Key Points
Dnmt3a ablation in HSCs predisposes mice to develop a spectrum of myeloid and lymphoid malignancies.
Dnmt3a-KO-derived myeloid malignancies and T-cell acute lymphocytic leukemia/lymphoma show distinct methylation aberrations.
Abstract
DNA methyltransferase 3A (DNMT3A) is mutated in hematologic malignancies affecting myeloid, mixed, and lymphoid lineages, and these mutations are associated with poor prognosis. Past studies in mice revealed Dnmt3a-knockout (KO) hematopoietic stem cells (HSCs) had increased self-renewal, but no leukemia was observed. Here, all lethally irradiated mice transplanted with Dnmt3a-deleted HSCs died within 1 year. Animals were diagnosed with a spectrum of malignancies similar to those seen in patients with DNMT3A mutations, including myelodysplastic syndrome, acute myeloid leukemia, primary myelofibrosis, and T- and B-cell acute lymphocytic leukemia. In some cases, acquired malignancies exhibited secondary mutations similar to those identified in patients. Loss of Dnmt3a led to disturbed methylation patterns that were distinct in lymphoid and myeloid disease, suggesting lineage-specific methylation aberrations promoted by Dnmt3a loss. Global hypomethylation was observed in all of the malignancies, but lymphoid malignancies also exhibited hypermethylation, particularly at promoter regions. This mouse model underscores the important role of Dnmt3a in normal hematopoietic development and demonstrates that Dnmt3a loss of function confers a preleukemic phenotype on murine HSCs. This model may serve as a tool to study DNMT3A mutation-associated malignancies and for developing targeted strategies for eliminating preleukemic cells for prevention and treatment of hematologic malignancies in the future.
Introduction
Since the initial reports of DNA methyltransferase 3A (DNMT3A) mutations in acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS) patients in 2010,1-3 mutations in DNMT3A have been reported frequently in hematologic malignancies of myeloid and lymphoid lineages.4-6 In AML, about 60% of patients exhibit heterozygous mutation at Arginine 882 (R882), which acts as a dominant negative, disrupting normal methylation function.7,8 The remaining patients often have biallelic involvement, with compound heterozygous mutations or loss of homozygosity. In T-cell acute lymphocytic leukemia/lymphoma (T-ALL), the R882 mutation is observed in 20% of patients with DNMT3A mutations, and about half of the remaining patients have biallelic mutations.6,9 Together, these observations suggest that DNMT3A functions as a classic tumor suppressor, where most or all of the protein function must be lost for malignancy development.
Mutation of DNMT3A has been found at high variant allele frequencies, suggesting that it is mutated in founding clones.10-12 In AML patients, DNMT3A mutations are also found in phenotypically normal hematopoietic stem cells (HSCs) that maintain multilineage differentiation capacity, suggesting that DNMT3A mutations can confer a preleukemic state.13,14 These preleukemic stem cells are clinically silent and are outcompeted by malignant cells during disease presentation,15 but preleukemic clones bearing DNMT3A mutations may survive treatment and expand during remission. The self-renewal capacity of preleukemic stem cells presumably allows for the acquisition of mutations that transform the preleukemic cells to malignant cells. These findings indicate that DNMT3A mutations arise early, predisposing cells to leukemia and enabling the selection of cells that have acquired additional mutations during transformation to leukemia.
That DNMT3A mutant HSCs in patients can maintain self-renewal capacity is consistent with observations that murine Dnmt3a-knockout (KO) HSCs appear phenotypically normal and exhibit increased self-renewal after transplant.16 In earlier experiments, purified HSCs were serially transplanted every 18 weeks along with wild-type (WT) bone marrow; leukemia was not observed, raising the possibility that Dnmt3a-KO HSCs would not serve as a model for human DNMT3A-associated malignancy development. Here, we sought to determine whether ablation of Dnmt3a in mice, in the absence of serial transplantation and with longer in vivo maintenance, could recapitulate the types of hematologic diseases observed in patients harboring DNMT3A mutations, despite the distinct mutation type (complete loss of function). Thus, we performed a long-term survival study to investigate the impact of loss of Dnmt3a on mouse HSCs, a strategy that allowed us to look in depth at the role of Dnmt3a in methylation patterns and mutation acquisition in hematologic diseases.
Materials and methods
Mice
Animal procedures were approved by the Institutional Animal Care and Use Committee and conducted in accordance with institutional guidelines. Dnmt3a-KO in C57Bl/6-CD45.2 Dnmt3afl/fl -Mx1-cre mice was induced by 6 intraperitoneal injections of polyinosinic-polycytidylic acid (300 μg per mouse in phosphate-buffered saline; Sigma) every other day. Bone marrow was harvested 8 weeks later for transplantation.
HSC transplantation
Femurs, tibiae, and iliac crests were obtained from donor mice, and bone marrow HSCs were purified using the Hoecsht 33342 side population17 combined with c-Kit magnetic enrichment and Sca1+ CD150+ and lineage− sorting (AutoMACS; Miltenyi Biotec; MoFlo [Beckman Coulter]; antibodies from Becton Dickinson or eBioscience). Cells were transplanted into C57Bl/6-CD45.1 recipients by retroorbital injection after 10.5-Gy split-dose irradiation.
Diagnosis/phenotype analysis
Mice were bled retroorbitally for complete blood counts (CBCs) and/or blood smears and flow cytometry analysis. CBCs were performed on a Hemavet 950 (Drew Scientific), and lineage analysis was performed as described previously.18 Additional immunophenotyping of hematopoietic organs was performed for diagnosis following the Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice and the Bethesda proposals for classification of lymphoid neoplasms in mice.19,20 See also supplemental Methods and supplemental Figure 1 on the Blood Web site for differential diagnosis criteria.
Histology
Fresh tissues were used for touch preparations (touch preps) or fixed for 24 hours in 10% formalin (Fischer Scientific), followed by overnight decalcification of bones in Richard-Allen Scientific Cal-Rite (Thermo Scientific), and processing in 70% ethanol. Tissues were stained with hematoxylin and eosin (Richard-Allen Scientific; Thermo Scientific), Wright-Giemsa (Hema 3 Stat Pack; Fisher Scientific), and reticulin stain (Chandler’s Precision; American MasterTech). Images were acquired with Olympus DP25 camera.
Sequencing
DNA was extracted with AllPrep Mini kit (Qiagen). Targeted sequencing of Notch1 used primers listed in supplemental Table 1, with M13F and M13R tags. Whole-exome capture and 50X sequencing was carried out by Otogenetics (Norcross, GA) and aligned to mm10 reference genome (GRCm38) using BWA-MEM 0.7.7.21 Polymerase chain reaction duplicate trimming was done by Picard Tools 1.84 (http://picard.sourceforge.net) and samtools.22 Further sequence processing (base quality score recalibration and indel realignment) adhered to best practice recommendations using Genome Analysis Toolkit v3.1.23 Discovery of somatic single nucleotide variants was performed using MuTect at the default setting,24 and indels detection was performed using FreeBayes variant caller.25 High-quality variants were filtered (genotype quality >20; minimum variant read count 4) and functionally annotated using SnpEff 3.6 GRCm38.74.26 Known variants from mouse dbSNP build138 were identified to arrive at novel somatic variants.
Retroviral transduction
Nras-G12D (Addgene 14725: pCGN N-Ras12D) was subcloned into MSCV-RFB-IRES-GFP. Plasmids were cotransfected with pCL-Eco into 293T cells to package virus. We collected supernatant 48 hours after transfection. Donor mice were treated with 5-fluorouracil (150 mg/kg; American Pharmaceutical Partners) 6 days before bone marrow harvest. Enrichment for Sca-1+ cells was performed on an autoMACS, and cells were adjusted to 5 × 105 cells/mL in medium containing StemPro-34 (Gibco), nutrient supplement, penicillin/streptomycin, glutamine (2 mM), stem cell factor (10 ng/mL; R&D Systems), mouse thromboprotein (100 ng/mL; R&D Systems) and polybrene (4 μg/mL; Sigma). Cells were spin-infected at 250g at room temperature for 2 hours, incubated for 1 hour at 37°C, then injected into recipients.
Reduced representation bisulfite sequencing (RRBS)
Illumina TruSeq libraries were generated from 500 ng of genomic DNA27 and 36 bp single-end sequenced on an Illumina HiSeq 2000. Adapter and base quality trimming was performed with Trim Galore. Reads were mapped (mm9) with BSMAP 2.74. Quantification and differential analyses were performed with methylKit 0.9.2.28 Methylation data were mapped to a 1-kb tiling region set, giving rise to 42 305 genomic regions for which quantitative data were available (supplemental Table 2). Chromatin and transcriptional regulator enrichment analysis of differentially methylated regions (DMRs) in peaks or binding sites was measured by Fisher’s exact test. Predictions of cis-regulatory function were performed with the GREAT 2.0 Web server. All other methylation data analyses were performed with the R statistics package.
Results
Mice transplanted with Dnmt3a-KO HSCs die prematurely
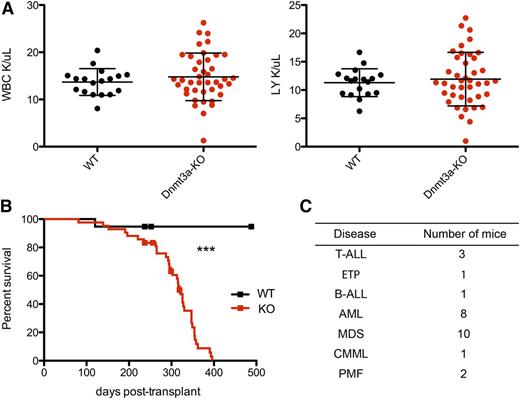
To test the long-term effects of loss of Dnmt3a in mouse HSCs, we deleted Dnmt3a by injecting Mx1-cre;Dnmt3af/f (Dnmt3a-KO) and Mx1-cre;Dnmt3a+/+ (WT) mice with polyinosinic-polycytidylic acid. Eight weeks later, we used these donors to establish a cohort of mice transplanted with either 500 Dnmt3a-KO or WT HSCs (side population c-Kit+ Sca-1+ lineage− CD150+), monitoring recipient peripheral blood by flow cytometry and CBCs every 1 to 2 months. Although there was never a significant difference in the average counts between groups, there was more variation in the total white blood cell (WBC) and lymphocyte counts in mice transplanted with Dnmt3a-KO HSCs than in the WT group, even at a relatively early time point when most of the mice were still alive (Figure 1A). Ultimately, all mice transplanted with Dnmt3a-KO HSCs appeared sick and became moribund, exhibiting blood counts outside of the expected range for C57Bl/6 mice (WBC: 16.7-19.7 × 103/μL, red blood cell: 9.08-9.86 × 106/μL, platelet: 656-976 × 103/μL). We observed a significantly shorter survival rate for mice transplanted with Dnmt3a-KO HSCs than mice receiving control HSCs, with a median survival of 321 days for Dnmt3a-KO and with more than half of the WT cohort still alive at day 482 (P < .0001) (Figure 1B). To identify malignancies in moribund mice, we performed necropsy, histology, and immunophenotyping of hematopoietic organs, diagnosing a range of myeloid and lymphoid diseases (Figure 1C), as detailed below.
Mice transplanted with Dnmt3a-KO HSCs have reduced survival. (A) WBC and lymphocyte (LY) count at 26 weeks posttransplant; n = 18 WT and n = 41 Dnmt3a-KO. Error bars represent mean plus standard deviation. (B) Kaplan-Meier survival curve of mice transplanted with WT (n = 20) or Dnmt3a-KO (n = 45) HSCs. Censored points (squares) represent 5 mice that were sacrificed for a nonhematopoietic phenotype. Median survival is 321 days for Dnmt3a-KO; undetermined for WT. The experiment was repeated with the same numbers of animals, with median survival times of 246 days for mice transplanted with Dnmt3a-KO HSCs and 467 days for those transplanted with WT. (C) Number of mice in the transplanted cohort that were definitively diagnosed with the indicated hematopoietic diseases. ***P < .0001. B-ALL, B-cell acute lymphocytic leukemia/lymphoma; CMML, chronic myelomonocytic leukemia; ETP, early thymic progenitor acute lymphoblastic leukemia; PMF, primary myelofibrosis.
Mice transplanted with Dnmt3a-KO HSCs have reduced survival. (A) WBC and lymphocyte (LY) count at 26 weeks posttransplant; n = 18 WT and n = 41 Dnmt3a-KO. Error bars represent mean plus standard deviation. (B) Kaplan-Meier survival curve of mice transplanted with WT (n = 20) or Dnmt3a-KO (n = 45) HSCs. Censored points (squares) represent 5 mice that were sacrificed for a nonhematopoietic phenotype. Median survival is 321 days for Dnmt3a-KO; undetermined for WT. The experiment was repeated with the same numbers of animals, with median survival times of 246 days for mice transplanted with Dnmt3a-KO HSCs and 467 days for those transplanted with WT. (C) Number of mice in the transplanted cohort that were definitively diagnosed with the indicated hematopoietic diseases. ***P < .0001. B-ALL, B-cell acute lymphocytic leukemia/lymphoma; CMML, chronic myelomonocytic leukemia; ETP, early thymic progenitor acute lymphoblastic leukemia; PMF, primary myelofibrosis.
T-ALL development in HSCs lacking Dnmt3a
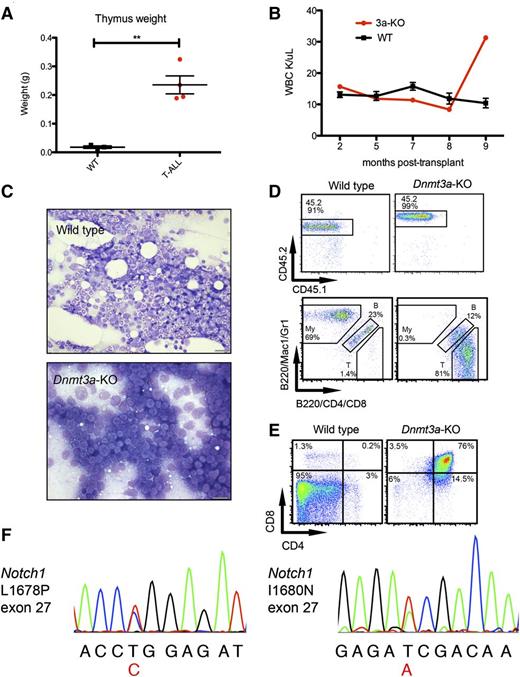
Four moribund mice transplanted with Dnmt3a-KO HSCs had enlarged thymuses (Figure 2A) and variable splenomegaly. WBC counts of these individual mice were similar to mice transplanted with WT HSCs initially but spiked shortly before the mouse became moribund (example in Figure 2B), contributing to the variation in WBC counts. Histology revealed bone marrow full of blast cells (Figure 2C), and immunophenotyping confirmed a donor-derived (CD45.2) T-cell phenotype, with many CD4-positive and/or CD8-positive cells and few B220+ B cells and Mac1+ and/or Gr1+ myeloid cells (Figure 2D). Further immunophenotyping revealed that most of the bone marrow cells of this mouse were CD4/CD8 double-positive (DP) T cells, in contrast to WT bone marrow, which contains virtually no DP T cells (Figure 2E). These phenotypes are consistent with T-ALL,20 which affected 3 of the moribund mice (supplemental Table 3). The other mouse with an enlarged thymus had a large CD44+CD25− population, indicating an early thymic progenitor leukemia (supplemental Table 3).
Development of T-ALL in mice transplanted with Dnmt3a-KO HSCs. (A) Thymus weights of mice transplanted with WT HSCs killed as age-matched controls while healthy, and 4 mice transplanted with Dnmt3a-KO HSCs that presented with enlarged thymuses on necropsy. Error bars represent mean ± SEM. (B) WBC count over time for 1 mouse receiving Dnmt3a-KO HSCs and 3 representative WT mice. Error bars represent mean ± SEM. (C) Bone marrow touch preps stained with Wright-Geimsa from a representative mouse transplanted with WT HSCs and from a mouse with T-ALL. Bars represent 20 μm. (D) Fluorescence-activated cell sorter analysis of the same Dnmt3a-KO recipient and a representative WT, showing (top row) that CD45.2 (donor) cells make up nearly all of the bone marrow cells, and (bottom row) myeloid (My), B cells (indicated with letter B), and T cells (T); parent gate is live cells. (E) Further immunophenotyping of these mice, bone marrow stained with CD4 and CD8; parent gate is live; CD45.2+. (F) Chromatogram traces showing Notch1 mutations identified in 2 T-ALL mice in exon 27. The variant base is listed below the sequence in red, and corresponding amino acid changes are indicated. **P < .001.
Development of T-ALL in mice transplanted with Dnmt3a-KO HSCs. (A) Thymus weights of mice transplanted with WT HSCs killed as age-matched controls while healthy, and 4 mice transplanted with Dnmt3a-KO HSCs that presented with enlarged thymuses on necropsy. Error bars represent mean ± SEM. (B) WBC count over time for 1 mouse receiving Dnmt3a-KO HSCs and 3 representative WT mice. Error bars represent mean ± SEM. (C) Bone marrow touch preps stained with Wright-Geimsa from a representative mouse transplanted with WT HSCs and from a mouse with T-ALL. Bars represent 20 μm. (D) Fluorescence-activated cell sorter analysis of the same Dnmt3a-KO recipient and a representative WT, showing (top row) that CD45.2 (donor) cells make up nearly all of the bone marrow cells, and (bottom row) myeloid (My), B cells (indicated with letter B), and T cells (T); parent gate is live cells. (E) Further immunophenotyping of these mice, bone marrow stained with CD4 and CD8; parent gate is live; CD45.2+. (F) Chromatogram traces showing Notch1 mutations identified in 2 T-ALL mice in exon 27. The variant base is listed below the sequence in red, and corresponding amino acid changes are indicated. **P < .001.
To examine whether the Dnmt3a-KO HSCs acquired common secondary mutations, we sequenced tumor cells for Notch1 mutations. NOTCH1 is frequently mutated in human T-ALL, and has been reported to co-occur with DNMT3A mutations.9,29 We performed Sanger sequencing of exons 26 to 28 and 34 of Notch1, and identified 2 different mutations in exon 27 (Figure 2F) affecting 2 of the 3 T-ALL mice. These mutations cause amino acid substitutions L1678P and I1680N, which correspond with mutations that co-occur with DNMT3A mutations in T-ALL patients.9 These are activating mutations in the heterodimerization domain, believed to facilitate cleavage and release of the intracellular domain of Notch1, increasing downstream signaling.30-32 These findings demonstrate that HSCs lacking Dnmt3a are predisposed to developing T-ALL with a high frequency of acquired Notch1 mutations.
Dnmt3a-KO HSCs give rise to MDS and myeloid malignancies
Moribund mice with normal or atrophic thymuses presented with abnormal WBC counts and hepatosplenomegaly or splenomegaly with variable hemorrhaging in the peritoneum and/or pleural cavity. We definitively diagnosed 21 of these mice with myeloid diseases using strict criteria (supplemental Methods; supplemental Figure 1)19 as described in Table 1 and as discussed below. The remaining 14 mice did not completely fulfill the criteria of any disease or were unable to be definitively diagnosed (supplemental Table 3).
Characteristics of mice diagnosed with AML and MDS
| Mouse . | Survival* (d) . | Necropsy findings . | WBC† (×103/μL) . | RBC† (×106/μL) . | PLT† (×103/μL) . | BM histology‡ . | Infiltration§ . | Diagnosis . |
|---|---|---|---|---|---|---|---|---|
| 226 | 139 | HSM, hem | 26.16 | 8.37 | 244 | Blasts | Spleen, liver | AML |
| 229 | 362 | SM, mottled | 19.92 | 8.07 | 1192 | Blasts | Spleen, liver | AML |
| 230 | 221 | Pale, HM | 3.82 | 6.72 | 325 | Blasts | Spleen, liver | AML |
| 231 | 347 | Pale, HM, hem, MF‖ | ND | ND | ND | Blasts | Spleen, liver, kidney | AML |
| 235 | 330 | HSM, pale, hem, MF | 3.08 | 2.69 | 126 | Blasts | Spleen, liver | AML |
| 246 | 326 | SM, hem, MF | ND | ND | ND | Blasts | Spleen, liver | AML |
| 257 | 328 | HM, hem | 0.46 | 6.61 | 556 | Blasts | Spleen, liver | AML |
| 263 | 263 | HSM, mottled, hem | 17.36 | 5.17 | 807 | Blasts | Spleen, liver | AML |
| 223 | 299 | HSM, hem | 4.04 | 4.15 | 424 | Dysplasia | NA | MDS |
| 227 | 358 | HSM, mottled, hem | ND | ND | ND | Dysplasia | NA | MDS |
| 232 | 355 | SM, hem, pale, MF | ND | ND | ND | Dysplasia | NA | MDS |
| 236 | 295 | HSM | 8.42 | 4.34 | 859 | Dysplasia | NA | MDS |
| 237 | 315 | HM, mottled, hem, pale | ND | ND | ND | Dysplasia | NA | MDS |
| 238 | 196 | SM, hem | 1.28 | 2.54 | 142 | Dysplasia | NA | MDS |
| 242 | 321 | HSM, mottled, hem | 2.44 | 1.49 | 146 | Dysplasia | NA | MDS |
| 252 | 319 | SM, hem, MF | 19.08 | 8.33 | 353 | Dysplasia | NA | MDS |
| 256 | 326 | Dark SP, hem | 4.1 | 6.18 | 556 | Dysplasia | NA | MDS |
| 258 | 316 | HM, hem | ND | ND | ND | Dysplasia | NA | MDS |
| Mouse . | Survival* (d) . | Necropsy findings . | WBC† (×103/μL) . | RBC† (×106/μL) . | PLT† (×103/μL) . | BM histology‡ . | Infiltration§ . | Diagnosis . |
|---|---|---|---|---|---|---|---|---|
| 226 | 139 | HSM, hem | 26.16 | 8.37 | 244 | Blasts | Spleen, liver | AML |
| 229 | 362 | SM, mottled | 19.92 | 8.07 | 1192 | Blasts | Spleen, liver | AML |
| 230 | 221 | Pale, HM | 3.82 | 6.72 | 325 | Blasts | Spleen, liver | AML |
| 231 | 347 | Pale, HM, hem, MF‖ | ND | ND | ND | Blasts | Spleen, liver, kidney | AML |
| 235 | 330 | HSM, pale, hem, MF | 3.08 | 2.69 | 126 | Blasts | Spleen, liver | AML |
| 246 | 326 | SM, hem, MF | ND | ND | ND | Blasts | Spleen, liver | AML |
| 257 | 328 | HM, hem | 0.46 | 6.61 | 556 | Blasts | Spleen, liver | AML |
| 263 | 263 | HSM, mottled, hem | 17.36 | 5.17 | 807 | Blasts | Spleen, liver | AML |
| 223 | 299 | HSM, hem | 4.04 | 4.15 | 424 | Dysplasia | NA | MDS |
| 227 | 358 | HSM, mottled, hem | ND | ND | ND | Dysplasia | NA | MDS |
| 232 | 355 | SM, hem, pale, MF | ND | ND | ND | Dysplasia | NA | MDS |
| 236 | 295 | HSM | 8.42 | 4.34 | 859 | Dysplasia | NA | MDS |
| 237 | 315 | HM, mottled, hem, pale | ND | ND | ND | Dysplasia | NA | MDS |
| 238 | 196 | SM, hem | 1.28 | 2.54 | 142 | Dysplasia | NA | MDS |
| 242 | 321 | HSM, mottled, hem | 2.44 | 1.49 | 146 | Dysplasia | NA | MDS |
| 252 | 319 | SM, hem, MF | 19.08 | 8.33 | 353 | Dysplasia | NA | MDS |
| 256 | 326 | Dark SP, hem | 4.1 | 6.18 | 556 | Dysplasia | NA | MDS |
| 258 | 316 | HM, hem | ND | ND | ND | Dysplasia | NA | MDS |
BM, bone marrow; dark Sp, dark spleen color; hem, hemorrhaging; HM, hepatomegaly; HSM, hepatosplenomegaly; MF, myelofibrosis; mottled, mottled liver; pale, pale paw pads and/or ears; PLT, platelet; RBC, red blood cell; SM, splenomegaly.
Survival indicates time from transplant to natural death or induced death if moribund.
WBC, RBC, and PLT counts are indicated for mice that had a CBC done at the time of death. Control ranges: WBC, 16.7-19.7 × 103/μL; RBC, 9.08-9.86 × 106/μL; PLT, 656-976 × 103/μL. ND indicates that a CBC was not done at the time of death. In these cases, blood smears and CBCs from previous time points were indicative of anemia and/or thrombocytopenia.
“Blasts” indicates >20% blasts in BM; “dysplasia” indicates that dysplasia was observed in 1 or more lineages (see also supplemental Figure 2).
Infiltration of indicated tissues was determined by histology and/or flow cytometry. “Spleen” indicates an increase specifically in nonlymphoid cells. NA indicates that this phenotype is not applicable to MDS.
MF determined by positive reticulin staining posttransplant.
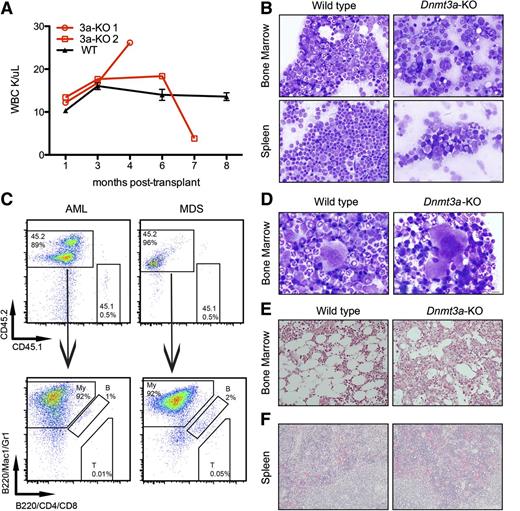
Mice diagnosed with AML (Figure 1C) had blood counts that spiked or dropped at the onset of symptoms and succumbed to disease within a month (Figure 3A; Table 1). We observed increased blasts (Figure 3B) in the bone marrow and extramedullary hematopoiesis in the spleen and liver, with an average of 97% (±2.6%) of bone marrow cells derived from the Dnmt3a-KO CD45.2 donor (Figure 3C). Immunophenotyping confirmed that most of these cells expressed myeloid markers Mac1 and/or Gr1 (Figure 3C). Ten of the remaining mice with increased donor-derived myeloid cells in the bone marrow (Figure 3C) were diagnosed with MDS, which was the most frequently diagnosed disease (Figure 1C). These mice presented with peripheral cytopenias and with dysplastic bone marrow in one or more lineages (Figure 3D; supplemental Figure 2; Table 1).
Mice transplanted with Dnmt3a-KO HSCs develop myeloid malignancies. (A) WBC counts of 2 mice transplanted with Dnmt3a-KO HSCs and 3 representative mice transplanted with WT HSCs. Error bars represent mean ± SEM. (B) Wright-Giemsa staining of bone marrow and spleen touch preps from a mouse transplanted with WT HSCs and a mouse transplanted with Dnmt3a-KO HSCs diagnosed with AML. (C) Immunophenotyping confirms donor-derived cells (CD45.2) in the bone marrow express myeloid markers (Mac1/Gr1). My, myeloid; B, B-cells; T, T-cells. (D) Wright-Giemsa–stained bone marrow touch preps from a mouse transplanted with WT HSCs and from a mouse transplanted with Dnmt3a-KO HSCs that was diagnosed with MDS; shown is a dysplastic binucleated megakaryocyte with coarse granules in the cytosol in the MDS mouse. (E) Reticulin staining of a bone marrow section from a mouse transplanted with WT HSCs, and from a mouse transplanted with Dnmt3a-KO HSCs, with black fibers indicative of fibrosis. (F) Hematoxylin and eosin staining of spleen sections from the 2 mice diagnosed with PMF demonstrating extramedullary hematopoiesis. Bars represent 20 μm (B,E-F) and 10 μm (D).
Mice transplanted with Dnmt3a-KO HSCs develop myeloid malignancies. (A) WBC counts of 2 mice transplanted with Dnmt3a-KO HSCs and 3 representative mice transplanted with WT HSCs. Error bars represent mean ± SEM. (B) Wright-Giemsa staining of bone marrow and spleen touch preps from a mouse transplanted with WT HSCs and a mouse transplanted with Dnmt3a-KO HSCs diagnosed with AML. (C) Immunophenotyping confirms donor-derived cells (CD45.2) in the bone marrow express myeloid markers (Mac1/Gr1). My, myeloid; B, B-cells; T, T-cells. (D) Wright-Giemsa–stained bone marrow touch preps from a mouse transplanted with WT HSCs and from a mouse transplanted with Dnmt3a-KO HSCs that was diagnosed with MDS; shown is a dysplastic binucleated megakaryocyte with coarse granules in the cytosol in the MDS mouse. (E) Reticulin staining of a bone marrow section from a mouse transplanted with WT HSCs, and from a mouse transplanted with Dnmt3a-KO HSCs, with black fibers indicative of fibrosis. (F) Hematoxylin and eosin staining of spleen sections from the 2 mice diagnosed with PMF demonstrating extramedullary hematopoiesis. Bars represent 20 μm (B,E-F) and 10 μm (D).
We also observed both primary myelofibrosis and myelofibrosis associated with AML/MDS. Bone marrow sections from mice with hypocellular marrow were subjected to silver stain to determine the presence of myelofibrosis (Figure 3E). Half of these mice (7/14) stained positive for reticulin fibers. Most (12/14) of these cases co-occurred with AML or MDS, but in 2 cases, myelofibrosis was the only identified disease; these were classified as PMF (Figure 1C). Mice diagnosed with PMF also displayed extramedullary hematopoiesis in the spleen (Figure 3F). Details of mice diagnosed with PMF and malignancies other than AML or MDS are summarized in supplemental Table 3.
Together, these data confirm that a lack of Dnmt3a in mouse HSCs confers an increased risk of development of hematologic diseases. The fact that the Dnmt3a-KO mouse mimics the range of diseases seen in humans, with identical Notch1 mutations identified in T-ALL, establishes the utility of this mouse as a model for development of DNMT3A mutation-associated hematologic disease.
Comutations that correlate with human disease counterparts
The variation in the cohort of mice transplanted with Dnmt3a-KO HSCs suggested that Dnmt3a-KO HSCs are predisposed to malignant transformation but that the specific disease progression could depend on acquired mutations. To obtain a global view of mutations in the myeloid diseases, we performed whole-exome sequencing. We extracted DNA from 4 mouse AML samples and 4 mouse MDS samples (whole bone marrow). DNA from tail tissue of 2 healthy individual Mx1-cre;Dnmt3af/f mice was also sequenced to exclude strain-specific polymorphisms.
To identify the most likely candidate driver genes, we focused on “high-confidence” variants that were identical in both control samples but had a variant in at least 1 disease sample. We also removed variants in genes that have no known human homolog, leaving 179 genes with identified single nucleotide variants. We also checked for the presence of indels but did not identify any leukemia-associated indels, such as Flt3-ITD (supplemental Table 4). We compared the variants identified in our mouse disease samples to mutations commonly reported in patients with hematologic malignancies. Several genes overlapped with those mutated in patients. For example, Npm1 was mutated in 1 mouse AML sample (Table 2), and NPM1 mutations frequently co-occur with DNMT3A mutations in human AML.1 However, only 1 of these murine mutations occurred at the equivalent amino acid position to its human counterpart. One animal with MDS exhibited a mutation in Kras at residue G12D, which has been reported in patients.3
Acquired variants identified by whole-exome sequencing
| Gene . | Mouse variant . | Reported mutation(s) in patients* . | Classification in mouse WES† . |
|---|---|---|---|
| Kras | p.G12D | p.G12D | Unique |
| Celsr1 | p.R2319W | p.P2600H | Unique |
| Kmt2a (MLL1) | p.T1311I | translocations, PTD | Recurrent |
| Ttn | p.G11436R | p.E12813K, p.I13568_ins | Recurrent |
| Brinp3 (FAM5C) | 3′UTR* | p.R719C, p.T119S | Not in “high confidence” |
| Npm1 | V60I | frameshift in last exon | Not in “high confidence” |
| Tead1 | 3′UTR | p.I12S | Not in “high confidence” |
| Gene . | Mouse variant . | Reported mutation(s) in patients* . | Classification in mouse WES† . |
|---|---|---|---|
| Kras | p.G12D | p.G12D | Unique |
| Celsr1 | p.R2319W | p.P2600H | Unique |
| Kmt2a (MLL1) | p.T1311I | translocations, PTD | Recurrent |
| Ttn | p.G11436R | p.E12813K, p.I13568_ins | Recurrent |
| Brinp3 (FAM5C) | 3′UTR* | p.R719C, p.T119S | Not in “high confidence” |
| Npm1 | V60I | frameshift in last exon | Not in “high confidence” |
| Tead1 | 3′UTR | p.I12S | Not in “high confidence” |
_ins, in-frame insertion; PTD, partial tandem duplication; 3′UTR*, change predicted by SnpEff to lead to increased nonsense-mediated decay; WES, whole-exome sequencing.
Mutations reported in patients with hematological malignancies.
Variants identified in 1 mouse disease sample (“unique”) or in multiple disease samples (“recurrent”) compared with control DNA, with predicted amino acid change. Not in “high confidence” variants are only identified when compared with one control sample individually.
To ensure that we did not miss mutations due to strict filtering based on matched sibling controls, we compared the sequences of genes that are mutated in patients from each disease sample to each control individually. With this strategy, we identified 3 more genes with variants in our mice but, as above, the variants we identified were not identical to those that have been associated with human disease (Table 2).
Acquired mutations in Ras genes cooperate with Dnmt3a-KO to drive malignancy
Because the Kras mutation identified by whole-exome sequencing was identical to the most common reported mutation in KRAS, which sometimes co-occurs with DNMT3A mutations, we asked whether mutant Ras expression would cooperate with Dnmt3a mutation in mouse HSCs. Of the RAS family, NRAS is most frequently mutated (70%) in hematologic diseases,33 so we focused on Nras.
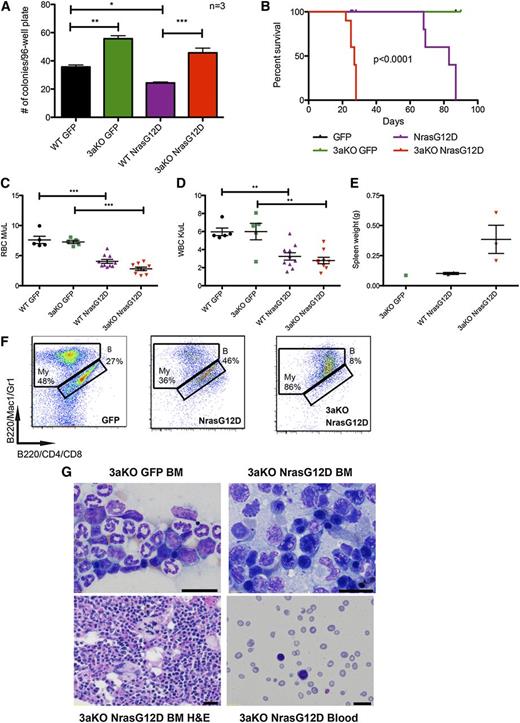
To determine whether lack of Dnmt3a would interact with Nras mutation and lead to a phenotypic change, we transduced Dnmt3a-KO and WT HSCs with NrasG12D or empty retroviral vector and sorted them into methylcellulose media. Expression of NrasG12D in a WT background reduced the number of colonies formed. As expected, Dnmt3a-KO HSCs produced more colonies than WT when transduced with green fluorescent protein alone. Dnmt3a-KO HSCs expressing NrasG12D produced significantly more colonies than WT HSCs expressing NrasG12D (Figure 4A), suggesting that the absence of Dnmt3a maintains the self-renewal of HSCs even in the presence of oncogenic mutations.
Lack of Dnmt3a accelerates Nras driven malignancy. (A) Number of colonies per plate after single green fluorescent protein (GFP)-positive stem and progenitor cells were sorted into individual wells of 96-well plates (N = 3 plates per genotype/transduction). Representative of 2 independent experiments. (B) Kaplan-Meier survival curve of mice transplanted with WT HSCs transduced with MSCV-IRES-GFP (n = 7) or MSCV-NrasG12D-IRES-GFP (n = 10) and mice transplanted with Dnmt3a-KO HSCs transduced with MSCV-IRES-GFP (n = 7) or MSCV-NrasG12D-IRES-GFP (n = 10) (median survival of WT-NrasG12D mice: 83 days; KO-NrasG12D: 27 days; P < .0001). Representative of 2 independent experiments. Red blood cell (RBC) counts (C) and WBC counts (D) 3 weeks after transplant of WT or Dnmt3a-KO HSCs transduced with GFP or NrasG12D (n = 10 mice for each genotype transplanted with NrasG12D; n = 7 mice for each genotype transplanted with GFP). (E) Spleen weights of mice sacrificed at 10 weeks posttransplant. Error bars represent mean ± SEM. (F) Immunophenotyping of bone marrow from mice transplanted with WT HSCs transduced with MSCV-IRES-GFP or MSCV-NrasG12D-IRES-GFP and Dnmt3a-KO HSCs transduced with MSCV-IRES-GFP, stained for myeloid (My) and B cells (indicated with letter B). Parent gate is live; CD45.2+GFP+. (G) Wright-Giemsa staining of bone marrow (BM) touch preps showing dysplastic erythroid precursors with irregular nuclear contours and peripheral blood smears, and a hematoxylin and eosin (H&E)-stained bone marrow section showing dysplastic megakaryocytes with bizarre-shaped nuclei from mice transplanted with Dnmt3a-KO HSCs transduced with MSCV-IRES-GFP or MSCV-NrasG12D-IRES-GFP. Bars represent 20 μm. *P < .05, **P < .01, ***P < .001.
Lack of Dnmt3a accelerates Nras driven malignancy. (A) Number of colonies per plate after single green fluorescent protein (GFP)-positive stem and progenitor cells were sorted into individual wells of 96-well plates (N = 3 plates per genotype/transduction). Representative of 2 independent experiments. (B) Kaplan-Meier survival curve of mice transplanted with WT HSCs transduced with MSCV-IRES-GFP (n = 7) or MSCV-NrasG12D-IRES-GFP (n = 10) and mice transplanted with Dnmt3a-KO HSCs transduced with MSCV-IRES-GFP (n = 7) or MSCV-NrasG12D-IRES-GFP (n = 10) (median survival of WT-NrasG12D mice: 83 days; KO-NrasG12D: 27 days; P < .0001). Representative of 2 independent experiments. Red blood cell (RBC) counts (C) and WBC counts (D) 3 weeks after transplant of WT or Dnmt3a-KO HSCs transduced with GFP or NrasG12D (n = 10 mice for each genotype transplanted with NrasG12D; n = 7 mice for each genotype transplanted with GFP). (E) Spleen weights of mice sacrificed at 10 weeks posttransplant. Error bars represent mean ± SEM. (F) Immunophenotyping of bone marrow from mice transplanted with WT HSCs transduced with MSCV-IRES-GFP or MSCV-NrasG12D-IRES-GFP and Dnmt3a-KO HSCs transduced with MSCV-IRES-GFP, stained for myeloid (My) and B cells (indicated with letter B). Parent gate is live; CD45.2+GFP+. (G) Wright-Giemsa staining of bone marrow (BM) touch preps showing dysplastic erythroid precursors with irregular nuclear contours and peripheral blood smears, and a hematoxylin and eosin (H&E)-stained bone marrow section showing dysplastic megakaryocytes with bizarre-shaped nuclei from mice transplanted with Dnmt3a-KO HSCs transduced with MSCV-IRES-GFP or MSCV-NrasG12D-IRES-GFP. Bars represent 20 μm. *P < .05, **P < .01, ***P < .001.
To determine whether this cooperation would have an impact in vivo, we transplanted freshly transduced HSCs into lethally irradiated recipients. In a WT background, NrasG12D expression drove a myeloid disease that killed mice, with a median survival of 64 days. In the absence of Dnmt3a, disease was accelerated, with a median survival of 27 days (Figure 4B). CBCs at 3 weeks posttransplant revealed that expression of NrasG12D caused peripheral cytopenias, regardless of the presence of Dnmt3a (Figure 4C-D) and an enlarged spleen in the Dnmt3a-KO+Nras (Figure 4E). Histology and immunophenotyping revealed that Dnmt3a-KO+Nras led to an AML-like phenotype characterized by bone marrow filled with myeloid cells (Figure 4F), immature blast morphology and dysplastic cells, and peripheral blood with marked pancytopenia and circulating blasts (Figure 4G).
These data demonstrate the cooperation of a known oncogene with loss of Dnmt3a. In vitro, RasG12D overexpression had a negative effect on progenitor growth; however, loss of Dnmt3a provided a distinct advantage, a behavior likely contributing to the rapid disease that developed in vivo.
Methylation changes are specific to disease lineage
Loss of Dnmt3a could have general effects on the methylation patterns across diseases or specific effects during the development of different diseases. The effects of DNMT3A mutations on methylation patterns and gene expression are still poorly understood, even within individual diseases. Although we have reported methylation analysis in Dnmt3a-KO HSCs,16 our previous analysis was on HSCs from healthy mice. To examine changes in methylation patterns of Dnmt3a-KO-derived malignancies, we performed enhanced reduced representation bisulfite sequencing on tumor and normal samples. We analyzed 4 AML, 4 MDS (whole bone marrow), and 3 T-ALL samples (thymus cells). For comparison, we purified LSK progenitor cells and DP T-cells from 3 mice transplanted with WT HSCs.
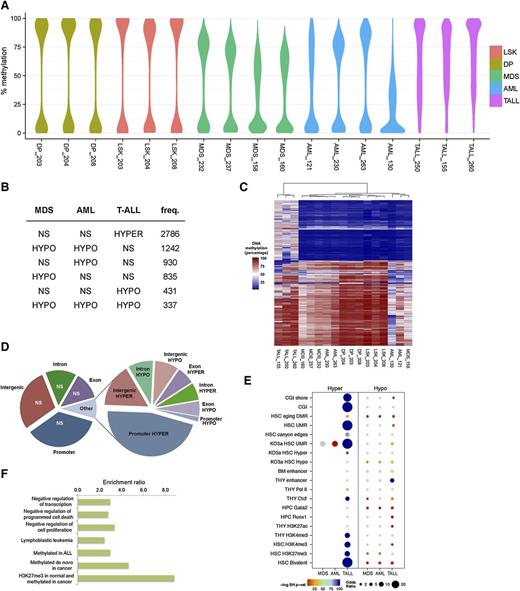
Using enhanced reduced representation bisulfite sequencing, we were able to interrogate approximately 42 Mb of the mouse methylome. Overall methylation was markedly reduced in the MDS and AML samples compared with normal LSK cells, as evidenced by 0.7 quantile CpG methylation values of 76% and 78% compared with 91%, respectively (supplemental Figure 3A). When comparing methylation maps between cell types, we observed the lowest correlations with T-ALL (supplemental Figure 3B). We investigated locus-specific differences in the methylomes via pairwise comparisons between MDS or AML and WT LSK cells, and T-ALL and DP T cells. Differentially methylated regions (DMRs) were defined as those having at least 25% methylation difference between disease and control. When comparing malignant and normal cell populations, we observed that hypomethylation was the predominant event in MDS and AML (Figure 5A), occurring most frequently in intergenic regions (supplemental Figure 4). A core set of 337 hypomethylated DMRs was common to all 3 diseases, representing approximately one-third of such regions in T-ALL. There was substantial overlap between MDS and AML, most of which was myeloid specific (1256 of 1596 DMRs) (Figure 5B). This observation indicated that in disease driven by Dnmt3a loss, methylation profiles are highly dependent on the lineage from which the disease originated. AML was characterized by the greatest frequency and magnitude of hypomethylation (Figure 5B). T-ALL deviated greatly from the myeloid malignancies with respect to gain of methylation (Figure 5A-C). Approximately 75% of T-ALL DMRs were hypermethylated, with a clear preference for promoter regions (Figure 5D). Hypermethylation was rarely observed in AML and MDS, accounting for ∼0.3% of DMRs in either disease.
DNA methylation changes in Dnmt3a-null driven malignancies. (A) Distribution of absolute DNA methylation levels in all experiments for genomic regions subject to differential methylation in MDS, AML, and/or T-ALL mice. The width of the y-axis indicates the frequency with which the value is observed. (B) The top 6 classes of DNA methylation changes (relative to control samples) across the diseases are shown. Hypermethylation in T-ALL is the most frequent, and shared hypomethylation between MDS and AML is the second most frequent. (C) The methylation levels of DMRs in 1 or more diseases were analyzed by hierarchical clustering using a Jensen-Shannon distance metric of methylation specificity. (D) Pie chart showing the proportion of DMRs in T-ALL mice by gee association. Left chart summarizes region coverage of intergenic, introns, exons, and promoters. Right chart summarizes DMRs by the same categories. (E) Chromatin and transcriptional regulator enrichment analysis of MDS, AML, and T-ALL DMRs. Enrichment of DMRs in peaks or binding sites curated in publicly available mouse functional genomics data sets for the indicated factors and cell/tissue lineages was quantified using odds ratios and P values (Benjamini-Hochberg testing correction) determined by Fisher’s exact test. Dot sizes are proportionate to the odds ratio. Color intensity represents the negative-log Benjamini-Hochberg–corrected P value. Nonsignificant results are in gray. (F) T-ALL hypermethylation recapitulates functions and ontologies of human leukemia. Potential cis-regulatory functions of promoter-proximal (10 kb up to 1 kb downstream of transcriptional start sites) hypermethylated DMRs in T-ALL were predicted with GREAT 2.0. All enriched terms have a multiple testing corrected q < 0.005. CGI, CpG islands; LSK, Lin-Sca-1+c-Kit+; NS, nonsignificant; TALL, T-ALL; THY, thymus; UMR, undermethylated regions.
DNA methylation changes in Dnmt3a-null driven malignancies. (A) Distribution of absolute DNA methylation levels in all experiments for genomic regions subject to differential methylation in MDS, AML, and/or T-ALL mice. The width of the y-axis indicates the frequency with which the value is observed. (B) The top 6 classes of DNA methylation changes (relative to control samples) across the diseases are shown. Hypermethylation in T-ALL is the most frequent, and shared hypomethylation between MDS and AML is the second most frequent. (C) The methylation levels of DMRs in 1 or more diseases were analyzed by hierarchical clustering using a Jensen-Shannon distance metric of methylation specificity. (D) Pie chart showing the proportion of DMRs in T-ALL mice by gee association. Left chart summarizes region coverage of intergenic, introns, exons, and promoters. Right chart summarizes DMRs by the same categories. (E) Chromatin and transcriptional regulator enrichment analysis of MDS, AML, and T-ALL DMRs. Enrichment of DMRs in peaks or binding sites curated in publicly available mouse functional genomics data sets for the indicated factors and cell/tissue lineages was quantified using odds ratios and P values (Benjamini-Hochberg testing correction) determined by Fisher’s exact test. Dot sizes are proportionate to the odds ratio. Color intensity represents the negative-log Benjamini-Hochberg–corrected P value. Nonsignificant results are in gray. (F) T-ALL hypermethylation recapitulates functions and ontologies of human leukemia. Potential cis-regulatory functions of promoter-proximal (10 kb up to 1 kb downstream of transcriptional start sites) hypermethylated DMRs in T-ALL were predicted with GREAT 2.0. All enriched terms have a multiple testing corrected q < 0.005. CGI, CpG islands; LSK, Lin-Sca-1+c-Kit+; NS, nonsignificant; TALL, T-ALL; THY, thymus; UMR, undermethylated regions.
Given the similar patterns of methylation loss in myeloid diseases and the distinct gains of methylation in T-ALL, we asked whether differential methylation in Dnmt3a-null-driven diseases impacts distinct transcriptional regulatory systems. To this end, we tested the overrepresentation of DMRs among the chromatin immunoprecipitation sequencing profiles of transcriptional regulators and epigenetic signatures in embryonic stem cells and hematopoietic cells, including data from our previous work on healthy Dnmt3a-KO HSCs, which could be considered “preleukemic.”16,34-37 Hypermethylation in T-ALL was enriched for HSC bivalent chromatin domains, consistent with previous reports of bivalent genes being frequently hypermethylated in cancers38-40 and the finding that age-associated DNA hypermethylation occurs predominantly at bivalent domains in HSCs and other tissues.41 Our present findings together with previous findings of others indicate that bivalent domains are particularly susceptible to DNA hypermethylation. Hypomethylated DMRs were enriched for thymus enhancers in T-ALL and for bone marrow enhancers in myeloid diseases (Figure 5E; supplemental Table 5).
Our analysis revealed that the most significant and the most disease-specific enrichments occurred in T-ALL cells at undermethylated regions and canyon edges as well as Gata2 and Runx1 TF binding sites in Dnmt3a-KO HSCs. A more detailed analysis of cis-regulatory function in promoter-proximal regions predicted that T-ALL hypermethylation affects genes involved in human lymphoblastic leukemia and also negatively regulates several hallmarks of cancer, including programmed cell death and cell proliferation (Figure 5F). In MDS the distinguishing methylation signature from T-ALL was hypomethylation of bone marrow enhancers distal to genes involved in myeloid cell differentiation (fold enrichment = 3.6, q = 5.98 × 10−4).
The differences in methylation in different diseases arising from the same pool of Dnmt3a-KO HSCs suggest that although lack of normal Dnmt3a function may result in a disturbed methylation landscape, the downstream effects depend on the specific lineage of the disease.
Discussion
Here, we show that lack of Dnmt3a in mouse HSCs confers a predisposition to malignant transformation, possibly by inducing plasticity in the DNA methylation pattern. As in patients with reported DNMT3A mutations, mice transplanted with Dnmt3a-KO HSCs develop a spectrum of diseases affecting both myeloid and lymphoid lineages, establishing our Dnmt3a-KO mouse as a valuable model for studying the effects of loss of Dnmt3a on HSCs.
Here, we are using an inducible Dnmt3a-null mutation, but the mutations observed in patients are distinct. In AML, ∼60% of patients have the R882 mutation, which is thought to act as a dominant negative,7,8 with some WT DNMT3A function remaining. In T-ALL, patients more frequently have compound heterozygous mutations, but the biochemical impact of these mutations is not known. Thus, in patients, some DNMT3A-associated DNA methylation activity may remain, and other protein-protein interactions involving DNMT3A may be altered. In our mouse model, such remaining Dnmt3a activity and potential interactions are not duplicated. Nevertheless, we have recapitulated at the phenotypic level many of the diseases seen in patients harboring DNMT3A mutations, establishing that loss of Dnmt3a is sufficient to predispose HSCs to malignant transformation, and that residual Dnmt3a is not required for disease predisposition.
Loss of Dnmt3a in HSCs was not associated with a consistent change in DNA methylation patterns across disease types; rather, the lack of Dnmt3a seems to confer plasticity to the methylome that may allow for transformation after acquisition of additional hits. The overrepresentation of hypomethylated DMRs in our myeloid disease samples is surprising, given recent reports of mouse models of MDS and MDS/AML that have promoter hypermethylation similar to that seen in patients.42 This difference could be explained by the null allele of Dnmt3a generated in our model. Our results underscore the notion that hypermethylation of promoters, which has been a focus of numerous cancer methylation studies, represents only a fraction of cancer-associated methylation changes. Differential methylation outside of CpG islands in gene promoters may play an important role in leukemogenesis, and a better understanding of the connection between nonpromoter methylation and gene expression changes and disease development is needed.
A few of the acquired mutations we identified in our Dnmt3a-KO HSC-derived diseases, such as Notch1 and Kras, were the same as mutations reported alongside DNMT3A mutations in patients. In addition, the murine system allows the introduction of specific mutations, in this case Dnmt3a loss followed by NrasG12D expression. This strategy can be used to study the functional effects of combinations of mutations.
Interestingly, many of the variants we identified were different than mutations reported in patients, and it was surprising that so few occur in disease-associated hotspots. This may reflect a difference between the mouse and human loci that are likely to be mutated, or a difference in the genes that cooperate with Dnmt3a mutations in mice compared with humans. Alternatively, epigenetic alterations may be sufficient to drive malignant transformation in mouse HSCs. This possibility is in contrast to the situation in patients, where almost all samples have multiple recurrent mutations. Further analysis of the murine diseases is required to resolve this issue.
Distinctions in study design and mouse strains may explain the differences between the results of this study and those reported recently by Peters et al43 in which mice lacking Dnmt3a in the HSPC compartment were predisposed to a chronic lymphocytic leukemia–like disease. Despite the differences in disease spectrum, both studies implicate Dnmt3a as an important player in preventing hematologic malignancy development in mice.
We previously uncovered a role for Dnmt3a in maintaining the balance of self-renewal and differentiation in HSCs16 ; however, in those studies we did not observe malignancy development. By performing serial transplantation every 18 weeks, the mice may have been killed while they still contained only clinically silent preleukemic stem cells. It is also possible that microenvironmental changes are required to promote disease development in the context of Dnmt3a mutations. In the current study, significant rates of disease occurred after 6 months, much longer than our previous strategy allowed. Environmental changes (eg, inflammation acquired with age) could impair normal hematopoiesis and contribute to myelofibrosis development, similar to transgenic BCR/ABL cells, as recently reported.44 Our current study design also eliminated the presence of WT HSCs, forcing the Dnmt3a-KO HSCs to divide extensively and contribute to blood regeneration, which could promote the acquisition of secondary effects. Ultimately, by extending the time frame and eliminating WT HSCs, we uncovered a predisposition to malignant development in the absence of Dnmt3a.
Overall, our data demonstrate that Dnmt3a loss of function confers a preleukemic phenotype on mouse HSCs. Our Dnmt3a-KO model may be useful for development of targeted therapeutic strategies for DNMT3A mutation-associated hematologic malignancies.
The reduced representation bisulfite sequencing data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE60264).
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank members of the Goodell Laboratory for helpful discussions; R. Nistal, Y. Zheng, M. Zhou, and A. Rosen for technical support; J. Sederstrom and L. White for their assistance; and the Cytometry and Cell Sorting Core and the DNA Sequencing Core laboratories at Baylor College of Medicine.
This work was supported by the Robert and Janice McNair Foundation (L.Y.); the Edward P. Evans Foundation; and National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases grants DK092883 and DK084259, National Human Genome Research Institute grants R01HG007538 and HG007538, and National Cancer Institute grants CA183252, CA183252, and CA126752.
Authorship
Contribution: A.M. and L.Y. designed and performed experiments, analyzed results, made figures, and wrote the paper; B.R. analyzed data, wrote the paper, and made a figure; T.Z., E.C., and C.V.C. analyzed data; G.A.C. designed and performed experiments; W.L., D.W., and V.I.R. helped analyze data; and M.A.G. designed the experiments and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Margaret Goodell, Department of Pediatrics, and Molecular and Human Genetics, Baylor College of Medicine, One Baylor Plaza, N1030, Houston, TX 77030; e-mail: goodell@bcm.edu.
References
Author notes
A.M. and L.Y. contributed equally to this study.