated. (G) Scatter diagrams showing RNA-sequence data. The signal levels of RefSeq genes (RPKM + 1 in log2) in Ezh2Δ/Δ BM CD8+ T-ALL cells and WT BM CD8+ T cells are plotted. The light gray lines represent the boundaries for twofold increase and twofold decrease, respectively. The number of genes upregulated and downregulated more than twofold in Ezh2Δ/Δ T-ALL cells compared with WT cells are indicated. The representative direct target genes of Notch1 are shown as red dots. FDR, false discovery rate; NES, normalized enrichment score; NOM, nominal.
Four mice developed T-ALL/lymphoma with the prominent expansion of CD8α+Tcrαβ+ T cells in the thymus (Figure 2E). They also showed variable levels of infiltration of tumor cells in the PB (Figure 2D-E) and other organs (data not shown). To understand the molecular basis of Ezh2Δ/Δ T-ALL, we performed RNA-sequence analysis of BM CD8+ T-ALL cells and WT BM CD8+ T cells. GSEA revealed positive enrichment of PRC2 targets defined in HSCs, including those marked with H3K27me3 alone (H3K27me3 only) as well as those with H3K27me3 and H3K4me3 (bivalent genes) at their promoters (Figure 2F; supplemental Table 3).33 It has been reported that 25% of T-ALL patients have loss-of-function mutations in the PRC2 gene, mostly in EZH2, and 65% of the patients with PRC2 gene mutations concurrently have activating NOTCH1 mutations.34 These findings suggest that EZH2 and NOTCH1 mutations directly or indirectly cooperate in T-ALL. In Ezh2Δ/Δ T-ALL, however, direct targets of Notch1 were not significantly activated (Figure 2G) and the Notch target gene set was negatively enriched (Figure 2F), suggesting that T-ALL that developed in the absence of Ezh2 in mice required activation of additional pathways other than Notch. In this regard, positive enrichment of the epidermal growth factor signaling gene sets is intriguing (Figure 2F), but requires extensive validation.
Most Ezh2Δ/Δ mice had CD19+B220midCD5+IgM+ B-1a B cells in the PB at variable levels (supplemental Figure 3A-B). Among these, some mice developed lymphoproliferative neoplasm of B-1a B cells. Although we did not precisely examine B-1a B cell expansion in primary recipients, we observed the expansion of B220low non-T (CD4−CD8−), nonmyeloid cells in the PB of 16 of 36 mice, which were thought to be B-1a B cells (supplemental Table 1). Most of the B-1a B cells were polyclonal by genomic PCR of the D-J rearrangement of the Igh gene. However, some mice showed clonal expansion, and were, thus, assumed to have developed into chronic lymphocytic leukemia (CLL) (supplemental Figure 3C; supplemental Tables 1-2). These results indicated that Ezh2 loss predisposed mice to heterogeneous hematologic malignancies.
Loss of Ezh2 reprogrammed the transcriptional landscape of HSPCs
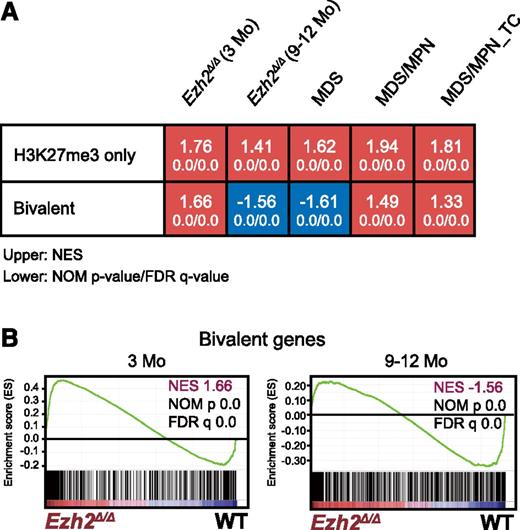
In order to understand the molecular mechanism underlying myelodysplastic disorders induced by Ezh2 insufficiency in more detail, we performed gene expression profiling by a microarray analysis using LSK cells collected from 3 to 5 mice randomly selected at early and late time points after the deletion of Ezh2 (3 and 9-12 months, respectively). LSK cells were also collected individually from MDS (n = 2), MDS/MPN (n = 1), and MDS/MPN_TC (n = 1) mice ∼9 to 12 months after the deletion of Ezh2.
We performed a GSEA. As expected, PRC2 targets defined in HSCs, including H3K27me3-only genes (marked only with H3K27me3) and bivalent genes (marked with H3K27me3 and H3K4me3) (supplemental Table 3),33 were again more likely to be derepressed and were more positively enriched in LSK cells 3 months after the deletion of Ezh2 than in WT LSK cells (Figure 3). Of note, only bivalent genes became negatively enriched with significance in LSK cells at later time points (9-12 months after the deletion of Ezh2) and in MDS LSK cells, whereas H3K27me3-only genes remained positively enriched. These results suggest that Ezh1, the only H3K27 methyltransferase in the absence of Ezh2, becomes functionally augmented in myelodysplastic disorders and largely complements Ezh2 loss, particularly at the promoters of bivalent genes.
Loss of Ezh2 reprogrammed the transcriptional landscape of HSPCs. (A) Gene expression alterations associated with disease progression. GSEA was performed using the microarray analysis data of WT and Ezh2Δ/Δ LSK cells from early and late time points after the deletion of Ezh2 (3 and 9-12 months, respectively). LSK cells were also collected individually from MDS (10F14 and 11F2), MDS/MPN (11F5), and MDS/MPN_TC mice (10F7) ∼10 months after the deletion of Ezh2. The NES from the gene expression profiles of LSK cells derived from GSEA is shown as a number in each cell. The NOM P value and FDR are shown below NES. Red and blue colors represent positive (upregulated in the given genotype relative to WT) and negative (upregulated in WT relative to the given genotype) enrichment, respectively. (B) Gene set enrichment plots for the bivalent gene set in HSCs in panel A comparing Ezh2Δ/Δ LSK cells to WT LSK cells 3 and 9 to 12 months after the deletion of Ezh2.
Loss of Ezh2 reprogrammed the transcriptional landscape of HSPCs. (A) Gene expression alterations associated with disease progression. GSEA was performed using the microarray analysis data of WT and Ezh2Δ/Δ LSK cells from early and late time points after the deletion of Ezh2 (3 and 9-12 months, respectively). LSK cells were also collected individually from MDS (10F14 and 11F2), MDS/MPN (11F5), and MDS/MPN_TC mice (10F7) ∼10 months after the deletion of Ezh2. The NES from the gene expression profiles of LSK cells derived from GSEA is shown as a number in each cell. The NOM P value and FDR are shown below NES. Red and blue colors represent positive (upregulated in the given genotype relative to WT) and negative (upregulated in WT relative to the given genotype) enrichment, respectively. (B) Gene set enrichment plots for the bivalent gene set in HSCs in panel A comparing Ezh2Δ/Δ LSK cells to WT LSK cells 3 and 9 to 12 months after the deletion of Ezh2.
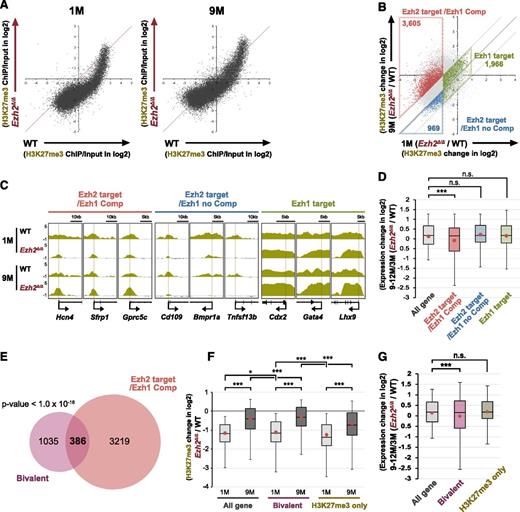
Western blot analysis, however, did not show any significant changes in global H3K27me3 levels between the early (1 month) and late (9 months) time points postdeletion of Ezh2 (supplemental Figure 4). Thus, we next performed chromatin immunoprecipitation (ChIP)-sequence analysis of H3K27me3. As evident in scatter plots of H3K27me3 signals, H3K27me3 levels at promoters significantly increased in LSK cells at 9 months compared with 1 month postdeletion of Ezh2 (Figure 4A). We then categorized genes with ChIP signals >1 against the input signals depending on the changes in H3K27me3 levels at 1 and 9 months postdeletion of Ezh2. We defined genes that lost H3K27me3 levels at least twofold at 1-month postdeletion of Ezh2 as “Ezh2 target genes.” Among these, 3605 genes that gained H3K27me3 levels at least twofold at 9 months compared with 1 month postdeletion of Ezh2 were defined as “Ezh2 targets with Ezh1 compensation (Ezh2 target/Ezh1 Comp).” In contrast, 969 genes that did not gain H3K27me3 levels were defined as “Ezh2 targets with no Ezh1 compensation (Ezh2 target/Ezh1 no Comp).” We also defined 1966 genes that did not lose H3K27me3 levels at least twofold at 1-month postdeletion of Ezh2 and did not show any significant changes thereafter as “Ezh1 target genes” (Figure 4B; supplemental Table 3). ChIP-sequence data of the representative gene loci of each category are visualized in Figure 4C. Of note, only Ezh2 target/Ezh1 Comp genes showed moderate but significant trend toward gene repression at 9 to 12 months compared with 3 months postdeletion of Ezh2 (Figure 4D).
Restoration of H3K27me3 levels at part of Ezh2 target gene promoters in Ezh2Δ/Δ HSPCs. (A) Scatter plots showing the correlation of the fold enrichment values (ChIP/input) (transcription start site ± 2.0 kb) of H3K27me3 against the input signals of RefSeq genes between WT and Ezh2Δ/Δ LSK cells from WT and Ezh2Δ/Δ mice at 1 and 9 months postdeletion of Ezh2. (B) A scatter plot showing the correlation of H3K27me3 levels in Ezh2Δ/Δ LSK cells against WT LSK cells (Ezh2Δ/Δ/WT) between 1 and 9 months postdeletion of Ezh2. The diagonal light gray lines represent the boundaries for twofold increase and twofold decrease, respectively. The vertical line represents the boundary for twofold reduction in fold enrichment of H3K27me3 in Ezh2Δ/Δ LSK cells at 1 month postdeletion of Ezh2. The genes with ChIP signals >1 over the input signals were categorized depending on the changes in H3K27me3 levels at 1 and 9 months postdeletion of Ezh2. The genes that lost H3K27me3 levels at least twofold at 1 month postdeletion of Ezh2 were defined as “Ezh2 targets.” Among these, the genes that gained H3K27me3 levels at least twofold at 9 months compared with 1 month postdeletion of Ezh2 were defined as “Ezh2 target/Ezh1 Comp.” In contrast, the genes that did not gain H3K27me3 levels were defined as “Ezh2 target/Ezh1 no Comp.” The genes that did not lose H3K27me3 levels twofold at 1 month postdeletion of Ezh2 and did not show any significant changes thereafter were defined as “Ezh1 target genes.” Each category was boxed. (C) Visualization of ChIP-sequence data of H3K27me3 levels of representative genes that belong to the indicated categories defined in panel A in WT and Ezh2Δ/Δ LSK cells at 1 and 9 months postdeletion of Ezh2 using the Integrative Genomics Viewer (IGV). Schematic diagrams of these gene loci indicate their genomic structures. Exons and untranslated regions are demarcated by large and small black boxes, respectively. (D) Box-and-whisker plots showing the expression changes of indicated genes in Ezh2Δ/Δ LSK cells relative to WT LSK cells (Ezh2Δ/Δ/WT) at 9 to 12 months compared with 3 months postdeletion of Ezh2. Boxes represent 25 to 75 percentile ranges. Vertical lines represent 10 to 90 percentile ranges. Horizontal bars represent median. Mean values are indicated by red dots. Microarray data in Figure 3 were applied to this analysis. ***P < .001 (Student t test). (E) Venn diagram showing the overlap between Ezh2 target/Ezh1 Comp genes in panel B and bivalent genes. P < 1.0 × 10−16. (F) Box-and-whisker plots showing the changes in H3K27me3 levels of indicated genes in Ezh2Δ/Δ LSK cells relative to WT LSK cells (Ezh2Δ/Δ/WT) at 1 and 9 months postdeletion of Ezh2. Mean values are indicated by red dots. *P < .05, ***P < .001 (Student t test). (G) Box-and-whisker plots showing the expression changes of indicated genes in Ezh2Δ/Δ LSK cells relative to WT LSK cells (Ezh2Δ/Δ/WT) at 9 to 12 months compared with 3 months postdeletion of Ezh2. Mean values are indicated by red dots. Microarray data in Figure 3 was applied to this analysis. ***P < .001 (Student t test). ns, not significant.
Restoration of H3K27me3 levels at part of Ezh2 target gene promoters in Ezh2Δ/Δ HSPCs. (A) Scatter plots showing the correlation of the fold enrichment values (ChIP/input) (transcription start site ± 2.0 kb) of H3K27me3 against the input signals of RefSeq genes between WT and Ezh2Δ/Δ LSK cells from WT and Ezh2Δ/Δ mice at 1 and 9 months postdeletion of Ezh2. (B) A scatter plot showing the correlation of H3K27me3 levels in Ezh2Δ/Δ LSK cells against WT LSK cells (Ezh2Δ/Δ/WT) between 1 and 9 months postdeletion of Ezh2. The diagonal light gray lines represent the boundaries for twofold increase and twofold decrease, respectively. The vertical line represents the boundary for twofold reduction in fold enrichment of H3K27me3 in Ezh2Δ/Δ LSK cells at 1 month postdeletion of Ezh2. The genes with ChIP signals >1 over the input signals were categorized depending on the changes in H3K27me3 levels at 1 and 9 months postdeletion of Ezh2. The genes that lost H3K27me3 levels at least twofold at 1 month postdeletion of Ezh2 were defined as “Ezh2 targets.” Among these, the genes that gained H3K27me3 levels at least twofold at 9 months compared with 1 month postdeletion of Ezh2 were defined as “Ezh2 target/Ezh1 Comp.” In contrast, the genes that did not gain H3K27me3 levels were defined as “Ezh2 target/Ezh1 no Comp.” The genes that did not lose H3K27me3 levels twofold at 1 month postdeletion of Ezh2 and did not show any significant changes thereafter were defined as “Ezh1 target genes.” Each category was boxed. (C) Visualization of ChIP-sequence data of H3K27me3 levels of representative genes that belong to the indicated categories defined in panel A in WT and Ezh2Δ/Δ LSK cells at 1 and 9 months postdeletion of Ezh2 using the Integrative Genomics Viewer (IGV). Schematic diagrams of these gene loci indicate their genomic structures. Exons and untranslated regions are demarcated by large and small black boxes, respectively. (D) Box-and-whisker plots showing the expression changes of indicated genes in Ezh2Δ/Δ LSK cells relative to WT LSK cells (Ezh2Δ/Δ/WT) at 9 to 12 months compared with 3 months postdeletion of Ezh2. Boxes represent 25 to 75 percentile ranges. Vertical lines represent 10 to 90 percentile ranges. Horizontal bars represent median. Mean values are indicated by red dots. Microarray data in Figure 3 were applied to this analysis. ***P < .001 (Student t test). (E) Venn diagram showing the overlap between Ezh2 target/Ezh1 Comp genes in panel B and bivalent genes. P < 1.0 × 10−16. (F) Box-and-whisker plots showing the changes in H3K27me3 levels of indicated genes in Ezh2Δ/Δ LSK cells relative to WT LSK cells (Ezh2Δ/Δ/WT) at 1 and 9 months postdeletion of Ezh2. Mean values are indicated by red dots. *P < .05, ***P < .001 (Student t test). (G) Box-and-whisker plots showing the expression changes of indicated genes in Ezh2Δ/Δ LSK cells relative to WT LSK cells (Ezh2Δ/Δ/WT) at 9 to 12 months compared with 3 months postdeletion of Ezh2. Mean values are indicated by red dots. Microarray data in Figure 3 was applied to this analysis. ***P < .001 (Student t test). ns, not significant.
Bivalent genes appeared to largely overlap with Ezh2 target/Ezh1 Comp genes (Figure 4E), thus showing an increase in the H3K27me3 levels at 9 months compared with 1 month postdeletion of Ezh2 (Figure 4F) and a moderate but significant trend toward gene repression over time (Figure 4G). In contrast, the expression profile of H3K27-only genes did not change over time (Figure 4G) as demonstrated in GSEA (Figure 3A). These findings indicate that the recovery of H3K27me3 levels takes place not globally but in a locus-specific manner over time and suggest repositioning of Ezh1 to Ezh2 targets to complement Ezh2 loss.
Ezh1 was essential for the maintenance of Ezh2 insufficient myelodysplastic disorders
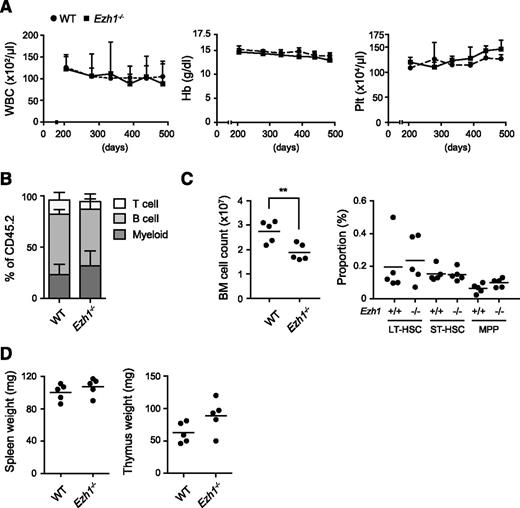
As we have previously reported7,24 and also supported in this study, Ezh1 may play a critical role in the maintenance of hematopoiesis and also in disease progression in the absence of Ezh2. To address this issue, we analyzed Ezh1 constitutive knockout mice.30,31 Mice lacking Ezh1 are viable, fertile, and healthy, and lack Ezh1 mRNA.31 We observed Ezh1−/− mice up to 16.5 months. They showed no hematologic abnormalities in the PB including cell blood counts (Figure 5A), lineage composition (Figure 5B), and cell morphology (data not shown). Although total BM cell numbers were significantly reduced, the absolute number of HSCs and progenitors in the BM was similar to that in the WT control (Figure 5C). The sizes of the thymus and spleen were also similar to those of the WT control (Figure 5D). These results clearly indicated that Ezh1 insufficiency alone did not cause hematologic malignancies. This finding corresponds well to the finding that no inactivating mutations in EZH1 have been identified in hematologic malignancies in contrast to EZH2.
Ezh1 loss did not cause any hematologic malignancies. (A) PB cell counts in Ezh1−/− mice up to 16.5 months after birth. WBC, Hb, and Plt counts in the PB from WT (n = 5) and Ezh1−/− (n = 5) mice are shown as the mean ± standard deviation (SD). (B) The proportions of myeloid (Mac-1+), B220+ B, and CD4+ or CD8+ T cells among CD45+ donor-derived hematopoietic cells in the PB at the time points indicated. Data are shown as the mean ± SD (n = 5). (C) Absolute numbers of total BM cells (left panel) and long-term (LT), short-term (ST), and multipotent progenitors (MPPs) in a unilateral pair of the femur and tibia from Ezh1−/− mice 16.5 months after birth. Data are plotted as dots and the mean values are indicated as bars (WT, n = 5; Ezh1−/−, n = 5). (D) Weights of the spleen and thymus from Ezh1−/− mice 16.5 months after birth. Data are plotted as dots and the mean values are indicated as bars (WT, n = 5; Ezh1−/−, n = 5). Statistical significance of difference was measured by unpaired 2-tailed Student t test. **P < .005.
Ezh1 loss did not cause any hematologic malignancies. (A) PB cell counts in Ezh1−/− mice up to 16.5 months after birth. WBC, Hb, and Plt counts in the PB from WT (n = 5) and Ezh1−/− (n = 5) mice are shown as the mean ± standard deviation (SD). (B) The proportions of myeloid (Mac-1+), B220+ B, and CD4+ or CD8+ T cells among CD45+ donor-derived hematopoietic cells in the PB at the time points indicated. Data are shown as the mean ± SD (n = 5). (C) Absolute numbers of total BM cells (left panel) and long-term (LT), short-term (ST), and multipotent progenitors (MPPs) in a unilateral pair of the femur and tibia from Ezh1−/− mice 16.5 months after birth. Data are plotted as dots and the mean values are indicated as bars (WT, n = 5; Ezh1−/−, n = 5). (D) Weights of the spleen and thymus from Ezh1−/− mice 16.5 months after birth. Data are plotted as dots and the mean values are indicated as bars (WT, n = 5; Ezh1−/−, n = 5). Statistical significance of difference was measured by unpaired 2-tailed Student t test. **P < .005.
To directly validate the role of Ezh1 in hematopoiesis in the absence of Ezh2, we then generated Cre-ERT;Ezh1−/−Ezh2fl/fl mice (CD45.2), transplanted BM cells into lethally irradiated recipient mice, and then deleted Ezh2 by injecting tamoxifen at 6 weeks posttransplantation. The deletion of Ezh2 induced cytopenia in the PB of Ezh1−/−Ezh2Δ/Δ mice, and Ezh1−/−Ezh2Δ/Δ cells, particularly myeloid cells, were progressively depleted. Instead, residual CD45.1+ host cells gradually overtook Ezh1−/−Ezh2Δ/Δ cells and largely restored the blood cell counts in the PB. (Figure 6A). We subsequently evaluated the capacity of Ezh1−/−Ezh2Δ/Δ HSCs to repopulate hematopoiesis by competitive repopulating assays. We transplanted Cre-ERT;Ezh1−/−Ezh2fl/fl BM cells along with twice more BM competitor cells into lethally irradiated recipients. At 6 weeks posttransplantation, Ezh2 was deleted by injecting tamoxifen. Ezh1−/−Ezh2Δ/Δ donor cells were rapidly outcompeted by the competitor cells and the donor cells were depleted from both the PB and BM (Figure 6B-C).
Ezh1 was essential for the maintenance of Ezh2 insufficient hematopoiesis. (A) The failure of Ezh1−/−Ezh2Δ/Δ hematopoietic cells to maintain hematopoiesis. BM cells from Cre-ERT and Cre-ERT;Ezh1−/−Ezh2fl/fl mice were transplanted into lethally irradiated recipient mice without rescue BM cells, and Ezh2 was then deleted by injecting tamoxifen at 6 weeks posttransplantation. Following tamoxifen injection, the recipient mice were fed with a tamoxifen diet (400 mg of tamoxifen citrate per kg diet) for 10 days. WBC counts, the chimerism of CD45.2+ donor-derived cells, including Mac-1+ myeloid cells, B220+ B cells, and CD4+ or CD8+ T cells, in the PB from WT (n = 5) and Ezh1−/−Ezh2Δzh (double knockout [DKO]) mice (n = 5) are shown as the mean ± SD. (B-C) Competitive repopulating assays. BM cells (CD45.2) from Cre-ERT, Ezh1−/−, Cre-ERT;Ezh2fl/fl, and Cre-ERT;Ezh1−/−Ezh2fl/fl mice were transplanted into lethally irradiated recipient mice (CD45.1) with twice more competitor BM cells (CD45.1), and Ezh2 was then deleted by injecting tamoxifen at 8 weeks posttransplantation. The chimerism of donor-derived CD45.2+ cells, Mac-1+ myeloid cells, B220+ B cells, and CD4+ or CD8+ T cells in the PB is shown as percentage of chimerism values prior to the treatment with tamoxifen in panel B. Donor chimerism in BM LSK cells at 12 weeks posttransplantation is shown in panel C. Data are plotted as dots and the mean values are indicated as bars (n = 3-4, each). (D) A quantitative reverse transcription PCR (RT-PCR) analysis of CDK inhibitors (p15Ink4b, p16Ink4a and p19Arf) in WT, Ezh2Δ/Δ, and Ezh1−/−Ezh2Δ/Δ LSK and LK cells 2 weeks after the deletion of Ezh2. mRNA levels were normalized to Hprt1 expression and relative expression levels are shown as the mean ± SD for triplicate analyses. The cells whose expression was arbitrarily set to 1 are indicated as “1”. Statistical significance of difference was measured by unpaired 2-tailed Student t test. *P < .05, **P < .01, ***P < .001.
Ezh1 was essential for the maintenance of Ezh2 insufficient hematopoiesis. (A) The failure of Ezh1−/−Ezh2Δ/Δ hematopoietic cells to maintain hematopoiesis. BM cells from Cre-ERT and Cre-ERT;Ezh1−/−Ezh2fl/fl mice were transplanted into lethally irradiated recipient mice without rescue BM cells, and Ezh2 was then deleted by injecting tamoxifen at 6 weeks posttransplantation. Following tamoxifen injection, the recipient mice were fed with a tamoxifen diet (400 mg of tamoxifen citrate per kg diet) for 10 days. WBC counts, the chimerism of CD45.2+ donor-derived cells, including Mac-1+ myeloid cells, B220+ B cells, and CD4+ or CD8+ T cells, in the PB from WT (n = 5) and Ezh1−/−Ezh2Δzh (double knockout [DKO]) mice (n = 5) are shown as the mean ± SD. (B-C) Competitive repopulating assays. BM cells (CD45.2) from Cre-ERT, Ezh1−/−, Cre-ERT;Ezh2fl/fl, and Cre-ERT;Ezh1−/−Ezh2fl/fl mice were transplanted into lethally irradiated recipient mice (CD45.1) with twice more competitor BM cells (CD45.1), and Ezh2 was then deleted by injecting tamoxifen at 8 weeks posttransplantation. The chimerism of donor-derived CD45.2+ cells, Mac-1+ myeloid cells, B220+ B cells, and CD4+ or CD8+ T cells in the PB is shown as percentage of chimerism values prior to the treatment with tamoxifen in panel B. Donor chimerism in BM LSK cells at 12 weeks posttransplantation is shown in panel C. Data are plotted as dots and the mean values are indicated as bars (n = 3-4, each). (D) A quantitative reverse transcription PCR (RT-PCR) analysis of CDK inhibitors (p15Ink4b, p16Ink4a and p19Arf) in WT, Ezh2Δ/Δ, and Ezh1−/−Ezh2Δ/Δ LSK and LK cells 2 weeks after the deletion of Ezh2. mRNA levels were normalized to Hprt1 expression and relative expression levels are shown as the mean ± SD for triplicate analyses. The cells whose expression was arbitrarily set to 1 are indicated as “1”. Statistical significance of difference was measured by unpaired 2-tailed Student t test. *P < .05, **P < .01, ***P < .001.
In order to understand the molecular mechanism underlying the impaired repopulating capacity of Ezh1−/−Ezh2fΔ/Δ HSCs in more detail, we examined the canonical polycomb targets, p15Ink4b, p16Ink4a, and p19Arf, which are critical for the maintenance of HSC activity.3,4,35,36 As expected, all of these tumor suppressor genes were markedly upregulated in Ezh1−/−Ezh2Δ/Δ LSK and Lin−c-Kit+ cells (Figure 6D), but not in Ezh1−/− and Ezh2Δ/Δ cells (Figure 6D; supplemental Figure 5).
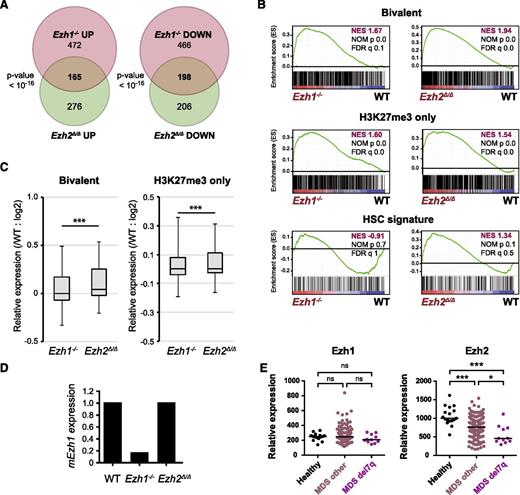
To check how Ezh1 can complement Ezh2 loss in hematopoiesis, we next performed RNA-sequence analysis of Ezh1−/− and Ezh2Δ/Δ LSK cells from recipient mice at 1-month postinjection of tamoxifen. Regardless of their modest hematologic phenotypes, Ezh1−/− LSK cells showed changes in gene expression profiles similar to Ezh2Δ/Δ LSK cells and a significant part of genes altered in Ezh1−/− and Ezh2Δ/Δ LSK cells overlapped (Figure 7A). GSEA revealed that Ezh1 loss also caused derepression of PRC2 target genes including H3K27me3-only genes and bivalent genes (Figures 7B), although the derepression levels were milder than those noticed with Ezh2 loss (Figure 7C). These findings indicate that the targets of Ezh1 and Ezh2 largely overlap. Of interest, the HSC gene signature defined by Chambers et al37 was positively and negatively enriched in Ezh2Δ/Δ and Ezh1−/− LSK cells, respectively, although statistically not significant (Figure 7B). This finding may partly account for a tumor suppressor function of Ezh2 but not Ezh1 in myelodysplastic disorders. RNA-sequence data revealed no significant change in Ezh1 mRNA levels in Ezh2Δ/Δ LSK cells (Figure 7D).
Gene expression profiles of Ezh1-deficient HSPCs. (A) Venn diagram showing the overlap between genes upregulated (at least twofold) and downregulated (at least twofold) in Ezh1−/− and Ezh2Δ/Δ LSK cells compared with WT LSK cells. RNA-sequence analysis was performed using LSK cells from WT, Ezh1−/−, and Ezh2Δ/Δ mice at 1 month postinjection of tamoxifen. (B) GSEA plots demonstrating enrichment levels of indicated gene sets in Ezh1−/− and Ezh2Δ/Δ LSK cells compared with WT LSK cells. NES, NOM P value, and FDR are indicated. (C) Box-and-whisker plots showing the expression levels of H3K27me3-only genes and bivalent genes in Ezh1−/− and Ezh2Δ/Δ LSK cells relative to WT LSK cells. Boxes represent 25 to 75 percentile ranges. Vertical lines represent 10 to 90 percentile ranges. Horizontal bars represent median. ***P < .001 (Student t test.). (D) Graphic presentation of RNA-sequence data. RPKM of Ezh1 in Ezh1−/− and Ezh2Δ/Δ LSK cells were shown relative to that of WT LSK cells. (E) Expression of EZH1 and EZH2 in CD34+ MDS cells. Relative expression of EZH1 and EZH2 in CD34+ cells from healthy controls, MDS patients, and MDS patients with 7q deletion. The data are presented as scatter diagrams with median values (bars). ***P < .001 (Student t test). The data from MDS patients were retrieved from published database.38
Gene expression profiles of Ezh1-deficient HSPCs. (A) Venn diagram showing the overlap between genes upregulated (at least twofold) and downregulated (at least twofold) in Ezh1−/− and Ezh2Δ/Δ LSK cells compared with WT LSK cells. RNA-sequence analysis was performed using LSK cells from WT, Ezh1−/−, and Ezh2Δ/Δ mice at 1 month postinjection of tamoxifen. (B) GSEA plots demonstrating enrichment levels of indicated gene sets in Ezh1−/− and Ezh2Δ/Δ LSK cells compared with WT LSK cells. NES, NOM P value, and FDR are indicated. (C) Box-and-whisker plots showing the expression levels of H3K27me3-only genes and bivalent genes in Ezh1−/− and Ezh2Δ/Δ LSK cells relative to WT LSK cells. Boxes represent 25 to 75 percentile ranges. Vertical lines represent 10 to 90 percentile ranges. Horizontal bars represent median. ***P < .001 (Student t test.). (D) Graphic presentation of RNA-sequence data. RPKM of Ezh1 in Ezh1−/− and Ezh2Δ/Δ LSK cells were shown relative to that of WT LSK cells. (E) Expression of EZH1 and EZH2 in CD34+ MDS cells. Relative expression of EZH1 and EZH2 in CD34+ cells from healthy controls, MDS patients, and MDS patients with 7q deletion. The data are presented as scatter diagrams with median values (bars). ***P < .001 (Student t test). The data from MDS patients were retrieved from published database.38
Finally, we checked the expression of EZH1 and EZH2 in human MDS patients.38 As expected, EZH2 expression was significantly downregulated in CD34+ cells from MDS patients and this trend was more significant in CD34+ cells from MDS patients with the 7q deletion, which often results in the loss of EZH2 at 7q36.1 (Figure 7E). In contrast, EZH1 expression was comparable between controls and MDS patients. These findings indicate that the compensatory function of EZH1 in the setting of EZH2 loss is not regulated by its expression, but support repositioning of EZH1 to EZH2 targets as suggested by ChIP-sequence data (Figure 4).
Discussion
In the present study, we demonstrated that the deletion of Ezh2 caused heterogeneous hematopoietic malignancies including myelodysplastic disorders (MDS and MDS/MPN) and lymphoid leukemia in mice. These results support the tumor suppressive role of EZH2 in these diseases. In contrast, as we reported previously,24,25 AML was not observed in our cohorts. This result confirmed the oncogenic role of EZH2 in AML, in which inactivating EZH2 mutations are rare,14,20,26,27 and the efficacy of EZH2 inhibitors was shown in preclinical studies.10,39
Corresponding to the recent finding that inactivating EZH2 mutations were recurrently identified in patients with MDS and MDS/MPN,13-20 Ezh2 loss alone in mice induced MDS and MDS/MPN after a long latency. In our previous study, we reported the development of MDS/MPN only.24 However, we also observed the development of an MDS-like disease in this much larger cohort. These findings strongly support the role of inactivating EZH2 mutations in the pathogenesis of myelodysplastic disorders. EZH2 mutations are acquired by the preceding HSC clones with founder mutations, such as TET2 and DNMT3A, and splicing factor gene mutations,16,40,41 which initiate and promote clonal hematopoiesis during aging.40-42 Once additional mutations are acquired in founder clones or subclones, concurrent mutations are thought to cooperate in the progression of myelodysplastic disorders. We previously demonstrated cooperation between the Tet2 hypomorph (Tet2KD/KD) and Ezh2 loss in a mouse model of myelodysplastic disorders.24 Although TET2 mutations are more frequently detected in myelodysplastic disorders than EZH2 mutations, the Tet2 hypomorph did not cause obvious myelodysplasia in mice.24 In contrast, Ezh2 loss induced myelodysplasia, particularly morphologic dysplasia, from the early time point in this study. Given that Asxl1 loss also leads to myelodysplasia through dysregulated H3K27me3 modification,43 PRC2 insufficiencies may be closely associated with myelodysplasia.
In this study, we analyzed only homozygous inactivation of Ezh2. However, the majority of mutations in EZH2 in human disease are heterozygous.13-15 Of interest, we have previously evaluated the effect of heterozygous inactivation of Ezh2 in Tet2 gene trap mice (Tet2KD/KD). The median survival was significantly shortened by heterozygous loss of Ezh2 (Tet2KD/KDEzh2Δ/Δ mice [n = 20], Tet2KD/KDEzh2Δ/+mice [n = 14], and Tet2KD/KDEzh2+/+ mice [n = 34] being 172 days, 243 days, and undetermined, respectively; Tet2KD/KDEzh2Δ/+mice vs Tet2KD/KDEzh2+/+ mice, P = .004 by log-rank test). These data clearly suggest a haploinsufficient effect of EZH2.
PRC2 gene mutations, including EZH2 mutations, are also frequently identified in adult T-ALL (25%)34,44,45 as well as childhood early T-cell precursor (ETP) ALL (42%) and non-ETP ALL (12%).46 We observed the development of T-cell receptor αβ (TCRαβ)-type T-ALL/lymphoma in the secondary transplantation of Ezh2-deficient hematopoietic cells. In contrast, TCRγδ-type T-ALL has been reported to develop much earlier in Ezh2-deficient mice.44 These discrepancies may be due to the different designs of gene targeting and/or the different deleter mice used. It would be intriguing to ask why diverse malignancies, such as MDS, MDS/MPN, and T-ALL, developed in Ezh2Δ/Δ mice. Our expression profiling in this study did not provide any definite insight into this question. Only the epidermal growth factor signaling gene sets appeared to be significantly enriched in CD8+ T-ALL cells (Figure 2F) but not in MDS or MDS/MPN LSK cells (data not shown). Further study is essential to understand the differential impacts of Ezh2 loss in hematologic malignancies.
Although inactivating EZH2 mutations have yet to be identified in B-cell malignancies including CLL,47 we often detected the expansion of B-1a B cells in Ezh2-deficient mice, some of which developed LPD or a CLL-like disease after a long latency. B-1 cells are innate-like lymphocytes that have important functions in immune defense, and are found at higher frequencies within the peritoneal and pleural cavities of mice.48 Ezh2 loss has been shown to markedly affect the differentiation of B cells in the BM.49 Consistent with these findings, we did not observe the development of any conventional B-cell malignancies in Ezh2-deficient mice. It is possible that the expansion of B-1 cells is secondary to the compromised differentiation of conventional B cells. However, Ezh2 may also be involved in the negative regulation of the differentiation of B-1 cells. The role of Dnmt3 in this regulation has been suggested by the recent finding that the loss of Dnmt3a alone or with Dnmt3b showed a strong propensity to induce B-1 CLL in FVB mice.50
In our gene expression analysis, bivalent genes appeared to be largely repressed in HSPCs over time in the absence of Ezh2. This could be partly due to replacement by DNA methylation following loss of H3K27me3 modification. However, ChIP-sequence data clearly showed that H3K27me3 levels at the promoters of PRC2 targets, particularly bivalent genes, significantly recovered over time in HSPCs. Recently, Ezh1 has been demonstrated to reposition to Ezh2 target genes upon depletion of Ezh2 in erythroid cells.51 Correspondingly, H3K27me3 levels recovered not globally but in a locus-specific manner in this study. Given that the expression of Ezh1 does not change in the absence of Ezh2, repositioning of Ezh1 to Ezh2 target genes might be a promising mechanism underlying the compensatory function of Ezh1 for Ezh2.
In this study, an essential role for Ezh1 was confirmed by the finding that Ezh1−/−Ezh2Δ/Δ HSCs could not support hematopoiesis. Ezh1 was found to be critical for transcriptional repression of p15Ink4b, p16Ink4a, and p19Arf in Ezh2Δ/Δ HSPCs. Indeed, conditional deletion of Ezh1 in adult BM HSPCs has been reported to cause derepression of p15Ink4b and p16Ink4a,5 although straight knockout of Ezh1 used in this study did not cause derepression of these genes. Given that the loss of Ezh1 has only a minimal impact on global H3K27me3 levels,5 more comprehensive analysis is needed to understand Ezh1 function in HSPCs. Taken together, the results of the present study indicate that Ezh1 acts to maintain pathological stem cells under Ezh2 insufficiency and correspond well with the fact that no recurrent somatic mutations in EZH1 have been identified in patients with hematologic malignancies. Our findings also suggest that patients with EZH2 mutations would be more sensitive to EZH1 inhibition compared with patients without EZH2 mutations. Although there is currently no EZH1-specific inhibitor, this possibility should be tested in the near future.
Accumulating evidence suggests that epigenetic abnormalities hold the key to the pathogenesis of hematologic malignancies.52,53 The results of the present study provide a new mouse model for hematologic malignancies. Inactivating mutations targeting PRC2 components, such as EED and SUZ12, have recently been identified even in solid tumors.54 Our mouse models may facilitate understanding of the tumor suppressor role of PRC2 not only in hematologic malignancies, but also in a broader range of cancers.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Ezh1 constitutive knockout mice were generated at the Research Institute of Molecular Pathology (Vienna, Austria) in 2000 by Dønal O’Carroll (Laboratory Thomas Jenuwein) with the help of Maria Sibilia (Laboratory Erwin Wagner) and will be reported elsewhere. Ezh2fl/fl mice were kindly provided by Haruhiko Koseki (RIKEN, Japan). The authors thank Ola Mohammed Kamel Rizq for critical review of the manuscript.
This work was supported in part by Grants-in-Aid for Scientific Research (#24249054) and Scientific Research on Innovative Areas “Stem Cell Aging and Disease” (#25115002) and “Cancer Stem Cell” (#25130702) from Ministry of Education, Culture, Sports, Science, and Technology (MEXT), Japan, and grants from the Naito Foundation, Uehara Memorial Foundation, and Takeda Science Foundation.
M.M.-K. is a research fellow supported by Japan Society for the Promotion of Science, MEXT, Japan.
Authorship
Contribution: M.M.-K. performed the experiments, analyzed results, made the figures, and actively wrote the manuscript; K.A., G.S., M.O., T.T., T.M., and C.W. assisted with the experiments; and A.I. conceived of and directed the project, secured funding, and actively wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Atsushi Iwama, Department of Cellular and Molecular Medicine, Graduate School of Medicine, Chiba University, 1-8-1 Inohana, Chuo-ku, Chiba, 260-8670 Japan; e-mail: aiwama@faculty.chiba-u.jp.

![Figure 6. Ezh1 was essential for the maintenance of Ezh2 insufficient hematopoiesis. (A) The failure of Ezh1−/−Ezh2Δ/Δ hematopoietic cells to maintain hematopoiesis. BM cells from Cre-ERT and Cre-ERT;Ezh1−/−Ezh2fl/fl mice were transplanted into lethally irradiated recipient mice without rescue BM cells, and Ezh2 was then deleted by injecting tamoxifen at 6 weeks posttransplantation. Following tamoxifen injection, the recipient mice were fed with a tamoxifen diet (400 mg of tamoxifen citrate per kg diet) for 10 days. WBC counts, the chimerism of CD45.2+ donor-derived cells, including Mac-1+ myeloid cells, B220+ B cells, and CD4+ or CD8+ T cells, in the PB from WT (n = 5) and Ezh1−/−Ezh2Δzh (double knockout [DKO]) mice (n = 5) are shown as the mean ± SD. (B-C) Competitive repopulating assays. BM cells (CD45.2) from Cre-ERT, Ezh1−/−, Cre-ERT;Ezh2fl/fl, and Cre-ERT;Ezh1−/−Ezh2fl/fl mice were transplanted into lethally irradiated recipient mice (CD45.1) with twice more competitor BM cells (CD45.1), and Ezh2 was then deleted by injecting tamoxifen at 8 weeks posttransplantation. The chimerism of donor-derived CD45.2+ cells, Mac-1+ myeloid cells, B220+ B cells, and CD4+ or CD8+ T cells in the PB is shown as percentage of chimerism values prior to the treatment with tamoxifen in panel B. Donor chimerism in BM LSK cells at 12 weeks posttransplantation is shown in panel C. Data are plotted as dots and the mean values are indicated as bars (n = 3-4, each). (D) A quantitative reverse transcription PCR (RT-PCR) analysis of CDK inhibitors (p15Ink4b, p16Ink4a and p19Arf) in WT, Ezh2Δ/Δ, and Ezh1−/−Ezh2Δ/Δ LSK and LK cells 2 weeks after the deletion of Ezh2. mRNA levels were normalized to Hprt1 expression and relative expression levels are shown as the mean ± SD for triplicate analyses. The cells whose expression was arbitrarily set to 1 are indicated as “1”. Statistical significance of difference was measured by unpaired 2-tailed Student t test. *P < .05, **P < .01, ***P < .001.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/126/10/10.1182_blood-2015-03-634428/4/m_1172f6.jpeg?Expires=1769089565&Signature=eItk36z9foKe3P5tfJGN4fQicjwcGlCudebrjhj8HlKCs31enB4do0RW3cd7itnMFqd9gA-yn3HBxKyUC6G-KG~REL-r5N99BewkElhqMnY-~BolCfGZsXbEALxEp2VWD3zCBDSt7x9sc5gMZy7WJNaDuzoJi2cAqno3rLs27kVkc~UcdDKb3s2frwP2-9lNYkgPqLbWNFu1C4lsg0WKMY4lAgJal0kFywapzC4op3~XriO5ZdhEmfYfr~BiA4ntvCjKMq1nBv9pbrKHO7nEdDCK8OEraTXYEPNH8uATipspTxz-5NH~YCgn6UCyqSCRRWWLZUqrLe04b086o2aeWA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)