Key Points
A half loss of RUNX1 activity leads to defects in primitive erythropoiesis, megakaryopoiesis, and proplatelet formation.
An almost complete loss of RUNX1 activity leads to the amplification of the granulomonocytic compartment with increased genomic instability.
Abstract
To explore how RUNX1 mutations predispose to leukemia, we generated induced pluripotent stem cells (iPSCs) from 2 pedigrees with germline RUNX1 mutations. The first, carrying a missense R174Q mutation, which acts as a dominant-negative mutant, is associated with thrombocytopenia and leukemia, and the second, carrying a monoallelic gene deletion inducing a haploinsufficiency, presents only as thrombocytopenia. Hematopoietic differentiation of these iPSC clones demonstrated profound defects in erythropoiesis and megakaryopoiesis and deregulated expression of RUNX1 targets. iPSC clones from patients with the R174Q mutation specifically generated an increased amount of granulomonocytes, a phenotype reproduced by an 80% RUNX1 knockdown in the H9 human embryonic stem cell line, and a genomic instability. This phenotype, found only with a lower dosage of RUNX1, may account for development of leukemia in patients. Altogether, RUNX1 dosage could explain the differential phenotype according to RUNX1 mutations, with a haploinsufficiency leading to thrombocytopenia alone in a majority of cases whereas a more complete gene deletion predisposes to leukemia.
Introduction
RUNX1 protein is the α subunit of the core-binding factor (CBF) transcriptional complex. The protein contains an N-terminal Runt homology domain that binds to DNA and to CBFβ, the β subunit of CBF, and a C-terminal transactivation domain. Germline alterations in the RUNX1 gene are responsible for the familial platelet disorder (FPD) with a predisposition to acute myeloid leukemia (AML; OMIM 601399),1 a rare constitutive disorder that associates a moderate thrombocytopenia with a variable propensity to develop acute leukemia. Whereas all the germline RUNX1 alterations found in FPD/AML lead to thrombocytopenia, evolution to leukemia depends on the type of mutations (ie, mutations maintaining CBFβ-binding properties) to generate dominant-negative (DN) proteins that favor leukemic evolution, whereas mutations inducing haploinsufficiency only rarely display leukemic development.2 RUNX1 alterations that predispose to leukemia, mainly DN-like mutants,3,4 deregulate critical hematopoietic stem cell (HSC) and progenitor target genes such as NR4A3,4 leading to the amplification of a pool of cells susceptible to acquiring additional somatic mutations, sometimes affecting the second RUNX1 allele.5
FPD/AML is a suitable model for studying the defects in megakaryopoiesis that lead to thrombocytopenia. This disease can also be used to explore the initial events in leukemogenesis, because somatic alterations in the RUNX1 gene are involved in sporadic myeloid malignancies. Chromosomal translocations that involve the RUNX1 gene are commonly observed in AML,6 whereas RUNX1 gene mutations are identified in 6% to 32% of AML.7-9 Mutations in RUNX1 are also detected in 8% to 15% of chronic myelomonocytic leukemias10,11 and 9% of early-stage myelodysplastic syndromes (MDSs).12 The fact that RUNX1 mutations are detected at an early stage in MDSs, before acute leukemia occurrence, argues for an early event in leukemic transformation.13
Analysis of mouse models indicated that RUNX1 is a key regulator of definitive hematopoiesis, including HSC emergence.14 In adult murine hematopoietic compartments, RUNX1 is dispensable for HSC maintenance, but it negatively regulates myeloid progenitors while promoting lymphopoiesis and megakaryopoiesis.15,16 Regarding primitive hematopoiesis, an active yolk sac–derived erythropoiesis14 and a normal number of primitive erythroid progenitors were observed in Runx1 knockout (KO) mice, but primitive erythrocytes had an abnormal morphology and a reduced expression of Ter119, Klf1, and Gata1.17 Conditional Runx1 KO led to the development of a myeloproliferative syndrome but failed to reproduce the leukemic development observed in 35% of FPD/AML patients.15
Induced pluripotent stem cells (iPSCs)18 offer a new opportunity to model inherited human diseases in vitro and allow the investigation of initial pathogenic events that may occur during embryogenesis. Here, we generated iPSCs from FPD/AML patients with 2 different pedigrees: one harbored the DN-like mutation R174Q,4 and the other harbored a monoallelic RUNX1 deletion producing a true haploinsufficiency.2 We first observed that RUNX1 played a crucial role in regulating the first wave of human primitive hematopoiesis, giving rise to erythroid and megakaryocytic (MK) cells. We noticed also that the phenotype induced by the R174Q mutant was similar to almost complete RUNX1 knockdown in human embryonic stem cells (hESCs), indicating that the R174Q RUNX1 mutation should affect the binding of the remaining wild-type (WT) RUNX1 protein to CBFβ to induce an almost complete loss of function.3 Most importantly, we noticed that increased genomic instability of the granulomonocytic cell population depended on the dosage of functional RUNX1, which was associated with decreased expression of GADD45A19 as well as other p53 targets. Overall, these results show that the residual activity of WT RUNX1 protein would impact the risk for development of leukemia in FPD/AML patients.
Material and methods
Cell culture
Human dermal fibroblasts (HDFs) were derived from skin biopsies of patients after informed consent in accordance with the Declaration of Helsinki. The study was approved by the Ethics Committee of INSERM RBM 01-14 for the project “Network on the inherited diseases of platelet function and platelet production.”
HDFs were cultured in F10 Glutamax medium (Invitrogen, Cergy-Pontoise, France) with 20% fetal bovine serum (Invitrogen, Cergy-Pontoise, France). H9 hESC lines (National Institutes of Health code WA09) were obtained from WiCell Research Institute. iPSCs and hESCs were maintained in an undifferentiated state on irradiated mouse embryonic fibroblasts in hESC medium, as previously described.20 The use of hESCs was approved by Agence de la Biomédecine, No. R04-0020 and No. C04-0019. OP9 cells (Riken Institute, Wako, Japan) were maintained on gelatinized dishes.
iPSC generation
HDF transduction with the STEMCCA lentivirus was performed at passage 4 or 5. HDFs were transduced twice at 12-hour intervals. HDFs (5 × 104) were then plated on mouse embryonic fibroblasts and gelatin; 24 hours later, the medium was replaced by hESC medium and was changed every day. The ESC-like colonies were picked up separately between day 30 and day 45 and cultivated as for hESC culture.21 One iPSC control cell line (C1/CHOPWT2.2) has already been described.22
Hematopoietic differentiation on OP9 cells
To differentiate iPSCs in hematopoietic cells, previously described protocols using coculture on OP9 and cytokines; vascular endothelial growth factor 20 ng/mL (Miltenyi Biotech) added at day 0; and stem cell factor 25 ng/mL (Biovitrum AB, Stockholm, Sweden), interleukin-3 10 ng/mL (Miltenyi Biotech), erythropoietin 2 U/mL (Amgen, Thousand Oaks, CA), and thrombopoietin 20 ng/mL (Kirin Pharma Company, Tokyo, Japan) added at day 7 of culture were used.20
Clonogenic progenitor assays
HSC progenitors were sorted on the expression of CD34 and CD43 at day 11 or day 14 and were plated in triplicate at a density of 1.5 × 103 cells per milliliter in human methylcellulose medium H4434 (StemCell Technologies, Vancouver, BC, Canada) to quantify erythroid and colony-forming unit–granulocyte macrophage (CFU-GM) progenitors at a density of 3 × 103 cells per milliliter in serum-free fibrin clots to quantify MK progenitors. MK progenitors were stained with an anti-CD41a monoclonal antibody (BD Biosciences, Le Pont de Claix, France), as previously described.20 Images were obtained with AxioVision 4.6 software. The same software was used to measure the CFU-GM colony size.
Gene expression analysis by conventional and quantitative real-time PCR
Genomic DNAs and RNAs were extracted by using QIAmp DNA Mini Kit or RNeasy Micro Kit (QIAGEN, Courtaboeuf, France). Reverse transcription of RNA was performed with SuperScript Vilo cDNA Synthesis Kit (Invitrogen, Cergy Pontoise, France). Quantitative real-time polymerase chain reactions (qRT-PCRs) and qPCRs were performed on a 7500 Real-Time PCR system using SYBR-Green PCR Master Mix (Applied Biosystems, Saint-Aubin, France). Expression levels of genes were normalized to housekeeping genes PPIA and L32. Primers are listed in supplemental Table 1 (available online on the Blood Web site).
Statistical analyses
Data are presented as means (± standard deviation) or with 95% confidence intervals of the mean. Statistical significance was determined by Student t test. P < .05 was considered statistically significant. For transcriptome analysis, an analysis of variance test was performed with a P value threshold of P < .001. Other methods are provided in supplemental Data.
Results
RUNX1 alterations induce a defect in erythroid and MK cell output
To compare the consequences of two germline RUNX1 alterations, one leading to an increase in leukemia predisposition (R174Q mutation, pedigree A) and the other to thrombocytopenia alone (monoallelic deletion, pedigree D), four iPSC lines were derived from FPD/AML patient skin fibroblasts. The 2 pedigrees have been described previously.2,4 The strategy of deriving iPSCs and their characterization is shown in supplemental Figures 1-3. We selected 1 iPSC clone derived from each of 2 distinct members of pedigree A (AII_1, AII_2) and 2 iPSC clones derived from pedigree D (D(a) and D(b)). Three independent control iPSC lines (C1, C2, and C3) were used as references for all experiments. All iPSC lines were passaged 15 to 20 times to remove memory of origin, which may interfere with differentiation to downstream lineages.23,24 Deep sequencing analysis identified 109 de novo mutations for AII_1, 82 for AII_2, and 9 for D(a). The mutations of genes that could play a role in hematopoiesis were confirmed by Sanger sequencing (supplemental Table 2). However, the results showing that the same gene was never mutated in more than 1 patient sample suggest that the common phenotype observed in all FPD/AML patients studied here is exclusively linked to the germline RUNX1 alteration and not to the randomly acquired mutations. The protocols of hematopoietic differentiation are described in supplemental Figure 4. Hence, we studied the impact of RUNX1 alterations on progenitors generated by culturing CD34+CD43+ cells obtained on day 11, focusing on erythroid and MK cells, and those occurring at day 14, focusing on granulomonocytic cells. When assessing the colony-forming potential of CD34+CD43+ cells sorted at day 11, we observed a decrease in the number of erythroid progenitors generated from the RUNX1 R174Q mutated as well as RUNX1 haploinsufficient iPSC clones (Figure 1A-B). We also detected a threefold decrease in MK progenitor number for both pedigrees (Figure 1C-D). CD34+CD43+ cells sorted at day 11 were also cultured on OP9 stromal cells in liquid medium (supplemental Figure 4A), demonstrating a decrease in the percentage of erythroid (GPA+) and MK (CD41+CD42+) populations at day 18 in cells derived from the 2 pedigrees (Figure 1E-F). The absolute numbers of erythroid (GPA+) and MK cells generated from 1 × 102 CD34+CD43+ progenitors were decreased for both the RUNX1 R174Q mutation and RUNX1 deletion iPSC clones (Figure 1G-H). Strikingly, these results demonstrate that, in humans, RUNX1 plays an important role in the first wave of hematopoiesis yielding primitive erythroid and MK cells.
Deep decrease in erythroid and MK potential of progenitors derived from FPD/AML iPSC lines. AII_1 and AII_2 are iPSC clones of 2 patients from pedigree A; D(a) and D(b) are 2 clones of 1 patient from pedigree D; and C1, C2, and C3 are control iPSC lines. Cell numbers were normalized in all experiments to those produced by the C2 clone, which was reported in each experiment as a fixed value of 100. (A-D) CD34+CD43+ cells sorted at day 11 and tested for their colony-forming potential. (A-B) Methylcellulose assay. (A) The histograms present the numbers of erythroid progenitors (Ery-P) in 1 representative experiment of 3, each in triplicate. (B) Pictures of primitive erythroid colonies (Ery-P). (C-D) Fibrin clot culture. (C) The histograms present the numbers of MK progenitors (MK-P) in 1 representative experiment of 4, each in triplicate. (D) Pictures of primitive MK colonies (MK-P) after CD41 immunostaining. (A,C) Error bars represent ± standard deviation (SD) of triplicate. (B,D) Scale bar = 50 μm. (E-H) Analysis of hematopoietic populations at day 18 from progenitors sorted at day 11. (E) May-Grünwald-Giemsa staining of erythroblasts (GPA+) and MKs (CD41+CD42+) derived from iPSC lines. Scale bar = 10 μm. (F) Fluorescence-activated cell sorter (FACS) analysis of erythroid cells and MKs from FPD/AML iPSC lines in 1 representative experiment. (G) Absolute number of generated erythroid cells normalized to 1 × 102 plated progenitors. The histograms show average of the cell number obtained in 5 independent experiments for pedigree A and 3 for pedigree D. (H) Absolute number of generated MKs normalized to 1 × 102 plated progenitors. The histograms show average of the cell number obtained in 3 independent experiments for both pedigrees. (G-H) Error bars represent ± SD of the average. *P < .05; **P < .01; ***P < .001; Student t test.
Deep decrease in erythroid and MK potential of progenitors derived from FPD/AML iPSC lines. AII_1 and AII_2 are iPSC clones of 2 patients from pedigree A; D(a) and D(b) are 2 clones of 1 patient from pedigree D; and C1, C2, and C3 are control iPSC lines. Cell numbers were normalized in all experiments to those produced by the C2 clone, which was reported in each experiment as a fixed value of 100. (A-D) CD34+CD43+ cells sorted at day 11 and tested for their colony-forming potential. (A-B) Methylcellulose assay. (A) The histograms present the numbers of erythroid progenitors (Ery-P) in 1 representative experiment of 3, each in triplicate. (B) Pictures of primitive erythroid colonies (Ery-P). (C-D) Fibrin clot culture. (C) The histograms present the numbers of MK progenitors (MK-P) in 1 representative experiment of 4, each in triplicate. (D) Pictures of primitive MK colonies (MK-P) after CD41 immunostaining. (A,C) Error bars represent ± standard deviation (SD) of triplicate. (B,D) Scale bar = 50 μm. (E-H) Analysis of hematopoietic populations at day 18 from progenitors sorted at day 11. (E) May-Grünwald-Giemsa staining of erythroblasts (GPA+) and MKs (CD41+CD42+) derived from iPSC lines. Scale bar = 10 μm. (F) Fluorescence-activated cell sorter (FACS) analysis of erythroid cells and MKs from FPD/AML iPSC lines in 1 representative experiment. (G) Absolute number of generated erythroid cells normalized to 1 × 102 plated progenitors. The histograms show average of the cell number obtained in 5 independent experiments for pedigree A and 3 for pedigree D. (H) Absolute number of generated MKs normalized to 1 × 102 plated progenitors. The histograms show average of the cell number obtained in 3 independent experiments for both pedigrees. (G-H) Error bars represent ± SD of the average. *P < .05; **P < .01; ***P < .001; Student t test.
MKs derived from FPD/AML iPSCs display a profound defect in proplatelet formation
When CD41+CD42+ MKs were sorted after 14 days of coculture and analyzed 5 days later (Figure 2A), the percentage of proplatelet-forming MKs was decreased at least 10-fold in patient iPSCs compared with control iPSC-derived MKs (Figure 2B-C). Since RUNX1 is a positive or negative regulator of cytoskeleton components involved in proplatelet formation (MYL9, MYH9)25 or in ploidization (MYH10),26 we examined the expression of these genes. As observed in MKs derived from patient progenitors,25 MYL9 expression level was decreased whereas MYH10 expression was increased in MKs obtained from patient iPSCs, whatever the RUNX1 alteration (Figure 2D), demonstrating that the defect in megakaryopoiesis is independent of the type of RUNX1 alteration.
Defect in proplatelet formation by MK cells derived from FPD/AML iPSC lines. (A) Schema of protocol used for MK differentiation. iPSC lines were seeded on OP9 cells with vascular endothelial growth factor (VEGF) and thrombopoietin (TPO) at day 0. After 7 days, stem cell factor (SCF) was added to the culture medium. MK (CD41+CD42+) cells were sorted at day 14 and further cultured in the presence of TPO and SCF. Analysis of proplatelet formation and qRT-PCR were performed at day 19. (B) Representative microscopic images representing control and patient proplatelet-forming MKs. (C) The percentage of proplatelet-forming MKs was estimated by counting MKs exhibiting 1 or more cytoplasmic processes with areas of constriction. A total of 200 cells per well was counted. The histograms show 1 representative experiment of 5, each in triplicate. Error bars represent ± SD of triplicate. (D) qRT-PCR analysis of MYL9 and MYH10 expression level in MKs. The histograms show 1 representative experiment of 4 for MYL9 and 5 for MYH10, each in triplicate. Data are normalized to PPIA and L32 transcript level, and expression is compared with control C3. Similar results were obtained but only results normalized to PPIA are shown. Error bars represent ± SD of triplicate. **P < .01; ***P < .001; Student t test.
Defect in proplatelet formation by MK cells derived from FPD/AML iPSC lines. (A) Schema of protocol used for MK differentiation. iPSC lines were seeded on OP9 cells with vascular endothelial growth factor (VEGF) and thrombopoietin (TPO) at day 0. After 7 days, stem cell factor (SCF) was added to the culture medium. MK (CD41+CD42+) cells were sorted at day 14 and further cultured in the presence of TPO and SCF. Analysis of proplatelet formation and qRT-PCR were performed at day 19. (B) Representative microscopic images representing control and patient proplatelet-forming MKs. (C) The percentage of proplatelet-forming MKs was estimated by counting MKs exhibiting 1 or more cytoplasmic processes with areas of constriction. A total of 200 cells per well was counted. The histograms show 1 representative experiment of 5, each in triplicate. Error bars represent ± SD of triplicate. (D) qRT-PCR analysis of MYL9 and MYH10 expression level in MKs. The histograms show 1 representative experiment of 4 for MYL9 and 5 for MYH10, each in triplicate. Data are normalized to PPIA and L32 transcript level, and expression is compared with control C3. Similar results were obtained but only results normalized to PPIA are shown. Error bars represent ± SD of triplicate. **P < .01; ***P < .001; Student t test.
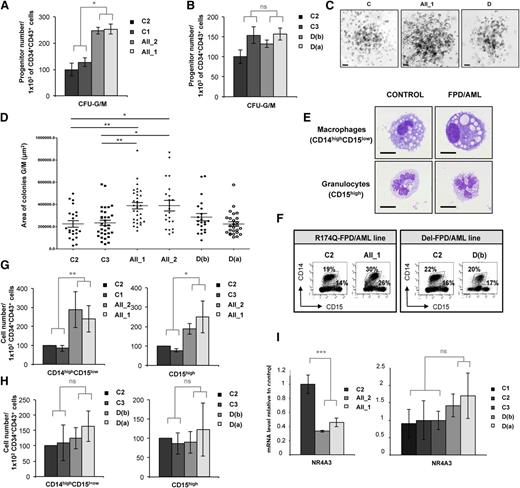
RUNX1 R174Q mutation specifically affects the output of granulomonocytes
When we assessed the GM colony-forming potential of CD34+CD43+ cells sorted at day 14 (supplemental Figure 4A), we observed a significant twofold increase in the number of CFU-GM colonies in the 2 patients with R174Q mutation compared with the 3 controls (Figure 3A), but not in the patient with a monoallelic RUNX1 deletion (Figure 3B). The size of GM colonies generated from R174Q iPSCs was also significantly increased (Figure 3C; median sizes: 389 420 µm2 for AII_2 and 388 953 µm2 for AII_1 compared with 225 078 µm2 for C2). This size increase was not seen for iPSCs carrying the RUNX1 deletion (Figure 3D), showing that RUNX1 haploinsufficiency did not affect the proliferation rate of GM lineage. Accordingly, the generation of mature monocytes and/or macrophages (CD14highCD15low) and granulocytes (CD15high) in liquid medium (Figure 3E-F) revealed an increase in the proportion of GM populations carrying the R174Q mutation but not the RUNX1 deletion. A significant twofold increase in the monocyte and/or macrophage output and a slight increase in granulocyte generation were detected for both patient lines with the R174Q mutation, but not for the patient lines with a RUNX1 monoallelic deletion (Figure 3G-H). In accordance with our previous results,4 the expression of the NR4A3 gene was decreased in CD34+CD43+ progenitors sorted at day 14 from the 2 R174Q mutated iPSC clones, although no significant difference was observed between control and RUNX1 monoallelic deleted cells (Figure 3I). These results indicate that only the R174Q RUNX1 mutation leads to deregulation of the granulomonocytic compartment, which correlates with the downregulation of NR4A3.
RUNX1 R174Q mutation but not RUNX1 deletion affects the output of granulomonocytes. AII_1, AII_2 are iPSC clones of 2 patients from pedigree A; D(a) and D(b) are 2 clones of 1 patient from pedigree D; and C1, C2 and C3 are control iPS cell lines. Cell numbers were normalized in all experiments to those produced by the C2 clone. (A-D) CD34+CD43+ cells were sorted at day 14 and tested for their colony-forming potential in a methylcellulose assay. The histograms show (A) 1 representative experiment of 3 for pedigree A and (B) 1 representative experiment of 7 for pedigree D, each in triplicate. Error bars represent ± SD of triplicate. (C) Photos of granulomonocytic colonies (CFU-GM). Scale bar = 100 μm. (D) Area of more than 30 CFU-GM colonies obtained from each iPSC line was analyzed by AxioVisio 4.6 software. (E-H) Analysis of hematopoietic populations generated at day 21 from progenitors sorted at day 14. (E) May-Grünwald-Giemsa staining of monocytes and/or macrophages (CD14highCD15low) and granulocytes (CD15high) derived from iPSCs. Scale bar = 10 μm. (F) FACS analysis of the GM cells in 1 representative experiment. (G-H) Absolute numbers of generated monocytes and/or macrophages (CD14highCD15low) and granulocytes (CD15high) normalized to 1 × 102 plated progenitors. (G) The histograms show average of the cell number obtained in 5 independent experiments for monocytes and/or macrophages and in 3 for granulocytes for pedigree A. (H) The histograms show average of the cell number obtained in 4 independent experiments for pedigree D. Error bars represent ± SD of the average. (I) qRT-PCR analysis of NR4A3 expression level in CD34+CD43+ cells in both A (left; n = 4) and D (right; n = 4) pedigrees. Data are normalized to PPIA transcript, and expression is compared with control C3. Error bars represent ± SD of triplicate. *P < .05; **P < .01; ***P < .001; Student t test. ns, nonsignificant.
RUNX1 R174Q mutation but not RUNX1 deletion affects the output of granulomonocytes. AII_1, AII_2 are iPSC clones of 2 patients from pedigree A; D(a) and D(b) are 2 clones of 1 patient from pedigree D; and C1, C2 and C3 are control iPS cell lines. Cell numbers were normalized in all experiments to those produced by the C2 clone. (A-D) CD34+CD43+ cells were sorted at day 14 and tested for their colony-forming potential in a methylcellulose assay. The histograms show (A) 1 representative experiment of 3 for pedigree A and (B) 1 representative experiment of 7 for pedigree D, each in triplicate. Error bars represent ± SD of triplicate. (C) Photos of granulomonocytic colonies (CFU-GM). Scale bar = 100 μm. (D) Area of more than 30 CFU-GM colonies obtained from each iPSC line was analyzed by AxioVisio 4.6 software. (E-H) Analysis of hematopoietic populations generated at day 21 from progenitors sorted at day 14. (E) May-Grünwald-Giemsa staining of monocytes and/or macrophages (CD14highCD15low) and granulocytes (CD15high) derived from iPSCs. Scale bar = 10 μm. (F) FACS analysis of the GM cells in 1 representative experiment. (G-H) Absolute numbers of generated monocytes and/or macrophages (CD14highCD15low) and granulocytes (CD15high) normalized to 1 × 102 plated progenitors. (G) The histograms show average of the cell number obtained in 5 independent experiments for monocytes and/or macrophages and in 3 for granulocytes for pedigree A. (H) The histograms show average of the cell number obtained in 4 independent experiments for pedigree D. Error bars represent ± SD of the average. (I) qRT-PCR analysis of NR4A3 expression level in CD34+CD43+ cells in both A (left; n = 4) and D (right; n = 4) pedigrees. Data are normalized to PPIA transcript, and expression is compared with control C3. Error bars represent ± SD of triplicate. *P < .05; **P < .01; ***P < .001; Student t test. ns, nonsignificant.
RUNX1 transgene targeting corrects hematopoietic defects independently of the type of mutation
To exclude a role of iPSC line heterogeneity in the above-reported observation, we reintroduced a WT copy of the hemagglutinin (HA)-tagged RUNX1 gene into one iPSC line of each pedigree and used zinc-finger nucleases to place this gene under control of CD43 gene promoter into the “safe harbor” locus AAVS1 (Figure 4A).27 Clones with homozygous integration were selected, and the overexpression of HA-RUNX1 transcripts in CD34+CD43+ progenitors was checked (Figure 4B). In R174Q mutant cells, overexpression of RUNX1 was further verified by western blotting (Figure 4C) and by measuring the expression of 2 direct RUNX1 targets, p19INK4d and MPL (Figure 4D). Reintroduction of WT RUNX1 induced a threefold increase in the number of MK progenitors for the deletion and a twofold increase for the R174Q mutant in comparison with the parent iPSC lines (Figure 4E). The gene correction also increased the number of erythroid progenitors produced by the FPD/AML iPSC lines by twofold for the deletion and by more than fivefold for the R174Q mutation compared with the parent iPSC lines (Figure 4F). Interestingly, the number of GM progenitors decreased to control levels in the transgene-targeted R174Q iPSC line, demonstrating reversal of the phenotype (Figure 4G).
Genetic rescue by RUNX1 WT overexpression in FPD/AML iPSC lines. AII_1 represents iPSC clones of 1 patient from pedigree A; D(b) represents 1 clone of 1 patient from pedigree D; AII_1_RUNX1 and D(b)_RUNX1 are the iPSC clones overexpressing WT RUNX1; and C1, C2, and C3 are control iPSCs. (A) Gene correction strategy. The constitutively active AAVS1 “safe harbor” locus is shown on the top line and the targeting construct is shown below. Complementary DNA (cDNA) expression cassette driving expression of WT RUNX1 cDNA under the leukosialin promoter (pCD43) was inserted by zinc finger-mediated homologous recombination into intron 1 of AAVS1. HA: homologous arms left (L) and right (R); SA-2A-Puro-PA: puromycin drug resistance cassette. (B) Overexpression of HA-RUNX1 measured by qRT-PCR using forward primer in HA epitope and reverse primer in RUNX1 cDNA. Error bars represent 95% confidence intervals (CIs). (C) Western blot analysis of total (endogenous and overexpressed) RUNX1. (D) qRT-PCR analysis of MPL and P19INK4D expression level in MKs (CD41+CD42+) sorted at day 14. Data are normalized to PPIA transcript level, and expression is compared with control C3 (n = 3). (E-G) Cell numbers were normalized in all experiments to those produced by the C2 clone. (E-F) CD34+CD43+ cells were sorted at day 11 and tested for their colony-forming potential. (E) Assessment of MK progenitors in fibrin clot cultures (n = 3) (F) Assessment of erythroid progenitors in methylcellulose culture (n = 3) (G) CD34+CD43+ cells of pedigree A were sorted at day 14 and tested for their CFU-GM colony-forming potential in methylcellulose assay (n = 3). (D-G) Error bars represent ± SD of triplicate. *P < .05; **P < .01; ***P < .001; Student t test.
Genetic rescue by RUNX1 WT overexpression in FPD/AML iPSC lines. AII_1 represents iPSC clones of 1 patient from pedigree A; D(b) represents 1 clone of 1 patient from pedigree D; AII_1_RUNX1 and D(b)_RUNX1 are the iPSC clones overexpressing WT RUNX1; and C1, C2, and C3 are control iPSCs. (A) Gene correction strategy. The constitutively active AAVS1 “safe harbor” locus is shown on the top line and the targeting construct is shown below. Complementary DNA (cDNA) expression cassette driving expression of WT RUNX1 cDNA under the leukosialin promoter (pCD43) was inserted by zinc finger-mediated homologous recombination into intron 1 of AAVS1. HA: homologous arms left (L) and right (R); SA-2A-Puro-PA: puromycin drug resistance cassette. (B) Overexpression of HA-RUNX1 measured by qRT-PCR using forward primer in HA epitope and reverse primer in RUNX1 cDNA. Error bars represent 95% confidence intervals (CIs). (C) Western blot analysis of total (endogenous and overexpressed) RUNX1. (D) qRT-PCR analysis of MPL and P19INK4D expression level in MKs (CD41+CD42+) sorted at day 14. Data are normalized to PPIA transcript level, and expression is compared with control C3 (n = 3). (E-G) Cell numbers were normalized in all experiments to those produced by the C2 clone. (E-F) CD34+CD43+ cells were sorted at day 11 and tested for their colony-forming potential. (E) Assessment of MK progenitors in fibrin clot cultures (n = 3) (F) Assessment of erythroid progenitors in methylcellulose culture (n = 3) (G) CD34+CD43+ cells of pedigree A were sorted at day 14 and tested for their CFU-GM colony-forming potential in methylcellulose assay (n = 3). (D-G) Error bars represent ± SD of triplicate. *P < .05; **P < .01; ***P < .001; Student t test.
Downregulation of RUNX1 in the H9 ESC line reproduced the dominant-negative phenotype
To determine whether the R174Q mutant has acquired a new function or leads to a loss of function, we transduced undifferentiated H9 ESCs with RUNX1 short hairpin RNA (shRNA) or control shRNA constructs. The use of shRUNX1_1 almost completely abolished the expression of RUNX1 at the protein level in the mesodermal cell population (Figure 5A). First we confirmed the central role of RUNX1 in hematopoiesis induction by showing a decrease in the generation of hematopoietic clones after RUNX1 knockdown28 (supplemental Figure 5). Then, we assessed the colony-forming potential of CD34+CD43+ cells sorted at day 11, which demonstrated a more than 10-fold decrease in the number of erythrocyte progenitors (Figure 5B) and a threefold decrease in MK progenitors after RUNX1 knockdown (Figure 5C). In liquid medium, RUNX1 downregulation led to a decreased production of GPA+ erythroid cells and CD41+CD42+ MKs at day 18 (Figure 5D-E). Next, we performed the same experiments with CD34+CD43+ cells sorted at day 14 and observed an increased number of CFU-GM colonies after RUNX1 knockdown (Figure 5F) and an increased number of CD14highCD15low mature monocytes and/or macrophages and CD15high granulocyte populations at day 21 in liquid medium (Figure 5G). These results were validated by using a second shRNA, shRUNX1_2 (supplemental Figure 6). To further confirm that a decrease in RUNX1 activity is crucial for the amplification of GM progenitors, we transduced the iPSC clone D(b) carrying a monoallelic RUNX1 deletion with the shRUNX1_1. The additional decrease in RUNX1 level led to an increased number of CFU-GM colonies (Figure 5H).
RUNX1 knockdown in H9 ESC line and in an FPD/AML iPSC line with monoallelic RUNX1 deletion leads to the same phenotype as the FPD/AML-DN iPSC line. (A-G) RUNX1 knockdown in H9 ESC line. (A) Analysis of RUNX1 protein expression in mesodermal population from control and shRUNX1_1 ESC lines. (B-C) CD34+CD43+ cells were sorted at day 11 and tested for their colony-forming potential in methylcellulose assay and fibrin clot culture. (B) Assessment of erythroid progenitors in methylcellulose culture. (C) Assessment of MK progenitors in fibrin clot cultures. (B-C) The histograms show 1 representative experiment of 4 for erythroid progenitors and of 3 for MK progenitors, each in triplicate. Error bars represent ± SD of triplicate. (D-E) Analysis of hematopoietic populations generated at day 18 from progenitors sorted at day 11. (D) Absolute number of generated erythroid cells (GPA+) normalized to 1 × 102 plated progenitors (n = 4). (E) Absolute number of generated MK cells (CD41+CD42+) normalized to 1 × 102 plated progenitors (n = 4). (D-E) The histograms show the average of 4 independent experiments. Error bars represent ± SD of average. (F) CD34+CD43+ cells were sorted at day 14 and were tested for their CFU-GM colony-forming potential in methylcellulose assay. The histograms show 1 representative experiment of 4, each in triplicate. Error bars represent ± SD of triplicate. (G) Analysis of hematopoietic populations generated at day 21 from progenitors sorted at day 14. Absolute numbers of generated monocytes and/or macrophages (CD14highCD15low; n = 4) and granulocytes (CD15high; n = 4) normalized to 1 × 102 plated progenitors. The histograms show the average of 4 experiments. Error bars represent ± SD of average. (H) RUNX1 knockdown in D(a) iPSC line with shRUNX1_1. CD34+CD43+ cells were sorted at day 14 and tested for their CFU-GM colony-forming potential in methylcellulose assay. The histograms show 1 representative experiment of 3, each in triplicate. Error bars represent ± SD of triplicate. (I-J) Transcriptome analysis of CD34+CD43+ progenitors derived from shControl and shRUNX1_1 transduced hESC line sorted at day 14 of culture. FC: fold change between cells transduced with shRUNX1_1 and shControl lentiviruses. Only genes with expression variation ± 1.5 and P < .001 (analysis of variance [ANOVA] test) are listed. (J) Variation in expression of genes involved in p53-dependent DNA damage response; P < .001 (ANOVA test). E, erythroid; G/M, granulo-monocyte; I, immunity; IN, inflammation; MK, megakaryocyte. *P < .05; **P < .01; ***P < .001, Student t test.
RUNX1 knockdown in H9 ESC line and in an FPD/AML iPSC line with monoallelic RUNX1 deletion leads to the same phenotype as the FPD/AML-DN iPSC line. (A-G) RUNX1 knockdown in H9 ESC line. (A) Analysis of RUNX1 protein expression in mesodermal population from control and shRUNX1_1 ESC lines. (B-C) CD34+CD43+ cells were sorted at day 11 and tested for their colony-forming potential in methylcellulose assay and fibrin clot culture. (B) Assessment of erythroid progenitors in methylcellulose culture. (C) Assessment of MK progenitors in fibrin clot cultures. (B-C) The histograms show 1 representative experiment of 4 for erythroid progenitors and of 3 for MK progenitors, each in triplicate. Error bars represent ± SD of triplicate. (D-E) Analysis of hematopoietic populations generated at day 18 from progenitors sorted at day 11. (D) Absolute number of generated erythroid cells (GPA+) normalized to 1 × 102 plated progenitors (n = 4). (E) Absolute number of generated MK cells (CD41+CD42+) normalized to 1 × 102 plated progenitors (n = 4). (D-E) The histograms show the average of 4 independent experiments. Error bars represent ± SD of average. (F) CD34+CD43+ cells were sorted at day 14 and were tested for their CFU-GM colony-forming potential in methylcellulose assay. The histograms show 1 representative experiment of 4, each in triplicate. Error bars represent ± SD of triplicate. (G) Analysis of hematopoietic populations generated at day 21 from progenitors sorted at day 14. Absolute numbers of generated monocytes and/or macrophages (CD14highCD15low; n = 4) and granulocytes (CD15high; n = 4) normalized to 1 × 102 plated progenitors. The histograms show the average of 4 experiments. Error bars represent ± SD of average. (H) RUNX1 knockdown in D(a) iPSC line with shRUNX1_1. CD34+CD43+ cells were sorted at day 14 and tested for their CFU-GM colony-forming potential in methylcellulose assay. The histograms show 1 representative experiment of 3, each in triplicate. Error bars represent ± SD of triplicate. (I-J) Transcriptome analysis of CD34+CD43+ progenitors derived from shControl and shRUNX1_1 transduced hESC line sorted at day 14 of culture. FC: fold change between cells transduced with shRUNX1_1 and shControl lentiviruses. Only genes with expression variation ± 1.5 and P < .001 (analysis of variance [ANOVA] test) are listed. (J) Variation in expression of genes involved in p53-dependent DNA damage response; P < .001 (ANOVA test). E, erythroid; G/M, granulo-monocyte; I, immunity; IN, inflammation; MK, megakaryocyte. *P < .05; **P < .01; ***P < .001, Student t test.
Altogether, these results suggest that the differences between R174Q mutation and a monoallelic RUNX1 deletion can be explained by RUNX1 dosage. An almost complete functional inhibition or deletion, but not a haploinsufficiency, leads to the deregulation of the GM compartment, which may contribute to leukemic development. Gene expression was then explored on hematopoietic CD34+CD43+ progenitors derived from shRUNX1_1 and shControl transduced hESCs and sorted at day14 of culture. On the 44 000 tested probe sets, 412 probe sets were deregulated at least 1.5-fold in shRUNX1_1 hESCs, with 327 probe sets decreased and 85 probe sets increased. A focus on hematopoietic genes (Figure 5I) identified 22 erythroid and 11 MK genes deregulated. In the GM compartment, we highlighted a global increase of the GM signature with an increase of 11 genes. These results confirmed that RUNX1 plays an important role in the commitment between erythrocyte/MK and GM lineages.
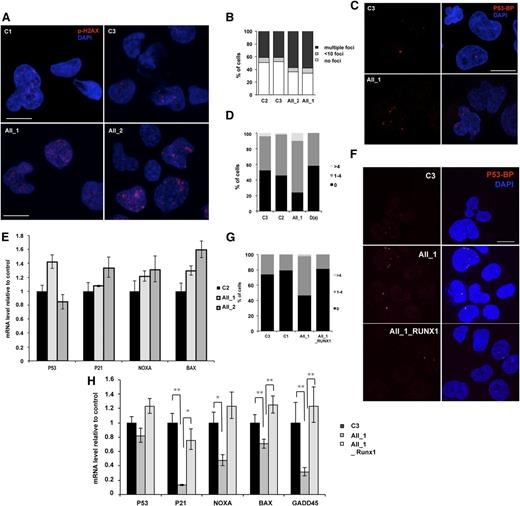
R174Q RUNX1 mutation but not haploinsufficiency leads to an increase in genomic instability
Gene expression analysis also demonstrated a slight decrease in 4 p53-dependent genes involved in DNA-damage response (Figure 5J). To explore the role of RUNX1 in genomic instability, we first sorted the CD43+ hematopoietic cells at day 10 of culture from 2 FPD/AML iPSC lines with R174Q mutation and control iPSC lines. The accumulation of DNA double-strain breaks was investigated by using an antibody recognizing the phospho-histone H2AX (P-H2AX). As shown in Figure 6A-B, an increase in the number of P-H2AX–positive cells was observed in CD43+ cells harboring the R174Q mutation compared with control cells, (from 50% for controls to about 70% for FPD/AML cells) as well as in the number of foci per cell. Because replication can induce P-H2AX foci formation, more particularly in highly proliferative erythroid cells that represent the majority of CD43+ cells, we used P53BP1 as a marker of double-strain breaks. The number of cells presenting P53BP1 foci in CD34+CD43+ progenitors was increased in the population of R174Q mutant cells (from 50% in control C3 and C2 cells to 80% in R174Q mutant cells) (Figure 6C-D), whereas no increase was detected for cells with RUNX1 deletion (Figure 6D). Then we evaluated the p53-dependent DNA-damage response pathway. In the entire CD34+CD43+ cell population, we did not detect any decrease in p21, NOXA, and BAX expression, whatever the RUNX1 alteration (Figure 6E). Because the R174Q mutant cell line is characterized by an expansion of the GM lineage, we focused on this population. As shown in Figure 6F-G, an increase in the number of CD14+CD15+ cells positive for P53BP1 foci (from 26% for control cells to 53%) and in the number of foci per cell was detected for R174Q mutant cells. These results were confirmed by qRT-PCR, showing a strong decrease of p21 and GADD45A and, to a lesser extent, NOXA and BAX gene expression in the CD14+CD15+ cell population (Figure 6H). Importantly, the number of cells showing P53BP1 foci was decreased to control levels. and the expression of all 4 genes was restored in FPD/AML iPSCs transduced with WT RUNX1 (Figure 6G-H). Altogether, our results demonstrate that R174Q RUNX1 mutation could predispose to leukemic transformation at least partly through NR4A3 downregulation and increased genomic instability in the GM compartment, which is not observed with cells harboring a RUNX1 haploinsufficiency. Moreover, the additional decrease of RUNX1 activity in haploinsufficient iPS cells by the shRUNX1_1 led not only to an increase in CFU-GM progenitor number but also to a decrease in p21 and GADD45A expression in the CD14+CD15+ cell population (supplemental Figure 7), attesting to the importance of the RUNX1 level in this phenotype.
RUNX1 DN-like mutant induces increased genomic instability compared with RUNX1 haploinsufficiency. AII_1 and AII_2 are iPSC clones of 2 patients from pedigree A; D(a) is 1 clone of 1 patient from pedigree D; AII_1_RUNX1 is the iPSC clone AII_1 overexpressing WT RUNX1; D(a)_shRUNX1_1 is the iPSC clone D(a) after RUNX1 knockdown by shRUNX1_1; and C2 and C3 are control iPSC lines. (A-B) CD43+ cells and (C-E) CD34+CD43+ cells were sorted at day 14 of culture. (A,C) Representative photos are shown; nucleus is stained with 4,6 diamidino-2-phenylindole (DAPI; blue) and double-strain breaks (DSBs) are stained (A) with P-H2AX (red) or (C) with p53-BP1 (red). The number of (B) P-H2AX and (D) P53-BP1 foci per 50 counted cells is shown. One representative experiment of 4 is shown (P < .0001 for the increase in P-H2AX–positive cells; P = .0164 for the increase in P53BP1-positive cells in pedigree A). (E) qRT-PCR analysis of different p53-dependent genes in CD34+CD43+ cells. Data are normalized to PPIA transcript level and expression is compared with control C2. The histograms show 1 representative experiment of 3, each in triplicate. Error bars represent 95% CIs. (F-H) CD14+CD15+ cells were sorted at day 15 of culture. (F) Representative photos are shown; nucleus is stained with DAPI (blue) and DSBs are stained with P53BP1 (red). (G) The number of P53BP1 foci per 100 counted cells is shown. One representative experiment of 3 is shown (P = .0017 for the increase in P53BP1-positive cells in AII_1). (H) qRT-PCR analysis of different p53-dependent genes in CD14+CD15+ cells. Data are normalized to PPIA transcript level and expression is compared with control C3. The histograms show 1 representative experiment of 3, each in triplicate. Error bars represent 95% CIs. *P < .05; **P < .01; ***P < .001; Student t test.
RUNX1 DN-like mutant induces increased genomic instability compared with RUNX1 haploinsufficiency. AII_1 and AII_2 are iPSC clones of 2 patients from pedigree A; D(a) is 1 clone of 1 patient from pedigree D; AII_1_RUNX1 is the iPSC clone AII_1 overexpressing WT RUNX1; D(a)_shRUNX1_1 is the iPSC clone D(a) after RUNX1 knockdown by shRUNX1_1; and C2 and C3 are control iPSC lines. (A-B) CD43+ cells and (C-E) CD34+CD43+ cells were sorted at day 14 of culture. (A,C) Representative photos are shown; nucleus is stained with 4,6 diamidino-2-phenylindole (DAPI; blue) and double-strain breaks (DSBs) are stained (A) with P-H2AX (red) or (C) with p53-BP1 (red). The number of (B) P-H2AX and (D) P53-BP1 foci per 50 counted cells is shown. One representative experiment of 4 is shown (P < .0001 for the increase in P-H2AX–positive cells; P = .0164 for the increase in P53BP1-positive cells in pedigree A). (E) qRT-PCR analysis of different p53-dependent genes in CD34+CD43+ cells. Data are normalized to PPIA transcript level and expression is compared with control C2. The histograms show 1 representative experiment of 3, each in triplicate. Error bars represent 95% CIs. (F-H) CD14+CD15+ cells were sorted at day 15 of culture. (F) Representative photos are shown; nucleus is stained with DAPI (blue) and DSBs are stained with P53BP1 (red). (G) The number of P53BP1 foci per 100 counted cells is shown. One representative experiment of 3 is shown (P = .0017 for the increase in P53BP1-positive cells in AII_1). (H) qRT-PCR analysis of different p53-dependent genes in CD14+CD15+ cells. Data are normalized to PPIA transcript level and expression is compared with control C3. The histograms show 1 representative experiment of 3, each in triplicate. Error bars represent 95% CIs. *P < .05; **P < .01; ***P < .001; Student t test.
Discussion
The current approaches used to explore FPD/AML pathogenesis remain limited, and access to patient primary cells is difficult because of the rarity of the disease. In addition, mouse models of Runx1 alterations do not completely reflect the phenotype observed in FPD/AML patients (eg, adult mice with a conditional Runx1 KO develop myeloproliferative syndrome/MDS rather than acute leukemia.15 Thus, to further explore the mechanisms of dysmegakaryopoiesis and sensitivity to leukemic transformation related to diverse RUNX1 alterations, we generated iPSC lines from 2 previously studied pedigrees: 1 with a DN-like mutation R174Q and 1 with a RUNX1 monoallelic deletion. The 2 pedigrees are both associated with thrombocytopenia, whereas only the first pedigree is associated with AML.
The generation of hematopoietic progenitors during embryonic life occurs initially in large blood vessels by the formation of clusters derived from endothelial cells, a process that strikingly relies on the presence of RUNX1.28,29 The number of human hematopoietic progenitors generated from FPD/AML iPSCs depends on the type of RUNX1 alteration.30 Here, we show that the knockdown of RUNX1 in ESCs limits the commitment to hematopoietic lineage at the clonal level.
The hematopoietic potential of different ESC31 and iPSC lines can be very heterogeneous32 ; therefore, the analysis of hematopoietic lineages obtained from iPSCs was normalized to the number of initial hematopoietic progenitors and not of iPSCs. To isolate myeloid hematopoietic progenitors, the CD34 marker was used in combination with CD43 (leukosialin), which is an early and specific hematopoietic marker appearing before CD45 during development.33
In accordance with previously reported data,34 we detected 2 discrete waves of primitive hematopoiesis with an increased capacity of the first CD34+CD43+ cells to generate primitive erythroid and MK progenitors compared with the CD34+CD43+ cells occurring later that demonstrate a granulomonocytic potential. By analyzing the clonogenic potential of hematopoietic CD34+CD43+ progenitors derived from FPD/AML iPSC clones, we observed a profound defect in the first wave giving rise to primitive erythroid and MK cells as well as in proplatelet formation from mature MKs, which is consistent with the dysmegakaryopoiesis observed in FPD/AML patients.25 In addition, the same deregulation of myosin expression was found in MKs derived from both iPSC and adult CD34+ cells,25 confirming the validity of the iPSC model. These defects were independent of the type of RUNX1 alteration (DN-like mutant or haploinsufficiency) and were partially restored after RUNX1 overexpression driven by the CD43 promoter in FPD/AML iPSC lines. Consistent with our results, defects in MK differentiation from iPSC lines derived from FPD/AML patients with Y260X,35 G172E, G143W, and N233fsX28330 RUNX1 mutations were also reported.
Regarding the second wave, DN-like R174Q RUNX1 mutation increased the proliferation rate and clonogenic potential of GM progenitors, which correlated with a decrease in NR4A3 expression. Because a similar phenotype was also observed after almost complete RUNX1 knockdown in hESCs, we can conclude that R174Q mutant is close to a complete loss of function. Conversely, the half decrease in RUNX1 dosage resulting from RUNX1 deletion did not affect the GM lineages and NR4A3 expression and further attest that DN-like R174Q mutant could contribute to the induction of a preleukemic state by increasing the GM progenitor compartment, the main cell target for AML. Even if our differentiation protocol allows us to study only primitive hematopoiesis, these results are in agreement with observations made in adult progenitors from FPD/AML patients harboring the R174Q mutation,4 in which the increase in the GM compartment has been associated with NR4A3 downregulation by RUNX1. Furthermore, the importance of NR4A3 dosage was already highlighted in the induction of myeloproliferative neoplasms and AML in NR4A3 KO mouse models.36-38 Contrary to our results showing the amplification of GM lineage after almost complete knockdown of RUNX1 or with DN-like R174Q mutation, this amplification was not observed with N233fsX283 mutation,30 which is highly associated with AML. However, this last mutation induces a deep defect in the generation of hematopoietic progenitors, which could in some way also be a preleukemic state similar to that in some aplastic anemias.
To the best of our knowledge, this is the first study that clarifies the differences in granulomonocytic lineage between a haploinsufficiency alone and the DN-like R174Q RUNX1 mutant. These differences are clearly dependent on the protein dosage. The R174Q mutant behaving as a DN may preclude the formation of active core-binding complexes. This hypothesis becomes even more plausible when we take the following into account: (1) the report that certain RUNX1 mutants, including R174Q, bind CBFβ more strongly than the WT,3 and mouse models of CBFβ deficiency recapitulate the phenotype observed with this RUNX1 mutant39 ; (2) the results obtained with the 2 shRUNX1 in hESCs in which abrogation of RUNX1 leads to a phenotype close to R174Q; and (3) the rescue of DN phenotype by overexpression of WT RUNX1 probably by changing the equilibrium of WT vs mutant binding to CBFβ. A recent study highlighted the complexity of RUNX1 dosage by reporting that a minimal dosage is necessary for MLL fusion leukemia.40 Here, we show that a very low level of RUNX1 increases the proliferative rate and genomic instability of GM progenitors that could be at the origin of the preleukemic state in patients, whereas a half-decrease in RUNX1 level leads to thrombocytopenia alone. However, it should be noted that leukemia development was described in 1 pedigree with hemizygous RUNX1 deletion,1 attesting that the simple dichotomy of DN vs haploinsufficient mutant for AML development is not so clear and that other genetic events could also contribute to the development of leukemia. However, our results are consistent with the importance of the transcription factor level in malignant disorders. This is underscored by the example of PU.1, which needs to be decreased to less than 80% for inducing AML41 whereas a haploinsufficiency has no effect.42,43
Finally, we demonstrate that a RUNX1 loss of function leads to an increase in genomic instability linked to a decreased expression of p53-dependent genes and to an increase in the number of P53BP1 foci. Moreover, in the presence of the R174Q mutant, 10-fold more mutations occurred during the reprogramming process as compared with monoallelic RUNX1 deletion and control cell lines.44 Consistent with these results, a novel role for RUNX1 during DNA-damage response has been recently identified. Indeed, it cooperates with p53 after DNA damage and is recruited onto p53 target gene promoters.45 In addition, a decreased expression of the direct RUNX1 target GADD45A, encoding for a sensor of DNA stress, was observed in bone marrow cells from MDS/AML patients harboring a RUNX1 mutation in the C-terminal domain.19 GADD45A was shown to be activated synergistically by RUNX1 and p53. Interestingly, Michaud et al46 previously reported a global genomic instability linked to RUNX1 impairment, and the incidence of RUNX1 mutations in therapy-related MDS and AML is particularly high, revealing again a potential role of RUNX1 in genomic instability.47
In conclusion, whereas analysis of mouse models failed to identify any defect in primitive hematopoiesis, we show for the first time that RUNX1 germline alterations affect human embryonic hematopoiesis, particularly the generation of primitive erythroid and MK cells. As already described by others,30,35 defects in the generation of mature MK cells do not depend on the type of RUNX1 variants. Conversely, a DN R174Q mutant of RUNX1 increases the proliferative rate and induces a genomic instability of GM progenitors, thereby contributing to development of a preleukemic state.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank P. Rameau and Y. Lécluse for flow cytometry cell sorting and analysis (Integrated Research Cancer Institute in Villejuif, Gustave Roussy, Villejuif, France), Dr G. Meurice for the comparative genome hybridization analysis (Plateforme de Bioinformatique, Gustave Roussy, Villejuif, France), Dr P. Opolon for the teratomas analysis (Plateforme d’ Histopathologie, Gustave Roussy, Villejuif, France), P. Nusbaum and A. Hubas for patient fibroblast culture (Banque de Cellules directed by Pr. J. Chelly, Hôpital Cochin, Paris, France), Pr. Jonveaux (Centre Hospitalier Universitaire de Nancy, Nancy, France) for his advice, Dr A. Foudi and Dr G. Mostoslavsky (Boston University and Boston Medical Center, Boston, MA) for providing lentiviral vector STEMCCA, Drs A. Galy and S. Charrier for providing lentiviral vector pRRLcpptPGKGFP-WPRE-Alb (Genethon, Evry, France), and the patients for participating in this study.
This work was supported by grants from the Agence Nationale de la Recherche (ANR-physiopathology, ANR-jeunes chercheurs) (H.R.), the Centre de Référence des Pathologies Plaquettaires, and the Association pour la Recherche sur le Cancer (projet libre, I.P. 2012 and W.V. 2009). The Comparative Genome Hybridization array was funded by the apprenticeship tax (TA2011). Labex Globule Rouge-Excellence (I.P., W.V.) was funded by the “Investissements d’avenir” program. I.A.-D. was supported by a doctoral fellowship from the Association pour la Recherche sur le Cancer. N.D. and W.V. are recipients of a research fellowship from Centre Hospitalier Universitaire Bordeaux (N.D.) and from the Institute Gustave Roussy-INSERM (W.V.).
Authorship
Contribution: I.A.-D., V.T.M., W.V., and H.R. designed experiments; I.A.-D., V.T.M., N.B., D.B., C.T., C.L., T.L., O.B., L.T., and J.A.M. performed experiments; I.A.-D., V.T.M., L.T., G.T., and H.R. analyzed data; B.L. and R.F. provided clinical and biological follow-up of patients; N.D., I.P., E.S., R.F., D.L.F., M.J.W., and W.V. discussed the techniques and results; H.R. supervised the work; and I.A.-D., V.T.M., W.V., and H.R. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hana Raslova, INSERM UMR1009, Institut Gustave Roussy, 114 rue Edouard Vaillant, 94805 Villejuif Cedex, France; e-mail: hraslova@igr.fr.
References
Author notes
I.A.-D. and V.T.M. contributed equally to this study.




![Figure 5. RUNX1 knockdown in H9 ESC line and in an FPD/AML iPSC line with monoallelic RUNX1 deletion leads to the same phenotype as the FPD/AML-DN iPSC line. (A-G) RUNX1 knockdown in H9 ESC line. (A) Analysis of RUNX1 protein expression in mesodermal population from control and shRUNX1_1 ESC lines. (B-C) CD34+CD43+ cells were sorted at day 11 and tested for their colony-forming potential in methylcellulose assay and fibrin clot culture. (B) Assessment of erythroid progenitors in methylcellulose culture. (C) Assessment of MK progenitors in fibrin clot cultures. (B-C) The histograms show 1 representative experiment of 4 for erythroid progenitors and of 3 for MK progenitors, each in triplicate. Error bars represent ± SD of triplicate. (D-E) Analysis of hematopoietic populations generated at day 18 from progenitors sorted at day 11. (D) Absolute number of generated erythroid cells (GPA+) normalized to 1 × 102 plated progenitors (n = 4). (E) Absolute number of generated MK cells (CD41+CD42+) normalized to 1 × 102 plated progenitors (n = 4). (D-E) The histograms show the average of 4 independent experiments. Error bars represent ± SD of average. (F) CD34+CD43+ cells were sorted at day 14 and were tested for their CFU-GM colony-forming potential in methylcellulose assay. The histograms show 1 representative experiment of 4, each in triplicate. Error bars represent ± SD of triplicate. (G) Analysis of hematopoietic populations generated at day 21 from progenitors sorted at day 14. Absolute numbers of generated monocytes and/or macrophages (CD14highCD15low; n = 4) and granulocytes (CD15high; n = 4) normalized to 1 × 102 plated progenitors. The histograms show the average of 4 experiments. Error bars represent ± SD of average. (H) RUNX1 knockdown in D(a) iPSC line with shRUNX1_1. CD34+CD43+ cells were sorted at day 14 and tested for their CFU-GM colony-forming potential in methylcellulose assay. The histograms show 1 representative experiment of 3, each in triplicate. Error bars represent ± SD of triplicate. (I-J) Transcriptome analysis of CD34+CD43+ progenitors derived from shControl and shRUNX1_1 transduced hESC line sorted at day 14 of culture. FC: fold change between cells transduced with shRUNX1_1 and shControl lentiviruses. Only genes with expression variation ± 1.5 and P < .001 (analysis of variance [ANOVA] test) are listed. (J) Variation in expression of genes involved in p53-dependent DNA damage response; P < .001 (ANOVA test). E, erythroid; G/M, granulo-monocyte; I, immunity; IN, inflammation; MK, megakaryocyte. *P < .05; **P < .01; ***P < .001, Student t test.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/6/10.1182_blood-2014-06-585513/4/m_930f5.jpeg?Expires=1769093671&Signature=i5~jKV-Oq3-rcZApBqkIOMRTDqQ5vN8uL9Bno1CUQ5rhDOsbOaGXCxlWmVvnPQf-TxgNbpcdmDzfvX~vQSNb6bNQ3j0HZSUoZrBdQcH0MtElhlTvgC4y~2ipfO3EqnQvot06qCqQcb5ZUEOUrlk3bbyV4wioCVuD~FzU-gfYimXlw7CfR7mR6IgP2qTE7dHYNxjPjFucKoJfRb6eVAXju3T87JeChJm7I268EpEGaZXpyLGJjIjGkcAJ86s7dkKN2~dTDC2EiUn4vljvGNy6Bzt8udJwsGvelf6ngLNVHiZ0wWHD5Pyk8J94MF7lWEjYNeJDpDsNfFFuFJ9iwt0kKw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal