Key Points
Germline gain-of-function mutations in STAT3 lead to lymphoproliferation and autoimmunity with prominent cytopenias.
Mutations in STAT3 cause altered regulatory T cells and cytokine signaling.
Abstract
Germline loss-of-function mutations in the transcription factor signal transducer and activator of transcription 3 (STAT3) cause immunodeficiency, whereas somatic gain-of-function mutations in STAT3 are associated with large granular lymphocytic leukemic, myelodysplastic syndrome, and aplastic anemia. Recently, germline mutations in STAT3 have also been associated with autoimmune disease. Here, we report on 13 individuals from 10 families with lymphoproliferation and early-onset solid-organ autoimmunity associated with 9 different germline heterozygous mutations in STAT3. Patients exhibited a variety of clinical features, with most having lymphadenopathy, autoimmune cytopenias, multiorgan autoimmunity (lung, gastrointestinal, hepatic, and/or endocrine dysfunction), infections, and short stature. Functional analyses demonstrate that these mutations confer a gain-of-function in STAT3 leading to secondary defects in STAT5 and STAT1 phosphorylation and the regulatory T-cell compartment. Treatment targeting a cytokine pathway that signals through STAT3 led to clinical improvement in 1 patient, suggesting a potential therapeutic option for such patients. These results suggest that there is a broad range of autoimmunity caused by germline STAT3 gain-of-function mutations, and that hematologic autoimmunity is a major component of this newly described disorder. Some patients for this study were enrolled in a trial registered at www.clinicaltrials.gov as #NCT00001350.
Introduction
The control of lymphocyte proliferation and tolerance is essential for both host defense and protection against self-directed immune attack. The discovery of autoimmune disorders with Mendelian inheritance that lead to a loss of tolerance has provided critical insights into biologic pathways important for regulation of the human immune response. For example, disorders of Fas-mediated apoptosis perturb peripheral lymphocyte tolerance and lead to the lymphoproliferation and hematologic autoimmunity seen in autoimmune lymphoproliferative syndrome (ALPS).1,2 Similarly, mutations in genes important for the development and/or function of regulatory T cells dramatically undermine peripheral tolerance, producing early-onset polyendocrine autoimmunity and immune dysregulation as seen in immunodeficiency polyendocrinopathy enteropathy x-linked (IPEX) and IPEX-like disorders.3 Mutations in AIRE, important for central T-cell tolerance, lead to a less severe syndrome of multiendocrine autoimmunity and mucocutaneous candidiasis called autoimmune polyendocrinopathy candidiasis and ectodermal dysplasia (APECED).4,5 An improved understanding of the mechanisms which regulate immune tolerance has the potential to improve the treatment of common autoimmune disorders by identifying and then targeting the relevant cellular pathways.
To further this process of discovery, we performed whole-exome sequencing of DNA from patients with early-onset autoimmunity and lymphoproliferative disease reminiscent of ALPS without a known genetic cause. We identified heterozygous signal transducer and activator of transcription 3 (STAT3) mutations in 13 individuals from 10 families, consistent with an apparent autosomal-dominant inheritance in certain kindreds and sporadic occurrences in others. Recently, a series of 5 patients with germline mutations in STAT3 associated predominantly with neonatal diabetes was reported by Flanagan et al,6 consistent with the hypothesis that such mutations can cause autoimmune disease. Here, we present genetic and functional data that reveal dysregulated cytokine signaling due to gain-of-function (GOF) STAT3 mutations with important implications for understanding lymphocyte tolerance and the pathogenesis of systemic autoimmunity.
Methods
Patients
Participants were enrolled independently at each institution into research studies using whole-exome sequencing to identify genetic causes of autoimmunity. Written and informed consent was obtained for living patients and their family members. These studies were approved by the institutional review boards of the authors’ institutions. Patients 2 through 4 and family 1 were enrolled in clinicaltrials.gov identifier NCT00001350.
Whole-exome sequencing
Genomic DNA was extracted from whole blood or saliva and coding regions enriched (SureSelect All Exon [Agilent Technologies] or as described7 for family 2) followed by high-throughput next-generation sequencing (Illumina) at individual institutions. Following quality control, alignment to GRCH37 or GRCH38, and variant calling, exome data were analyzed using a variety of institution-specific pipelines to identify novel or rare variants, including de novo variants, and cross-referenced between patients to focus on candidate genes. Putative disease-causing STAT3 variants were confirmed by dideoxy-based sequencing of polymerase chain reaction (PCR) amplicons in each proband, and relevant family members where available. The STAT3 mutation in patient 6 was detected by targeted dideoxy-based sequencing of the coding regions of STAT3 (Correlagen). Subsequent whole-exome sequencing of this patient’s DNA did not identify another candidate disease-causing variant.
Expression plasmids
Expression plasmids with the observed mutations were generated using a plasmid containing the wild-type (WT) STAT3 complementary DNA (cDNA) (NM_139276; Origene) that was mutated with QuikChange site-directed mutagenesis (Agilent Technologies). Primers used are reported in supplemental Table 1 (see supplemental Data available at the Blood Web site). Targeted sequences were confirmed by dideoxy-based sequencing. Plasmids expressing a loss-of-function (LOF) STAT3 mutation (p.K658E) observed in hyper-immunoglobulin E (IgE) patients (Dr S. Holland, National Institutes of Health (NIH), Bethesda, MD, written communication, April 9, 2013) and a GOF STAT3 mutation (p.N647I) observed in large granular lymphocytic leukemia8 were provided by Dr S. Holland, NIH, Bethesda, MD, and generated from the same WT plasmid.
Transfection experiments
WT, GOF, LOF, or mutant STAT3 plasmids were transfected into the STAT3-deficient A4 human colon cancer cell line9 (derived from DLD cells, kindly provided by Dr G. Stark, Cleveland Clinic, Cleveland, OH,) by transient transfection (Lipofectamine; Life Technologies). The parental DLD cell line was used as a control for some assays as shown. Total STAT3 protein and phospho-STAT3 (Y705) were measured by western blot (clones 79D7 and D3A7, respectively; Cell Signaling Technology) with or without simulation with interleukin-6 (IL-6) (10 ng/mL; Miltenyi Biotec) for 15 minutes. To confirm expression of mutant A703T, total STAT3 was detected using clone 124H6 (Cell Signaling Technology). For STAT3 luciferase studies, cells were cotransfected with 250 ng of the indicated STAT3 plasmid and a STAT3-responsive firefly luciferase reporter mixed with a control renilla plasmid (CMV promoter) per the manufacturer’s instructions (Cignal STAT3; Qiagen), followed by measurement of firefly and renilla luciferase (Dual-Luciferase Reporter Assay; Promega). No change in expression of renilla luciferase was seen when cells were transfected with WT, LOF, or GOF STAT3 (data not shown). In some experiments, cells were stimulated with recombinant human IL-6 (10 ng/mL) or interferon-α (IFN-α) (50 ng/mL, both Miltenyi Biotec) for 12 hours. Results represent the ratio of reporter (firefly) to control (renilla) luciferase or are normalized as fold-change in the ratio as compared with WT plasmid. Cytokine stimulation was conducted in serum-containing media.
Detection of T-cell subsets
Flow cytometry was conducted on patient samples at 4 separate facilities. Typically, peripheral blood mononuclear cells (PBMCs) were isolated by density centrifugation and resuspended in RPMI 1640 supplemented with penicillin, streptomycin, and l-glutamine along with 10% fetal calf serum (FCS). Double-negative T cells were measured by flow cytometric analysis of PBMCs from patients at individual institutions and results represent the percentage of CD4negCD8neg T cells in the CD3+TCRα/β+ T-cell subset. For Th17 cells, PBMCs were costimulated with phorbyl 12-myristate 13-acetate and calcimycin or ionomycin for between 4.5 hours and overnight and flow cytometric analysis of intracellular IL-17 performed after fixation and permeabilization. Results shown represent healthy controls and patients performed in parallel at different institutions.
Regulatory T-cell (Treg) analysis was performed at individual institutions. For patients 2, 3, 4, and 9, PBMCs were first stained with Live/Dead Fixable Aqua viability dye (Invitrogen), CD3 and CD25 (both BD Biosciences). Cells were then fixed and permeabilized with a FOXP3 staining kit per the manufacturer’s instructions (eBioscience). Cells were next stained with FOXP3 (clone 236A/E7; eBioscience) and CD4 (BD Biosciences). Tregs from patient 1 were assayed in the Clinical Laboratory Improvement Amendments–certified Clinical Immunodiagnostic and Research Laboratory of the Medical College of Wisconsin, and fresh cells were stained for cell surface CD4 and CD127 and intracellular FOXP3. For patient 6, cells were washed and stained with a viability dye to exclude dead cells (Ghost Viability dye; Tonbo). Cells were next surface stained with CD4 (eBioscience) and CD25 (BD Biosciences), and then fixed and permeabilized using a FOXP3 staining kit per the manufacturer’s instructions (eBioscience). Cells were then stained with intracellular antibodies CD3 (Biolegend) and FOXP3 (clone 259D/C7; BD Biosciences) for 1 hour before washing and acquisition. Data were analyzed with FlowJo software.
EBV-transformed cell lines and phosphoflow
Epstein-Barr virus (EBV)–transformed lymphoblastoid cell lines were generated from available patients (patients 2, 3, 4, 5, and 9) and healthy controls by standard methods. PBMCs were isolated by density centrifugation and resuspended in RPMI 1640 supplemented with penicillin, streptomycin, and l-glutamine along with 10% FCS. For phosphoflow for patients 2, 3, 4 and 9, cells were rested in serum-free RPMI 1640 prior to cytokine stimulation. Cytokine stimulations were then performed in serum-free RPMI 1640 for 20 minutes at 37°C with IL-2 (80 U/mL), IL-21 (100 ng/mL), IL-27 (75 ng/mL), or IFN-γ (500 ng/mL). After stimulation, cells were fixed in 1.6% paraformaldehyde for 10 minutes at room temperature and then resuspended in 100% cold methanol and incubated overnight at −20°C. Cells were then washed and stained with fluorochrome-conjugated phospho-STAT antibodies (BD Biosciences). For phosphoflow for patient 1, cells were stimulated with IL-6 (10 ng/mL) or IL-21 (5 ng/mL) for the indicated times. Cells were then fixed with Cytofix and permeabilized with Phosphoflow Perm Buffer III (both BD Biosciences) per the manufacturer’s instructions. Cells were incubated with phospho-STAT3 antibodies at room temperature followed by flow cytometric analysis. For phosphoflow for patient 5, cells were stimulated with IL-2 (1000 U/mL) or IFN-γ (1000 ng/mL) at 37°C for times indicated. Cells were then washed, fixed with Cytofix, and permeabilized with Phosphoflow Perm Buffer III (BD Biosciences). After washing, cells were incubated with phospho-STAT antibodies at 4°C followed by flow cytometric analysis.
Data were analyzed with FlowJo software.
Real-time quantitative RT-PCR
Suppressor of cytokine signaling 3 (SOCS3) transcript was measured by real-time quantitative reverse transcription-polymerase chain reaction (RT-PCR) in EBV-transformed patient-derived cell lines. Cells were serum starved followed by 16-hour incubation in RPMI 1640 + 2% FCS with or without IL-21 (50 ng/mL). Transcript levels of SOCS3 were normalized to endogenous β-actin control (both TaqMan primer/probe sets were purchased from Life Technologies). Results represent fold-change compared with an unstimulated healthy control.
Statistics
Results were analyzed with the Mann-Whitney or unpaired 2-tailed Student t test using GraphPad Prism software, and P < .05 was considered statistically significant.
Results
Whole-exome sequencing identifies STAT3 variants in patients with lymphoproliferation and early-onset autoimmune disease
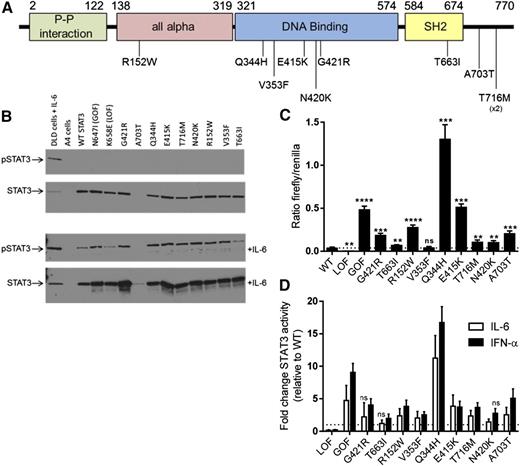
Whole-exome sequencing was performed at multiple centers on a cohort of patients with prominent lymphoproliferation, including lymphadenopathy and/or hepatosplenomegaly, as well as early-onset multisystem autoimmunity for whom a genetic diagnosis had not been established (Table 1). Upon comparing results in a subset of the cohort, it was noted that mutations in STAT3 were observed in 13 patients from 10 families (Table 1, supplemental Table 2). All were missense variants that were predicted to be deleterious by a variety of algorithms (supplemental Table 2) and absent from public databases of genetic variation (dbSNP and COSMIC), apart from c.1243G>A (p.E415K), which was recently reported as a somatic variant in liver cancer (COSMIC database). In contrast to the somatic GOF mutations seen in malignancy which cluster in the Src homology 2 (SH2) domain,8 the germline mutations identified here were located in multiple domains including the all-alpha, DNA binding, SH2, and C-terminal transactivation domains (Figure 1A). A single variant, c.2147C>T (p.T716M), was observed in 3 individuals with autoimmune enteropathy, 2 of whom were related (family 2, previously reported7 ). Interestingly, this same mutation was observed in a patient with early-onset type I diabetes.6
Patient characteristics
| Patient . | Age at onset, sex . | Current age . | STAT3 variant* . | Autoimmunity . | Lymphoproliferation . | Postnatal short stature† . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Hematologic . | Endocrine . | GI . | Other . | LAD . | Other . | |||||
| 1 | 4y, M | 10y | p.G421R | AIHA | No | Hepatitis | Scleroderma, polyarthritis | Yes | HSM | Yes |
| 2 | 7y, M | 31y | p.T663I | AIHA, AITP | No | No | No | Yes | HSM | No |
| 3 | 3y, M | 25y | p.R152W | AIHA, AITP | IDDM | No | Alopecia, lung nodules | Yes | HSM | No |
| 4 | 13y, M | 32y | p.V353F | AIHA, AITP, AIN | No | No | Inflammatory lung disease | Yes | No | No |
| 5 | 3y, F | 5y | p.Q344H | AIHA | No | Enteropathy | LIP | Yes | HSM | Yes |
| 6 | 5y, F | 9y | p.E415K | none | IDDM | Enteropathy, achalasia | Atopic dermatitis | Yes | HSM | Yes |
| 7 | <1y, F | 23y | p.T716M | AIHA, AITP, AIN | Hypothyroid | Enteropathy | No | No | No | Yes |
| 8 | 3y, F | Dec 11y | p.N420K | AIHA, AITP, AIN | No | No | Polyarthritis | Yes | No | No |
| Family 1 | ||||||||||
| 9, Proband | <1y, F | 26y | p.A703T | AIHA, AITP, AIN | No | Small bowel thickening | LIP, atopic dermatitis, alopecia | Yes | HSM | Yes |
| 10, Father | 15y, M | Dec 28y | p.A703T | AIHA, AIN | No | No | LIP | Yes | HSM | n/a |
| 11, Sibling | 12y, F | 24y | p.A703T | AITP, AIN | No | No | No | Yes | HSM | n/a |
| Family 2 | ||||||||||
| 12, Proband | <1y, M | 4y | p.T716M | none | No | Enteropathy | No | No | No | Yes |
| 13, Father | EO, M | 32y | p.T716M | AITP | No | Enteropathy | No | No | Hodgkin lymphoma | Yes‡ |
| Patient . | Age at onset, sex . | Current age . | STAT3 variant* . | Autoimmunity . | Lymphoproliferation . | Postnatal short stature† . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Hematologic . | Endocrine . | GI . | Other . | LAD . | Other . | |||||
| 1 | 4y, M | 10y | p.G421R | AIHA | No | Hepatitis | Scleroderma, polyarthritis | Yes | HSM | Yes |
| 2 | 7y, M | 31y | p.T663I | AIHA, AITP | No | No | No | Yes | HSM | No |
| 3 | 3y, M | 25y | p.R152W | AIHA, AITP | IDDM | No | Alopecia, lung nodules | Yes | HSM | No |
| 4 | 13y, M | 32y | p.V353F | AIHA, AITP, AIN | No | No | Inflammatory lung disease | Yes | No | No |
| 5 | 3y, F | 5y | p.Q344H | AIHA | No | Enteropathy | LIP | Yes | HSM | Yes |
| 6 | 5y, F | 9y | p.E415K | none | IDDM | Enteropathy, achalasia | Atopic dermatitis | Yes | HSM | Yes |
| 7 | <1y, F | 23y | p.T716M | AIHA, AITP, AIN | Hypothyroid | Enteropathy | No | No | No | Yes |
| 8 | 3y, F | Dec 11y | p.N420K | AIHA, AITP, AIN | No | No | Polyarthritis | Yes | No | No |
| Family 1 | ||||||||||
| 9, Proband | <1y, F | 26y | p.A703T | AIHA, AITP, AIN | No | Small bowel thickening | LIP, atopic dermatitis, alopecia | Yes | HSM | Yes |
| 10, Father | 15y, M | Dec 28y | p.A703T | AIHA, AIN | No | No | LIP | Yes | HSM | n/a |
| 11, Sibling | 12y, F | 24y | p.A703T | AITP, AIN | No | No | No | Yes | HSM | n/a |
| Family 2 | ||||||||||
| 12, Proband | <1y, M | 4y | p.T716M | none | No | Enteropathy | No | No | No | Yes |
| 13, Father | EO, M | 32y | p.T716M | AITP | No | Enteropathy | No | No | Hodgkin lymphoma | Yes‡ |
LAD, lymphadenopathy; EO, early childhood onset; Dec, deceased; AIHA, autoimmune hemolytic anemia; AITP, autoimmune thrombocytopenia; AIN, autoimmune neutropenia; IDDM, insulin-dependent diabetes mellitus; LIP, lymphocytic interstitial pneumonia; HSM, hepatosplenomegaly; n/a, not available.
Amino acid substitution based on coding sequence of NM_139276.2.
Less than fifth percentile.
Percentile n/a.
STAT3 mutations confer a GOF. (A) Schematic of human STAT3 protein showing the location of 9 different STAT3 missense mutations and the resulting amino acid changes. Mutations were located throughout the protein in the all alpha, DNA binding, SH2, and C-terminal (transactivation) protein domains. The numbers represent the amino acid location based on STAT3 transcript variant 1 (protein model based on the Pfam protein family database). The p.T716M variant was detected in 2 different families (patient 7 and family 2). (B) Western blot of STAT3 expression. WT, GOF, LOF, and the 9 different mutant STAT3 transcripts were expressed in the STAT3-deficient A4 cell line and expression of pSTAT3 and total STAT3 protein was determined using western blot without (top) and with (bottom) IL-6 (15 minutes, 10 ng/mL). IL-6–treated DLD cells (parental A4 line) are shown as a control. Expression of all mutants led to the detection of STAT3 protein, with the exception of p.A703T, a mutation near the binding site of the antibody used here (clone 79D7). This variant was detectable after IL-6 activation and with a different antibody clone (supplemental Figure 2). Mutant STAT3 proteins were not phosphorylated at baseline, but were phosphorylated after stimulation with IL-6. (C-D) STAT3 binding activity as measured by luciferase assay. A4 cells were transfected with WT or the indicated STAT3 mutants and STAT3-driven firefly luciferase and control renilla reporters. (C) Luciferase was assayed at 48 hours and STAT3 activity is shown as a ratio of firefly/control for each construct. All constructs with the exception of V353F had significantly increased activity at baseline compared with WT. Results represent the mean ± SEM of 5 to 10 independent experiments; the dotted line represents WT (**P < .01, ***P < .001, ****P < .0001). (D) STAT3 activity following 12-hour activation with IL-6 (10 ng/mL, white) or IFN-α (50 ng/mL, black). All mutations demonstrated significantly increased STAT3 activity after at least 1 of the cytokine stimulations (P < .05), typically after both unless indicated. Results shown represent fold-change vs WT for each cytokine stimulus; the dotted line represents a fold-change of 1 (no change from WT). Results represent the mean ± SEM of 5 independent experiments. ns, not significant; pSTAT3, phosphorylated STAT3.
STAT3 mutations confer a GOF. (A) Schematic of human STAT3 protein showing the location of 9 different STAT3 missense mutations and the resulting amino acid changes. Mutations were located throughout the protein in the all alpha, DNA binding, SH2, and C-terminal (transactivation) protein domains. The numbers represent the amino acid location based on STAT3 transcript variant 1 (protein model based on the Pfam protein family database). The p.T716M variant was detected in 2 different families (patient 7 and family 2). (B) Western blot of STAT3 expression. WT, GOF, LOF, and the 9 different mutant STAT3 transcripts were expressed in the STAT3-deficient A4 cell line and expression of pSTAT3 and total STAT3 protein was determined using western blot without (top) and with (bottom) IL-6 (15 minutes, 10 ng/mL). IL-6–treated DLD cells (parental A4 line) are shown as a control. Expression of all mutants led to the detection of STAT3 protein, with the exception of p.A703T, a mutation near the binding site of the antibody used here (clone 79D7). This variant was detectable after IL-6 activation and with a different antibody clone (supplemental Figure 2). Mutant STAT3 proteins were not phosphorylated at baseline, but were phosphorylated after stimulation with IL-6. (C-D) STAT3 binding activity as measured by luciferase assay. A4 cells were transfected with WT or the indicated STAT3 mutants and STAT3-driven firefly luciferase and control renilla reporters. (C) Luciferase was assayed at 48 hours and STAT3 activity is shown as a ratio of firefly/control for each construct. All constructs with the exception of V353F had significantly increased activity at baseline compared with WT. Results represent the mean ± SEM of 5 to 10 independent experiments; the dotted line represents WT (**P < .01, ***P < .001, ****P < .0001). (D) STAT3 activity following 12-hour activation with IL-6 (10 ng/mL, white) or IFN-α (50 ng/mL, black). All mutations demonstrated significantly increased STAT3 activity after at least 1 of the cytokine stimulations (P < .05), typically after both unless indicated. Results shown represent fold-change vs WT for each cytokine stimulus; the dotted line represents a fold-change of 1 (no change from WT). Results represent the mean ± SEM of 5 independent experiments. ns, not significant; pSTAT3, phosphorylated STAT3.
In contrast to the recently reported patients with STAT3 mutations who predominantly had type I diabetes and enteropathy,6 within our cohort, hematologic autoimmunity was most prevalent including autoimmune hemolytic anemia, neutropenia, and/or thrombocytopenia (Table 1). Similar solid-organ autoimmunity was present in some of our patients, including autoimmune enteropathy (supplemental Table 3). Others also exhibited arthritis, lung disease consistent with lymphocytic interstitial pneumonia, hepatitis, atopic dermatitis, alopecia, and/or scleroderma.
Several patients also had recurrent and/or severe infections (Table 2), which was not a prominent feature of the 5 patients recently reported.6 We noted 4 patients with fungal infections. Whether this particular finding may become more common as more patients are described with this disorder remains to be seen. In several of the patients with recurrent infections, hypogammaglobulinemia was noted (Table 2) but only patient 4 fulfilled criteria for common variable immune deficiency (supplemental Table 4). Of note, the chronic immune suppression required in these patients has the potential to cause some, but not all, of these phenotypes.
Other immunologic laboratory parameters and infection history
| Patient . | STAT3 . | Laboratory parameters . | Severe or recurrent infections . | |||
|---|---|---|---|---|---|---|
| Hypogam . | % DNTs . | FAS-apoptosis defect . | Other (ALC nadir/mm3) . | |||
| 1 | p.G421R | Yes | 3.3 | n/a | T lymphopenia (208) | No |
| 2 | p.T663I | No | 1.6-2.6 | No | (3100) | Viral pneumonia, molluscum, ocular herpes |
| 3 | p.R152W | Yes* | 2.7-3.5 | No | (1300) | Recurrent herpes zoster |
| 4 | p.V353F | Yes | 2.9 | No | B lymphopenia, poor memory B cells (710) | Onychomycosis, skin HPV, herpes zoster, dermatophytic infection |
| 5 | p.Q344H | Yes | n/a | n/a | Disseminated Pseudomonas, Candida, and Serratia, UTI | |
| 6 | p.E415K | Transient | n/a | n/a | Neonatal sepsis, recurrent URI, esophageal candidiasis | |
| 7 | p.T716M | No | <1 | n/a | T lymphopenia (610) | Lung infections |
| 8 | p.N420K | No | 1.5, 3.4 | No | T lymphopenia (913) | Recurrent respiratory infections |
| Family 1 | ||||||
| 9, Proband | p.A703T | No | 1.4 | Yes | (2500) | No |
| 10, Father | p.A703T | Yes | n/a | n/a | Bacterial and fungal soft tissue, osteomyelitis | |
| 11, Sibling | p.A703T | No | n/a | Yes | n/a | |
| Family 2 | ||||||
| 12, Proband | p.T716M | No | n/a | n/a | High B cells | No |
| 13, Father | p.T716M | n/a | n/a | n/a | No | |
| Patient . | STAT3 . | Laboratory parameters . | Severe or recurrent infections . | |||
|---|---|---|---|---|---|---|
| Hypogam . | % DNTs . | FAS-apoptosis defect . | Other (ALC nadir/mm3) . | |||
| 1 | p.G421R | Yes | 3.3 | n/a | T lymphopenia (208) | No |
| 2 | p.T663I | No | 1.6-2.6 | No | (3100) | Viral pneumonia, molluscum, ocular herpes |
| 3 | p.R152W | Yes* | 2.7-3.5 | No | (1300) | Recurrent herpes zoster |
| 4 | p.V353F | Yes | 2.9 | No | B lymphopenia, poor memory B cells (710) | Onychomycosis, skin HPV, herpes zoster, dermatophytic infection |
| 5 | p.Q344H | Yes | n/a | n/a | Disseminated Pseudomonas, Candida, and Serratia, UTI | |
| 6 | p.E415K | Transient | n/a | n/a | Neonatal sepsis, recurrent URI, esophageal candidiasis | |
| 7 | p.T716M | No | <1 | n/a | T lymphopenia (610) | Lung infections |
| 8 | p.N420K | No | 1.5, 3.4 | No | T lymphopenia (913) | Recurrent respiratory infections |
| Family 1 | ||||||
| 9, Proband | p.A703T | No | 1.4 | Yes | (2500) | No |
| 10, Father | p.A703T | Yes | n/a | n/a | Bacterial and fungal soft tissue, osteomyelitis | |
| 11, Sibling | p.A703T | No | n/a | Yes | n/a | |
| Family 2 | ||||||
| 12, Proband | p.T716M | No | n/a | n/a | High B cells | No |
| 13, Father | p.T716M | n/a | n/a | n/a | No | |
ALC, absolute lymphocyte count; HPV, human papillomavirus; Hypogam, hypogammaglobulinemia; n/a, not available; URI, upper respiratory infection; UTI, urinary tract infection.
Hypogammaglobulinemia after starting immunosuppression.
Similar to the previously reported patients,6 we noted postnatal short stature <5th percentile in a majority of our patients (7 of 13 patients). Several patients exhibited profound growth failure as represented by their growth curves (supplemental Figure 1A-D). The growth hormone axis was assessed in 3 patients; 2 patients were treated with growth hormone with good response (supplemental Table 5).
Additional immunologic laboratory features among our cohort were variable but included moderate T-cell lymphopenia, hypogammaglobulinemia, and elevated double-negative (CD4negCD8neg) CD3+TCRα/β+ T cells (DNTs) in many patients (Table 2). The observation of elevated DNTs is not only a consistent feature of ALPS and FAS pathway defects, but has also been observed in pediatric patients with more common autoimmune diseases and likely reflects a lymphoproliferative state.1,2,10 FAS-mediated apoptosis was assayed in 6 patients, two of whom showed a defect (Table 2; supplemental Table 6). Targeted sequencing in this patient excluded defects associated with classic ALPS.
In 2 families (patient 4 and family 1), there were family members carrying a STAT3 mutation who had a less severe phenotype or were asymptomatic (father of patient 4, not included here). In family 1, patient 11 experienced a single childhood episode of lymphoproliferation and hematologic autoimmunity which resolved without sequelae. A third sibling in family 1 also carries the STAT3 mutation but is asymptomatic. Therefore, not only is the clinical phenotype of these patients diverse (Tables 1 and 2; supplemental Table 7), but there also appear to be carriers of these mutations who display incomplete penetrance.
The observed STAT3 mutations lead to enhanced transcriptional activity but not constitutive phosphorylation of STAT3
STAT3 encodes signal transducer and activator of transcription 3, 1 of a family of transcription factors that play critical roles in affecting cytokine-induced changes in gene expression.11-13 Following receptor ligation, Janus kinase (JAK) kinases recruit, phosphorylate, and activate STATs. STATs then translocate to the nucleus and interact with specific DNA elements as part of transcriptional regulatory complexes, which, depending on the cytokine and STAT molecule involved, can promote both proinflammatory and antiinflammatory pathways.
The stereotyped clinical phenotype of our patient cohort with multisystem autoimmunity and lymphoproliferation (Table 1 and 2; supplemental Table 7) was quite distinct from that associated with germline STAT3 mutations shown to confer a LOF (ie, hyper-IgE syndrome).14,15 Analogous to the contrasting allelic disorders caused by GOF and LOF mutations in STAT1,13,16,17 we hypothesized that the changes observed here in STAT3 lead to a GOF. Mechanistically, GOF STAT3 mutations could be predicted to lead to autoimmunity because of the known involvement of STAT3 in intracellular signaling pathways relevant to immune dysregulation, including proinflammatory signaling, inhibition of Tregs, and enhancement of Th17 cell fate determination.18-21 Furthermore, somatic GOF STAT3 mutations are reported in association with large granular lymphocytic leukemias and acquired aplastic anemia, both of which involve a prominent autoimmune component.8,22 Indeed, the recent series of 5 patients with autoimmunity and STAT3 mutations included evidence that the mutations conferred a GOF.6
To test the effect of the mutations found in our cohort on STAT3 function, constructs containing the variants were transfected into a STAT3-deficient cell line (A4 cells).9 Each variant produced detectable immunoreactivity via western blot (Figure 1B), however, the p.A703T mutation was detectable only with a different anti-STAT3 antibody, presumably due to a loss of the antibody binding site in the presence of the mutation (Figure 1B; supplemental Figure 2). STAT3 was not significantly phosphorylated at baseline in any mutant, but was phosphorylated normally in response to IL-6 stimulation (Figure 1B). At baseline, all mutants with the exception of p.V353F conferred significantly increased transcriptional activity compared with WT (Figure 1C). The cytokines IL-6 and IFN-α, both activators of STAT3, were used to stimulate the transfected cells. Statistically significant increases in STAT3 activity compared with WT were seen for each mutant with either 1 or the other cytokine and typically for both (Figure 1D).
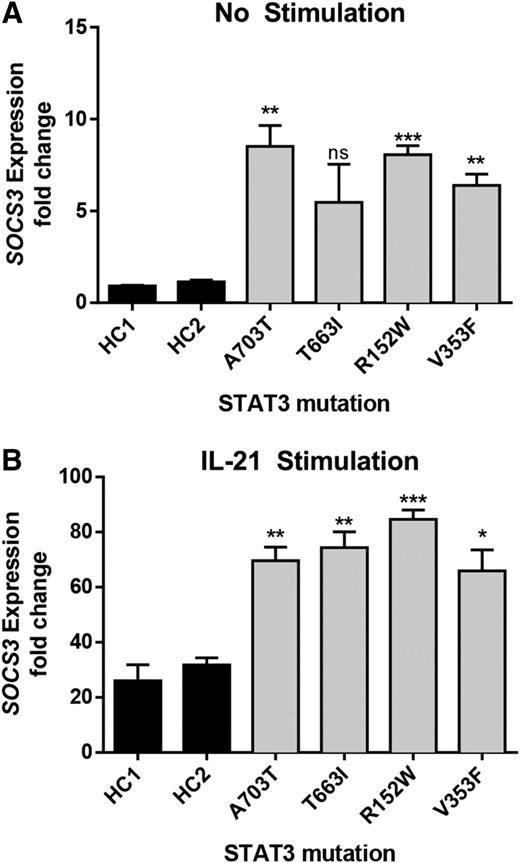
A major downstream target of STAT3 is SOCS3. Transcript levels of SOCS3 were elevated both at baseline and after IL-21 stimulation in EBV-transformed cell lines from 4 patients (Figure 2), including for mutant p.V353F, which did not have baseline activity higher than WT by luciferase. This finding highlights differences in these 2 assays that are measuring different outcomes, transfection and expression in a STAT3-deficient cell line and endogenous expression from a patient-derived cell line. It is possible that despite serum starvation prior to the outset of the experiments, there are still residual STAT3 agonist signals present in the STAT3 mutant cell line leading to a change in baseline activity. In combination, the findings in Figures 1-2 provide strong evidence that the observed mutations result in a gain of STAT3-mediated transcriptional function at baseline and/or with cytokine activation.
Elevated expression of SOCS3 in patient-derived EBV-transformed cell lines. Expression of the STAT3-target SOCS3 was determined by quantitative RT-PCR in resting (A) and (B) IL-21–activated (50 ng/mL for 21 hours) cell lines. Increased SOCS3 transcript was detected in patient-derived cells compared with healthy controls (HC1 and HC2). Results represent the fold-increased expression in SOCS3 transcript compared with unstimulated healthy control 1 (HC1) and are normalized to β-actin. Results represent the mean ± SEM of 3 independent experiments. *P < .05; **P < .01; ***P < .001.
Elevated expression of SOCS3 in patient-derived EBV-transformed cell lines. Expression of the STAT3-target SOCS3 was determined by quantitative RT-PCR in resting (A) and (B) IL-21–activated (50 ng/mL for 21 hours) cell lines. Increased SOCS3 transcript was detected in patient-derived cells compared with healthy controls (HC1 and HC2). Results represent the fold-increased expression in SOCS3 transcript compared with unstimulated healthy control 1 (HC1) and are normalized to β-actin. Results represent the mean ± SEM of 3 independent experiments. *P < .05; **P < .01; ***P < .001.
The effects of GOF STAT3 mutations on the phosphorylation of STAT3 in patients with autoimmune disease are unknown.6 Our transfection experiments suggested there was no constitutive phosphorylation of STAT3. To examine the effects of GOF STAT3 mutations on STAT3 phosphorylation in lymphocytes, we used EBV-transformed cell lines from 4 patients bearing different STAT3 mutations. Cell lines allowed us to minimize the variability of cellular activation state and the effects of concurrent treatment with immunosuppressants in primary patient samples. Similar to transfected cells, there was no phosphorylation of STAT3 at baseline, and phosphorylation of STAT3 in response to IL-21 was normal in all 4 patient-derived cell lines as compared with cell lines derived from healthy controls (supplemental Figure 3A). This finding was corroborated in primary cells from patient 1 (supplemental Figure 3B).
Impaired STAT5 and STAT1 phosphorylation with GOF STAT3 mutations
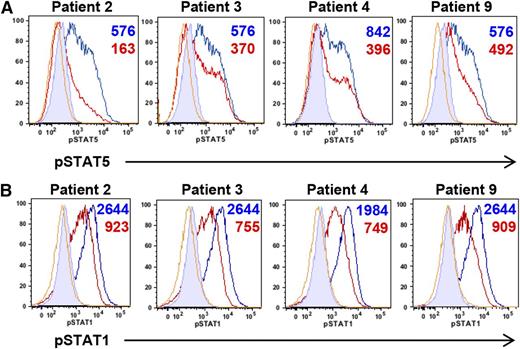
Our patients shared multiple clinical characteristics with STAT5b-deficient patients.23 Because phosphorylation of several STATs, including STAT5, can be negatively regulated by the STAT3-target SOCS3,24 we examined cytokine-induced phosphorylation of STAT5 in patient-derived EBV-transformed cell lines. In response to IL-2, STAT5 phosphorylation was lower than healthy controls (Figure 3A), suggesting that STAT3 GOF diminishes the STAT5 response. This was also found in primary cells from available patients (supplemental Figure 4). Phosphorylation of STAT5 remained decreased over the examined time course (supplemental Figure 5A). Similarly, STAT1 phosphorylation after IFN-γ was decreased in patient-derived cell lines across time points (Figure 3B, supplemental Figure 5B), as well as in available primary cells (supplemental Figure 6). These data suggest that upregulation of STAT3 transcriptional activity may have consequences for other cytokine signaling pathways.
Increased STAT3 activity in patient-derived EBV-transformed cell lines leads to decreased phosphorylation of other STAT molecules. (A) Impaired STAT5 phosphorylation in EBV-transformed cell lines derived from GOF STAT3 patients. Cells from patients (red) and healthy controls (blue) were stimulated with IL-2 (80 U/mL) and pSTAT5 measured by flow cytometry after 20 minutes. Numbers indicate the difference in MFI of phospho-STAT5 obtained by subtracting the MFI of unstimulated from stimulated samples. Yellow lines represent unstimulated patient samples; filled histograms, unstimulated healthy control samples. (B) Impaired STAT1 phosphorylation in EBV-transformed cell lines derived from GOF STAT3 patients. Cells from patients (red) and healthy controls (blue) were stimulated with IFN-γ (500 ng/mL) and pSTAT1 measured by flow cytometry after 20 minutes. Numbers indicate the difference in MFI of phospho-STAT1 obtained by subtracting the MFI of unstimulated from stimulated samples. Yellow lines represent unstimulated patient samples and filled histograms unstimulated healthy control samples. MFI, mean fluorescence intensity.
Increased STAT3 activity in patient-derived EBV-transformed cell lines leads to decreased phosphorylation of other STAT molecules. (A) Impaired STAT5 phosphorylation in EBV-transformed cell lines derived from GOF STAT3 patients. Cells from patients (red) and healthy controls (blue) were stimulated with IL-2 (80 U/mL) and pSTAT5 measured by flow cytometry after 20 minutes. Numbers indicate the difference in MFI of phospho-STAT5 obtained by subtracting the MFI of unstimulated from stimulated samples. Yellow lines represent unstimulated patient samples; filled histograms, unstimulated healthy control samples. (B) Impaired STAT1 phosphorylation in EBV-transformed cell lines derived from GOF STAT3 patients. Cells from patients (red) and healthy controls (blue) were stimulated with IFN-γ (500 ng/mL) and pSTAT1 measured by flow cytometry after 20 minutes. Numbers indicate the difference in MFI of phospho-STAT1 obtained by subtracting the MFI of unstimulated from stimulated samples. Yellow lines represent unstimulated patient samples and filled histograms unstimulated healthy control samples. MFI, mean fluorescence intensity.
Impaired Treg compartment in patients with GOF STAT3 mutations
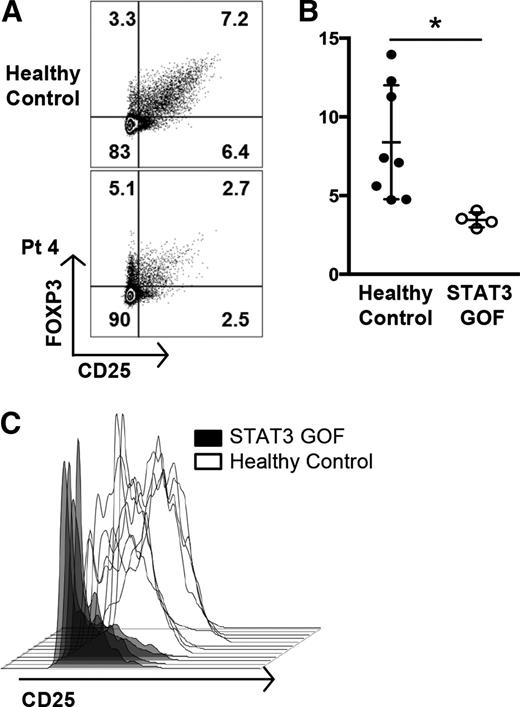
STAT3 is known to play an important role in influencing T-cell linage decisions as they integrate environmental cues.20,21 Specifically, SOCS3, which is upregulated by STAT3, can suppress Treg function.25,26 The Treg compartment in 2 of the recently described patients with GOF mutations in STAT3 was reportedly reduced.6 Analysis of peripheral blood cells from our patients within the same institution (Figure 4A) showed diminished numbers of Tregs as well as decreased expression of CD25, which is required for Treg function (Figure 4B-C). Two additional patients examined at other institutions also exhibited decreased expression of FOXP3 and/or CD25 (supplemental Figure 7).
STAT3 GOF patients have decreased Treg numbers and functional markers. (A) Flow cytometric analysis of FOXP3+CD25+ Treg populations from patient 4, gated on CD4+ T cells, compared with a healthy control. (B) Decreased Treg percentages in STAT3 GOF patients. Treg populations were determined by flow cytometry. The percentages of CD4+ T cells that were FOXP3+CD25+ Tregs from patients were decreased compared with 6 healthy controls and 2 patients with ALPS who had FAS defects (*P < .05). (C) Tregs from STAT3 GOF patients have decreased expression of CD25. Treg populations (CD4+ FOXP3+CD127low) were determined by flow cytometry and the CD25 expression further analyzed by histogram. Tregs from patients demonstrate lower levels of CD25 expression than healthy controls.
STAT3 GOF patients have decreased Treg numbers and functional markers. (A) Flow cytometric analysis of FOXP3+CD25+ Treg populations from patient 4, gated on CD4+ T cells, compared with a healthy control. (B) Decreased Treg percentages in STAT3 GOF patients. Treg populations were determined by flow cytometry. The percentages of CD4+ T cells that were FOXP3+CD25+ Tregs from patients were decreased compared with 6 healthy controls and 2 patients with ALPS who had FAS defects (*P < .05). (C) Tregs from STAT3 GOF patients have decreased expression of CD25. Treg populations (CD4+ FOXP3+CD127low) were determined by flow cytometry and the CD25 expression further analyzed by histogram. Tregs from patients demonstrate lower levels of CD25 expression than healthy controls.
PBMCs from a patient with a GOF STAT3 mutation exhibit delayed kinetics of dephosphorylation
The mechanism by which STAT3 GOF produces downstream effects such as increased SOCS3 transcription could not be explained by hyperphosphorylation (supplemental Figure 3), as has been reported for GOF mutations in STAT1.17 However, after stimulation with IL-6, PBMCs from patient 1 exhibited delayed dephosphorylation of pSTAT3 compared with healthy controls (supplemental Figure 8). This may provide an alternative explanation for increased STAT3 activity in some of these patients.
Two patients with germline GOF STAT3 mutations received hematopoietic stem cell transplants
All of the symptomatic patients reported here required significant immunosuppression to control their autoimmune diseases and 2 patients (patients 7 and 8) underwent hematopoietic stem cell transplantation (HSCT) for refractory autoimmunity as detailed in supplemental Table 8. Patient 8 died 138 days out from transplant of severe graft-versus-host disease with disseminated adenoviral infection. Patient 7 is alive and well with improved growth and complete remission of her autoimmune disease, and in particular she has no signs of the previously severe autoimmune enteropathy.
Blockade of the IL-6 pathway in a patient with a GOF STAT3 mutation resulted in clinical improvement and decreased Th17 cells
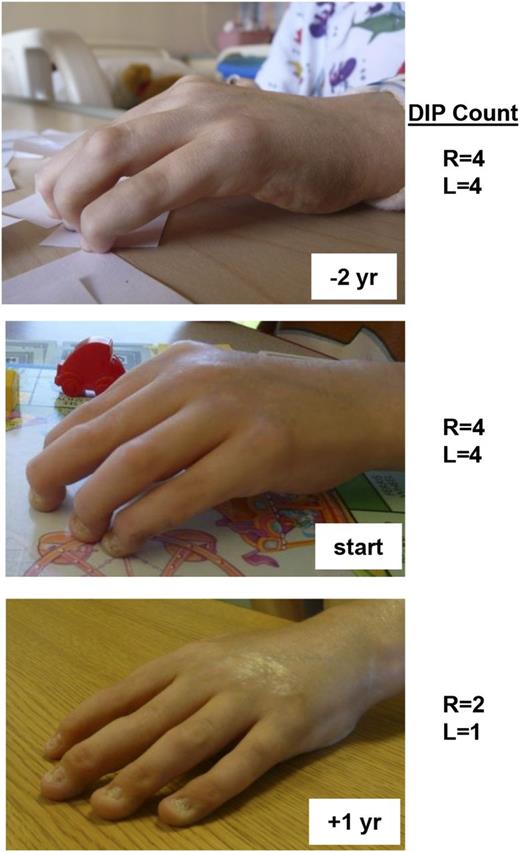
Over the span of 4 years, the arthritis and scleroderma-like skin changes in patient 1 failed to respond to treatment with anti–tumor necrosis factor α (TNFα), anti–IL-1, and anti–B-cell therapies, although his severe autoimmune hemolytic anemia responded well to rituximab. IL-6 is one of the primary cytokines that utilizes STAT3 for signal transduction. When considering additional therapeutic options for this patient after discovering his GOF STAT3 mutation, we hypothesized that IL-6 blockade might ameliorate his disease and so he was treated with the anti-IL6R monoclonal antibody therapy tocilizumab. Within months of starting tocilizumab, he exhibited a dramatic improvement of previously fixed flexion contractures of his distal interphalangeal joints (Figure 5). Similarly, his inability to fully open his hands due to a combination of metacarpal-phalangeal joint contractures and skin tightening also improved as measured by 2 pediatric rheumatologists. In keeping with the expected outcome of modulating STAT3 activity with treatment, Th17 cells were initially elevated in this patient and normalized after anti-IL6 treatment (supplemental Figure 9). Th17 frequencies were measured in 6 other patients but did not appear to be elevated (supplemental Figure 9), although concurrent immunosuppressive regimens preclude definitive conclusions regarding the potential role of Th17 cells in this patient cohort.
Patient response to anti-IL6R monoclonal antibody therapy. Patient 1 received therapy with the anti-IL6R monoclonal antibody tocilizumab. He had notable improvement of his long-standing polyarthritis and skin tightening as shown in the photographs (top, 2 years before treatment; middle, at initiation of treatment; and bottom, after one year of treatment). Indicated are the number of fixed DIP (distal interphalangeal) contractures of both his right and left hands at the designated times with respect to anti-IL-6R therapy. L, left; R, right; yr, year.
Patient response to anti-IL6R monoclonal antibody therapy. Patient 1 received therapy with the anti-IL6R monoclonal antibody tocilizumab. He had notable improvement of his long-standing polyarthritis and skin tightening as shown in the photographs (top, 2 years before treatment; middle, at initiation of treatment; and bottom, after one year of treatment). Indicated are the number of fixed DIP (distal interphalangeal) contractures of both his right and left hands at the designated times with respect to anti-IL-6R therapy. L, left; R, right; yr, year.
Discussion
Here, we describe GOF STAT3 mutations in a cohort of patients with lymphoproliferation and childhood-onset autoimmunity. We demonstrate that these mutations confer increased STAT3 transcriptional activity, impair cytokine signaling via other STAT molecules, and diminish the Treg compartment.
Our findings further support a role for human STAT3 in immune regulation and extend the spectrum of STAT3-associated disease. Patients bearing germline LOF STAT3 mutations have immunodeficiency and exhibit infections quite distinct from those observed in this cohort, along with impaired Th17 differentiation, markedly higher levels of IgE, and far more common connective tissue abnormalities.14,15,21 Somatic GOF mutations in STAT3 are associated with lymphoproliferative neoplasms, acquired aplastic anemia, and myelodysplastic syndrome, but again, appear to be distinct from the phenotype in this cohort, especially given the relative dearth of neoplastic disease among these patients with congenital GOF mutations.8,11,22
Although this cohort shares common clinical characteristics, including autoimmune cytopenias, lymphoproliferation, and short stature, affected individuals varied in their age at presentation and extent of solid-organ autoimmunity. Four of the 5 patients recently reported by Flanagan et al with early-onset autoimmunity and STAT3 mutations had neonatal diabetes.6 Among them was a young patient with type I diabetes and autoimmune enteropathy bearing the same c.2147C>T mutation (p.T716M) observed in patient 7 and family 2. Although our patients with p.T716M mutations did not have type I diabetes, 2 of our cohort with other STAT3 mutations did. This partial overlap in phenotype suggests that germline GOF mutations in STAT3 produce a strong susceptibility to immune dysregulation. However, the specific target organs involved can differ even between patients with the same mutation, similar to other monogenic disorders of autoimmunity.5
In addition to an ALPS-like phenotype, patients in our cohort share many clinical features of patients with STAT5b deficiency. Mutations in STAT5b can lead to endocrine abnormalities, short stature, recurrent infection, eczema, autoimmunity, and lymphocytic interstitial pneumonia.27 This is presumably due, in part, to defects in responses to growth hormone and in Treg number and function, both of which depend upon normal STAT5b signaling.27,28 Furthermore, the immune dysregulatory phenotype seen in some of our cohort resembles that of patients with CD25 or FOXP3 mutations leading to the loss of Tregs.3 As we observed defects in both pSTAT5 responses and the Treg compartment in our patients, we hypothesize that a possible mechanism for autoimmunity in STAT3 GOF is impaired absolute Treg function as a result of diminished STAT5 signaling, possibly related to increased SOCS3 activity. An absolute reduction in function could be reflective of diminished numbers of Tregs as we show. This could also be related to a defect in Treg function, which we are not able to assess due to leukopenia and decreased surface expression of CD25, which have precluded sorting the CD25+ Treg population in our patients.
The mechanism underlying the GOF in STAT3 has not yet been elucidated, but we provide evidence for at least 1 potential mechanism by showing delayed phosphorylation kinetics after stimulation of primary patient cells from patient 1 with IL-6. This may be the result of enhanced DNA binding (this mutation is in the DNA binding domain), leading to prolonged nuclear retention of STAT3. It is probable that the mechanism of GOF STAT3 mutations will be different depending upon the particular mutation and protein domain affected, similar to what has been reported with LOF STAT3 mutations.29
Two patients with GOF STAT3 mutations received HSCT for their refractory autoimmunity, and one of those patients had complete remission of her autoimmune disease. Due to the low numbers of transplanted patients, it is not possible to give an estimate of the likelihood of success of HSCT in this condition. However, 16 children with similarly severe and complex autoimmune disease have also undergone HSCT in Newcastle; 14 are alive and 12 are in complete remission from their disease (A.J.C., Newcastle University, Newcastle upon Tyne, United Kingdom, written communication, October 15, 2014). These data suggest that replacement of the hematopoietic compartment may alleviate tissue-specific autoimmune disease.
One of the patients here was treated with anti-IL6R monoclonal antibody therapy with significant improvement of his otherwise refractory polyarthritis and skin disease. Although this observation was made in a single patient, this patient’s response suggests that treatment of other patients with STAT3-associated autoimmunity with therapies targeting the STAT3 pathway is worth investigating. In addition to inhibitors of the IL-6 pathway, small-molecule inhibitors of STAT3 are under clinical investigation for malignancy.30,31
In summary, we describe a relatively large cohort of patients with a monogenic disorder of early-onset autoimmunity and lymphoproliferation caused by GOF germline STAT3 mutations. These findings significantly expand the clinical spectrum of this newly described monogenic disorder and provide evidence for a potential mechanism whereby this GOF leads to altered immune cell tolerance via interference with the activation of other STAT molecules. These findings suggest a broad range of autoimmune and lymphoproliferative disorders may be explained by inborn errors of cytokine signaling. They also provide a new model for studying the biology and clinical consequences of the complex regulatory networks involved in STAT-mediated signaling. Furthermore, we provide a rationale for the use of inhibitors of this pathway, including anti-IL-6R therapy as shown here or, potentially, small-molecule inhibitors of STAT3, to achieve substantial therapeutic benefit in this and related conditions.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs F. S. Cole, S. Holland, G. Stark, A. Weymann, J. Tarbox, M. Lenardo, K. Butler, and A. Laurence for reagents, patient care, and/or helpful discussion.
This work was supported in part by the intramural research program of the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH). Work in M.A.C.’s laboratory was supported by The Children’s Discovery Institute and St. Louis Children’s Hospital, The Scleroderma Foundation, the Rheumatic Diseases Core Center at Washington University (P30AR048335), and NIH training grant 2T32AR007279 (T.P.V.). Analysis of Washington University exome data was performed by the Human Genetics Facility of the Rheumatic Diseases Core Center (E.D.O.R.), supported by National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) P30AR048335. Work in S.H.’s laboratory was supported by the Sir Jules Thorn Charitable Trust. The Baylor-Hopkins Center for Mendelian Genomics was supported by the NIH National Human Genome Research Institute (U54HG006542 and U54HG003273). Genomic analysis by the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine was partially supported by National Cancer Institute (NCI) Cancer Center support grant P30 CA91842 to the Siteman Cancer Center and Institute of Clinical and Translational Sciences/Clinical & Translational Science Awards (ICTS/CTSA) grant UL1TR000448 from the National Center for Research Resources, a component of the NIH, and the NIH Roadmap for Medical Research. The work at the Children’s Mercy Center for Pediatric Genomic Medicine was supported by the WT Kemper Foundation, the Marion Merrell Dow Foundation, Black & Veatch, the Pat and Gil Clements Foundation, the Claire Giannini Foundation, and U19HD077693 from the National Institute of Child Health and Human Development (NICHD).
This publication is solely the responsibility of the authors and does not necessarily represent the official view of the NIH.
Authorship
Contribution: M.A.C., J.D.M., T.P.V., L.F., and S.H. wrote the manuscript, designed and performed research, analyzed and interpreted data, and provided study material and patients; C.A.M. and A.S.-P. designed and performed research, and analyzed and interpreted data; J.E.N., J.J.L., Y.Z., M.P.O., J.M., J.D.H., N.T., E.D.O.R., H.C.S., J.W.V., T.D., K.N., G.M., E.M.M., A.V.-H., H.S.S., K.R.E., Y.X., and S.F.K. performed research and analyzed and interpreted data; S.P., M.B., V.S., D.L.D., C.E.C., C.J.S., and S.S. analyzed and interpreted data and provided study material and patients; J.W. and D.S. analyzed and interpreted data; M.S.-K. designed the research; and H.M., V.K.R., H.C.S., A.J.W., N.V., K.L.M., L.B.K., G.P., and A.J.C. provided study material and patients.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Joshua D. Milner, NIH, Building 10-CRC, Room 5-3950, Bethesda, MD 20892; e-mail: jdmilner@niaid.nih.gov; and Megan A. Cooper, Department of Pediatrics, 660 S. Euclid Ave, Box 8208, Washington University School of Medicine, St. Louis, MO 63110; e-mail: cooper_m@kids.wustl.edu.
References
Author notes
J.D.M., T.P.V., L.F., and C.A.M. contributed equally to this work.