Abstract
The analyses carried out using 2 different bioinformatics pipelines (SomaticSniper and MuTect) on the same set of genomic data from 133 acute myeloid leukemia (AML) patients, sequenced inside the Cancer Genome Atlas project, gave discrepant results. We subsequently tested these 2 variant-calling pipelines on 20 leukemia samples from our series (19 primary AMLs and 1 secondary AML). By validating many of the predicted somatic variants (variant allele frequencies ranging from 100% to 5%), we observed significantly different calling efficiencies. In particular, despite relatively high specificity, sensitivity was poor in both pipelines resulting in a high rate of false negatives. Our findings raise the possibility that landscapes of AML genomes might be more complex than previously reported and characterized by the presence of hundreds of genes mutated at low variant allele frequency, suggesting that the application of genome sequencing to the clinic requires a careful and critical evaluation. We think that improvements in technology and workflow standardization, through the generation of clear experimental and bioinformatics guidelines, are fundamental to translate the use of next-generation sequencing from research to the clinic and to transform genomic information into better diagnosis and outcomes for the patient.
Introduction
Whole-genome sequencing (WGS) and whole-exome sequencing (WES) are widely used for mutational analysis in cancer samples. These new technologies are changing our understanding of the genomic landscape of cancer, including acute myeloid leukemia (AML). Indeed, in the last few years, WGS and WES have allowed the identification of key mutated genes in AML, such as DNMT3A1 and IDH1.2 However, proper identification of DNA mutations, such as single nucleotide variants (SNVs), small insertions and deletions (InDels), and copy number variants, relies on the sequencing technology, data quality, and statistical methods (bioinformatics pipelines) used to analyze sequencing data. Despite the development of many statistical methods and much effort to give standard rules for the assessment of good-quality next-generation sequencing (NGS) testing,3 no clear guidelines exist on how to practically analyze genome-sequencing data, as the optimization of somatic variant identification constitutes an important challenge in computational biology.4
To assess the quality of a statistical approach, the identified mutations are usually validated by an independent method, evaluating the percentage of false positives (specificity analysis). The evaluation of false negatives (sensitivity analysis), instead, is more difficult, as true somatic mutations in a given sample are for the most part unknown. Thus, the fraction of true mutations missed in an analysis is generally undetermined.
Moreover, one of the main challenges in analyzing WES and WGS data is the identification of somatic variants present at low frequency (under 10%), as they are more difficult to distinguish from sequencing errors. Low-frequency mutations may also occur due to the presence, in the tumor sample, of highly variable percentages of normal cells or multiple tumor clones, whose relative representation might also vary greatly.5 Because the detection of rare variants mainly depends on sequence coverage, this problem is even worsened in WGS studies, which usually have lower coverage than WES studies.
SomaticSniper6 and MuTect7 are 2 widely used bioinformatics pipelines to identify SNVs in cancer genomes. A comparative analysis of algorithms for somatic SNV detection in cancer samples rated SomaticSniper as one of the best performers.8 Its sensitivity was evaluated on WGS data from a cell line (coverage of 30 times) and reported to be ∼99.8% (calling 496 of the 497 previously validated sites6 ) This method, however, does not consider possible variations in sensitivity due to changes in the allelic fraction of the mutations (ie, low-frequency mutations) because cell lines have presumably lower biological heterogeneity than primary tumor samples. On the contrary, the more recently published MuTect7 describes benchmarking approaches to evaluate its sensitivity and specificity as a function of the allelic fraction, and it is designed to recognize low allele fraction events (≥0.03). Sensitivity performance on different datasets spans from 66.4% to 99.9% and, when tested on the dataset used for SomaticSniper, MuTect shows equal sensitivity.
Discrepant results from 2 independent analyses of the same AML genomic data
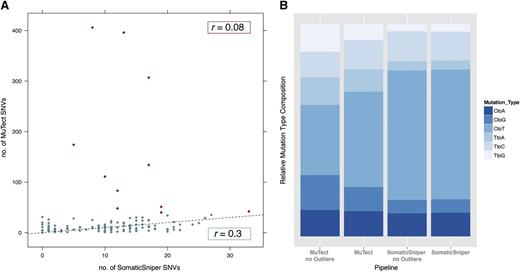
Two recent publications9,10 have reported the genomic analyses of 200 and 134 samples of AML from The Cancer Genome Atlas (TCGA) using SomaticSniper6 or MuTect.7 Although WES data from 133 AML samples are in common between the 2 publications, the identified exonic mutation rate per patient is quite different (Figure 1A; supplemental Table 1, see supplemental Data available on the Blood Web site): 11 vs 24 (0.03-1.7 per Mb vs 0-13.53 per Mb). In particular, the maximum number of mutations per patient is 33 and 406, respectively, with 5 samples in the second analysis being “hypermutated” AMLs because they have a number of somatic mutations in the coding regions which is dramatically above the median mutation rate of AMLs (as defined in Govindan et al11 ). The Pearson correlation coefficient between the numbers of mutations per patient reported by the 2 publications is low: it equals 0.08, considering the entire dataset, and 0.3, after removal of the outliers (samples having a number of mutations larger than the upper quartile by at least 1.5 times the interquartile range) (Figure 1A). In addition, the percentages of the possible base changes are surprisingly different between the outputs of the 2 methods (Figure 1B; supplemental Table 1), indicating that the identified mutated genes are quite different. Because the Nature article10 does not report the list of the identified mutated genes in these tumors, we reproduced their results applying MuTect to the AML WES raw data available in the TCGA data portal. By analyzing together all of the SNVs identified by TCGA using SomaticSniper and by MuTetc, we characterized a more comprehensive AML mutational landscape. By considering only missense, nonsense, loss of stop, silent variants, and variants affecting splice sites, we confirmed the presence of many more SNVs in the AML tumor samples than previously reported (from 1383 to 10533 variants). Interestingly, 65% (5983 of 9150) have variant allele frequencies (VAFs) <10%. We retrieved the number of mutations in driver genes (as defined by the Cancer Gene Census12 ) obtained using either the TCGA results or the union of TCGA and MuTect outputs. The recurrently (ie, in at least 2 patients) mutated driver genes in the analyzed cohort were 25 for the TCGA results and 104 for the union of TCGA and MuTect outputs. We observed only a slight increase in the percentage of patients harboring mutations in leukemia driver genes (eg, DNMT3A, FLT3, IDH2, RUNX1; supplemental Table 2); however, we identified many novel SNVs in genes implicated in the progression of many cancer types,12 although in no more than 5% of patients. Interestingly, 43% of patients harbored at least 1 of these novel mutations in cancer genes. This resembles a typical genomic landscape, defined as “mountains and hills” landscape, of solid tumors such as breast cancer, with few highly mutated genes and hundreds of low-frequency mutations. Importantly, we identified, in the same position in multiple patients, SNVs for the following genes: RNF2, TP53BP2, and RASA1. These genes encode for proteins involved in the regulation of cell proliferation or differentiation and are thus relevant for cancer progression. Finally, the identified SNVs can partly explain the hypermutated phenotype observed in some patients. Indeed, we have identified nonsilent SNVs in mismatch repair genes in 3 of 7 hypermutated patients: in the MSH6 gene in 2 patients and in the PMS1 gene in a third patient. However, our analysis is restricted to SNVs, so we cannot exclude the presence of additional mutations as InDels or structural variants in mismatch repair genes in the remaining patients that could explain hypermutation.
Comparison between the mutational analyses performed with the MuTect and SomaticSniper bioinformatics pipelines in 133 AMLs.9,10 (A) The same 133 AML samples were analyzed by WES and subsequently by MuTect7 or SomaticSniper6 bioinformatics pipelines: for every patient, the number of mutations identified with the 2 methods is reported on the x (SomaticSniper) and y (MuTect) axes; red points correspond to outliers (patients having a very high number of mutations in 1 or both methods). The Pearson correlation coefficient (r) was calculated for all samples (r = 0.08) or after removal of the outliers (r = 0.3). Both r values indicate a significant discordance between the 2 methods. The black dashed trend line indicates the expected number of identified mutations assuming that the analysis with the 2 pipelines gives exactly the same results. (B) For both methods, the percentage of mutations for every possible base change is reported. We considered both the complete set of patients and the set after outlier removal (no Outliers). The different distributions of the various types of mutations indicate that the 2 methods of analysis describe 2 different mutational landscapes for the same 133 AML patients.
Comparison between the mutational analyses performed with the MuTect and SomaticSniper bioinformatics pipelines in 133 AMLs.9,10 (A) The same 133 AML samples were analyzed by WES and subsequently by MuTect7 or SomaticSniper6 bioinformatics pipelines: for every patient, the number of mutations identified with the 2 methods is reported on the x (SomaticSniper) and y (MuTect) axes; red points correspond to outliers (patients having a very high number of mutations in 1 or both methods). The Pearson correlation coefficient (r) was calculated for all samples (r = 0.08) or after removal of the outliers (r = 0.3). Both r values indicate a significant discordance between the 2 methods. The black dashed trend line indicates the expected number of identified mutations assuming that the analysis with the 2 pipelines gives exactly the same results. (B) For both methods, the percentage of mutations for every possible base change is reported. We considered both the complete set of patients and the set after outlier removal (no Outliers). The different distributions of the various types of mutations indicate that the 2 methods of analysis describe 2 different mutational landscapes for the same 133 AML patients.
Comparison and validation of the 2 bioinformatics pipelines
To investigate their performances, we compared the behavior of SomaticSniper and MuTect on 20 leukemia samples from our series (19 primary AMLs, including 8 acute promyelocytic leukemias [APLs] and 1 secondary APL).13 All cases were analyzed by WES (supplemental Table 3) and mutations were called comparing the tumor and the corresponding remission samples for every patient. For the purpose of this study, we only report the results of the SNV analyses because the identification of InDels requires the use of additional independent bioinformatics tools.
The SNV landscapes identified by the 2 pipelines were quite different: a total of 194 SNVs and 178 mutated genes were detected by SomaticSniper, against 463 and 412 by MuTect, with only 161 SNVs and 150 mutated genes in common. Likewise, the frequency of SNVs per patient differed significantly: 9.7 vs 23.2 (0-0.6 per Mb vs 0.1-4.37 per Mb) (supplemental Table 4). As in the Nature article by Lawrence et al,10 MuTect could identify a hypermutated sample. In addition, the analysis of SNVs that have been causally implicated in tumors (mutations in genes overlapping with the Cancer Gene Census12 ) identified 21 genes using MuTect and only 14 using SomaticSniper (supplemental Table 4).
The 2 bioinformatics pipelines have different capacities to identify low-frequency SNVs. In our samples, SomaticSniper can identify SNVs with a VAF ≥10%; our data are in accordance with an independent study that challenged the ability of different mutation callers to detect SNVs at different allele frequencies, simulating WES data from 10 normal tumor pairs.14 MuTect instead allows, in our dataset, the identification of SNVs with VAFs as low as 2%. We therefore compared the mutations identified by the 2 pipelines according to their frequencies (≥10% and <10%). Notably, for those SNVs called by both pipelines (ie, ≥10%), the assigned frequency to each SNV is very similar (the average difference of frequency assignment was ∼1.1% in a range from 0% to 7%). The Pearson correlation coefficient of VAFs for the common mutations is, respectively, 0.99 in the tumor and 0.81 in the normal samples (supplemental Figure 1).
We observed 194 SNVs with a frequency ≥10% in the SomaticSniper analyses and 245 in the MuTect analyses (9.7 and 12.3 SNVs per patient, respectively); 161 of these SNVs were detected by both pipelines. Common SNVs accounted for around 83% and 66% of the total SNVs identified by each pipeline (Table 1), with the remaining SNVs specifically identified by only 1 of the 2 methods (“SomaticSniper only” and “MuTect only” SNVs were 33 and 84, respectively).
Total number of SNVs identified and validated in our dataset of 20 AMLs using either the SomaticSniper or the MuTect bioinformatics pipeline
| Frequencies . | All mutations . | Mutations with VAFs ≥10% . | Mutations with VAFs between 2% and 10% . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| All . | Tested . | Validated, n (%) . | x ≥10% . | Tested . | Validated, n (%) . | x <10% . | Tested . | Validated, n (%) . | |
| SomaticSniper-ALL | 194 | 94 | 84 (89) | 194 | 94 | 84 (89) | 0 | 0 | 0 |
| MutTect-ALL | 463 | 157 | 143 (91) | 245 | 97 | 95 (98) | 218 | 60 | 48 (80) |
| COMMON | 161 | 80 | 78 (98) | 161 | 80 | 78 (98) | 0 | 0 | 0 |
| SomaticSniper-ONLY | 33 | 14 | 6 (43) | 33 | 14 | 6 (43) | 0 | 0 | 0 |
| MuTect-ONLY | 302 | 77 | 65 (84) | 84 | 17 | 17 (100) | 218 | 60 | 48 (80) |
| Total (SomaticSniper+MuTect) | 496 | 171 | 149 (87) | 278 | 111 | 101 (91) | 218 | 60 | 48 (80) |
| Frequencies . | All mutations . | Mutations with VAFs ≥10% . | Mutations with VAFs between 2% and 10% . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| All . | Tested . | Validated, n (%) . | x ≥10% . | Tested . | Validated, n (%) . | x <10% . | Tested . | Validated, n (%) . | |
| SomaticSniper-ALL | 194 | 94 | 84 (89) | 194 | 94 | 84 (89) | 0 | 0 | 0 |
| MutTect-ALL | 463 | 157 | 143 (91) | 245 | 97 | 95 (98) | 218 | 60 | 48 (80) |
| COMMON | 161 | 80 | 78 (98) | 161 | 80 | 78 (98) | 0 | 0 | 0 |
| SomaticSniper-ONLY | 33 | 14 | 6 (43) | 33 | 14 | 6 (43) | 0 | 0 | 0 |
| MuTect-ONLY | 302 | 77 | 65 (84) | 84 | 17 | 17 (100) | 218 | 60 | 48 (80) |
| Total (SomaticSniper+MuTect) | 496 | 171 | 149 (87) | 278 | 111 | 101 (91) | 218 | 60 | 48 (80) |
For each category, we report the number of SNVs that have been tested and validated on the same DNAs. Rows from top: total number of mutations identified by each of the 2 different pipelines (ALL), number of mutations identified by both pipelines (COMMON), number of mutations uniquely identified by 1 of the 2 pipelines (ONLY). The validations have been performed through MiSeq, Ion Torrent, and Sanger sequencing and we considered validated every variant that was confirmed by at least 1 validation method.
We next validated in the same DNA sample 111 SNVs with a frequency ≥10%: 80 “common” and 31 “SomaticSniper or MuTect only,” using the Sanger, the Ion Semiconductor Sequencing Platform (Life Technologies), or the Illumina MiSeq Sequencing System (Table 1). Strikingly, the overall validation rate was extremely high (89% corresponding to 84 of 94 validated SomaticSniper SNVs and 98% corresponding to 95 of 97 validated MuTect SNVs), unveiling that a significant number of mutations are missed by 1 pipeline and identified by the other, or vice versa. Thus, when we examine the calling of SNVs at >10% frequency, the ability to minimize false positives is extremely good with both pipelines. On the other hand, the ability of the 2 pipelines to minimize false negatives appears relatively poor, with a minimum of 84 missed SNVs with SomaticSniper and of 33 with MuTect.
As mentioned, MuTect also allows identification of SNVs with a low VAF, up to the limit of 2% in our analyses. Indeed, it identified 218 SNVs with a frequency between 10% and 2% in our 20 AML samples (∼12 per patient; Table 1). Forty-eight of 60 (∼80%) of these SNVs were validated in the same DNAs, using semiconductor sequencing with the Ion Torrent Personal Genome Machine and the Illumina MiSeq Sequencing System. As the detection threshold of the Sanger technology, under our experimental conditions, exceeds ∼25% (data from C.R. and L.R.), we decided to use both the Ion Torrent and Illumina MiSeq technologies to also validate a subset of the lower-frequency mutations. We chose to apply these 2 independent sequencing platforms to ensure reliable results. The majority (29 of 34) of the mutations tested by both technologies gave concordant results, with an estimated empirical error rate of <1% (see supplemental Materials and methods for further details). The discordant positions were usually due to insufficient coverage of the MiSeq experiment, which, as a consequence, was unable to identify variants at very low VAFs. Therefore, we considered as validated any mutation that was confirmed by at least 1 technological platform.
Notably, 13 genes reported as relevant in cancer (The Cancer Gene Census),12 including CHEK2, ERBB2, ETV6, FGFR2, FLT3, KIAA1549, KRAS, MLL3, NRAS, PDE4DIP, RUNDC2A, TRIM33, WT1, harbored at least 1 mutation with a frequency of <10% in our patients. The median VAFs for the low-frequency mutations determined through our validation techniques were found to correspond to 6% in Illumina and 2% in Ion Torrent (supplemental Materials and Methods, supplemental Figure 2). The identification of low-frequency subclones can be very important in predicting the clinical behavior of leukemia, as recently suggested by their role in clonal evolution following chemotherapy.15 Assuming a condition of SNV heterozygosity and tumor homogeneity of the population (ie, 100% of leukemia blasts in the sample), a sensitivity of SNV calling equal to 2% corresponds to the presence of 4% of tumor cells harboring that SNV.
In conclusion, the analysis of the SNV-calling efficiency of the 2 different bioinformatics pipelines tested on 20 AML samples showed significantly different performances, underscoring the need to further refine them. In particular, compared with a relatively high specificity (ie, a low number of false-positive SNV callings), sensitivity was poor, as demonstrated by the high rate of false negatives (ie, those SNVs uniquely called by each of the 2 pipelines).
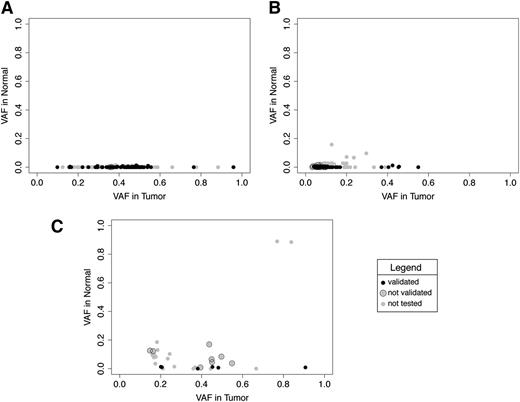
The reason for this inefficiency is not immediately obvious. The calling of an SNV relies on the detection of a variant base in the tumor sample, when compared with the same position in the control sample. It happens, however, that the same variant base detected in the tumor sample is also present in the control, though at lower frequency. This may be due to technical reasons (eg, sequence errors), tissue mosaicisms or, particularly in the case of leukemias, to contamination of the normal sample with tumor DNA given by the minimal residual disease (especially when the control sample is represented by DNA extracted from the peripheral blood of disease-remission state patients). We noticed that the frequency of variant bases in normal samples is significantly higher for those SNVs uniquely called by either pipeline, compared with the “common” SNVs, with an average frequency of 0.001 in the “Common,” 0.004 in the “MuTect-only” (Welch 2-tailed test, P value: .0004), and 0.1 in the “SomaticSniper-only” (P value: .005; Figure 2).
Comparison of variant base frequency in the SNVs and corresponding positions in the matching normal samples. (A) Common. (B) MuTect only. (C) SomaticSniper only. Each single dot corresponds to the variant base frequency in the tumor and normal samples for: (A) SNVs identified in AMLs by both pipelines (SomaticSniper and MuTect) and corresponding positions in the normal samples (“common”; B-C), and SNVs identified exclusively by MuTect or SomaticSniper and corresponding positions in the normal samples (“MuTect only”; “SomaticSniper only”). Black dots correspond to validated SNVs, gray dots with black circle to not-validated SNVs, and gray dots are not tested SNVs. We noticed that the majority of not-validated SNVs in MuTect appear at very low frequencies. Two not-validated SNVs in SomaticSniper hit known driver genes of AML (TET2 and DNMT3A); we think that the presence of these variants in the normal sample can be due to the contamination with tumor DNA given by the minimal residual disease.
Comparison of variant base frequency in the SNVs and corresponding positions in the matching normal samples. (A) Common. (B) MuTect only. (C) SomaticSniper only. Each single dot corresponds to the variant base frequency in the tumor and normal samples for: (A) SNVs identified in AMLs by both pipelines (SomaticSniper and MuTect) and corresponding positions in the normal samples (“common”; B-C), and SNVs identified exclusively by MuTect or SomaticSniper and corresponding positions in the normal samples (“MuTect only”; “SomaticSniper only”). Black dots correspond to validated SNVs, gray dots with black circle to not-validated SNVs, and gray dots are not tested SNVs. We noticed that the majority of not-validated SNVs in MuTect appear at very low frequencies. Two not-validated SNVs in SomaticSniper hit known driver genes of AML (TET2 and DNMT3A); we think that the presence of these variants in the normal sample can be due to the contamination with tumor DNA given by the minimal residual disease.
Notably, we observed that SomaticSniper allows mutation calling even if the variant allele is present in the normal sample at frequencies comparable with those of the tumor, both at high and low VAFs (Figure 2C). This can result in an augmented false-positive rate but can also allow the identification of some biologically relevant mutations in the set of variants defined as false positive according to our validation parameters. Indeed, in our dataset, SomaticSnipers has identified 2 mutations in the TET2 and DNMT3A genes. Although also present at high frequency in the normal sample, they were called SNVs because the difference in tumor and normal VAFs was statistically significant (Figure 2C; supplemental Table 5). We know from the literature that both TET2 and DNMT3A play a key role in leukemia development,16 and can be present at low frequency in the remission sample (especially DNMT3A), representing a typical case of contamination of the normal sample with residual tumor cells. These results underline the need to be aware of possible sources of contamination derived by minimal residual disease in normal samples, which can greatly challenge the efficiency of bioinformatics pipelines in SNV calling, as previously described.14 This is a particularly important issue for leukemic samples, for which the normal sample is usually represented by the remission sample (as in our study) and might be contaminated, depending on the patient, with different percentages of the tumor DNA.
Furthermore, we also observed some differences in the VAFs of mutations identified only by SomaticSniper and the VAFs for the same positions estimated by MuTect (supplemental Figure 3). This might be due to the fact that the 2 pipelines use different approaches to filter the reads used in the mutation-calling phase. MuTect, for example, is more lenient in filtering reads in the normal sample than in the tumor sample, resulting in an augmented strictness in calling mutations which are present at low allelic frequencies in the normal. SomaticSniper, instead, filters out poor-quality reads, both in normal and tumor samples, on the bases of a parameter given by the user. In addition, MuTect does not allow the identification of mutations that differ from the reference genome in the normal sample, although mutated in the tumor sample.
Conclusions
The presence of false negatives in the analysis of both pipelines has important consequences, as this may lead to an undervaluation of the occurrence of somatic mutations in AMLs, which might be critical for the identification of relevant targets, definition of clonality patterns,17 and identification of prognostic markers, which in turn may impact on therapeutic decisions (actionable mutations18 ).
Most of our knowledge regarding the genomic landscape of AML comes from the analysis with SomaticSniper of sequencing data of 200 cases of AML.9 We decided to compare SomaticSniper and MuTect because these 2 programs have been used to analyze and characterize the same AML genomes.9,10 Our goal was to reconcile the somehow contradictory results obtained analyzing the same set of data with these 2 techniques and, in particular, to test the putative presence of mutations at low frequency in AML genomes.
Our study raises the possibility that there might be a more complex landscape for AML genomes than previously reported. In addition, though we are unable to tell whether the mutations identified by MuTect in the 5 samples classified as “hypermutated” are all real or whether there may be sequencing artifacts, these findings raise new scientific questions on the possible causes that lead some AML samples to be hypermutated, and whether this characteristic is connected to particular clinical features.
However, SomaticSniper and MuTect are only 2 of the many existing mutation callers. For this reason, our analysis might underestimate the presence of false negatives, as the use of other mutation callers could increase the false-negative rate. The scientific community is becoming increasingly aware of the problems related to somatic mutations calling and is trying to take measures to overcome them. For example, the TCGA Consortium has begun to use multiple mutation callers for reporting somatic mutations in their projects in order to decrease the number of false negatives, a problem recently recognized by the consortium. Notably, a recent Dialogue for Reverse Engineering Assessments and Methods (DREAM) initiative (the International Cancer Genome Consortium-TCGA DREAM) has launched a mutation-calling challenge to improve standard methods for identifying cancer-associated mutations in genome-sequencing data.4
Concerning data production, a major issue of standard NGS platforms is the intrinsic error rate (around 1%), too high to allow detection of rare variants. NGS technologies refinement is a possible way to overcome the false-negatives problem: for example, introducing new applications for library preparation, such as the recently published method termed duplex sequencing.19 By tagging all the reads derived from the same DNA fragment and discarding any change between the 2 different strands of DNA, this method is reported to reduce the error rate of NGS up to 10 million fold. Therefore, it might shed some light on very rare variants and enable the identification of false positives and false negatives in cancer genomes, accurately describing the mutational landscape of individual tumors.
Improvement in sequencing technologies, increase in sequence coverage, development of precise guidelines in mutation calling, and increase in sensitivity will advance our understanding of the clonal substructure of human cancers, allow for the revision of the catalog of somatic mutation, and help elucidate mechanisms of mutational processes from which it is possible to infer patterns of DNA damage and repair processes.20 Conservatively, for now, it is recommended that WES data be analyzed with more than 1 pipeline. We also believe that workflow standardization through the generation of clear bioinformatics guidelines is fundamental to translate the use of NGS from research to the clinics and to transform genomic information into improved diagnosis and treatment.
The online version of this article contains a data supplement.
Acknowledgments
The authors thank Luca Rotta, Salvatore Bianchi, and Thelma Capra from the Genomic Unit for their help with the experimental validation. The results published here are in part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/.
This work was supported by grants from Fondazione Cariplo (L.R.). A.R. is supported by a triennial fellowship from Fondazione Italiana per la Ricerca sul Cancro (FIRC).
Authorship
Contribution: M.B. performed analysis and wrote the manuscript; L.R. and P.G.P. wrote the manuscript, designed research, and oversaw results; C.R. contributed to the design and interpretation of the study and wrote the manuscript; L.G., A.R., G.E.M.M., and L.L. contributed to the design and interpretation of the study; D.S., S.V., and L.B. performed experimental validation; and S.K.H., T.O., S.L., F.L.-C., E.T., A. Candoni, R.F., G.M., I.I., C.T., and A. Cignetti provided clinical data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Laura Riva, Center for Genomic Science of IIT@SEMM, Fondazione Istituto Italiano di Tecnologia, Via Adamello, 16, 20139 Milan, Italy; e-mail: laura.riva@iit.it; and Pier Giuseppe Pelicci, Department of Experimental Oncology, European Institute of Oncology, Via Adamello 16, 20139 Milan, Italy; e-mail: piergiuseppe.pelicci@ieo.eu.