Key Points
High-throughput RNAi screening identified lenalidomide sensitizer genes, including RSK2, RAB, peroxisome, and potassium channel family members.
Knockdown or inhibition of RSK2 synergized with lenalidomide to induce myeloma cytotoxicity and downregulation of interferon regulatory factor 4 and MYC.
Abstract
To identify molecular targets that modify sensitivity to lenalidomide, we measured proliferation in multiple myeloma (MM) cells transfected with 27 968 small interfering RNAs in the presence of increasing concentrations of drug and identified 63 genes that enhance activity of lenalidomide upon silencing. Ribosomal protein S6 kinase (RPS6KA3 or RSK2) was the most potent sensitizer. Other notable gene targets included 5 RAB family members, 3 potassium channel proteins, and 2 peroxisome family members. Single genes of interest included I-κ-B kinase-α (CHUK), and a phosphorylation dependent transcription factor (CREB1), which associate with RSK2 to regulate several signaling pathways. RSK2 knockdown induced cytotoxicity across a panel of MM cell lines and consistently increased sensitivity to lenalidomide. Accordingly, 3 small molecular inhibitors of RSK2 demonstrated synergy with lenalidomide cytotoxicity in MM cells even in the presence of stromal contact. Both RSK2 knockdown and small molecule inhibition downregulate interferon regulatory factor 4 and MYC, and provides an explanation for the synergy between lenalidomide and RSK2 inhibition. Interestingly, RSK2 inhibition also sensitized MM cells to bortezomib, melphalan, and dexamethasone, but did not downregulate Ikaros or influence lenalidomide-mediated downregulation of tumor necrosis factor-α or increase lenalidomide-induced IL-2 upregulation. In summary, inhibition of RSK2 may prove a broadly useful adjunct to MM therapy.
Introduction
The immunomodulatory drugs (IMiDs) thalidomide, lenalidomide, and pomalidomide are broadly used in the treatment of patients with multiple myeloma (MM) and have produced significant clinical advances in both newly diagnosed and advanced disease.1,2 However, only 30% of relapsed MM patients respond to single agent IMiD therapy and most patients eventually develop drug resistance. The underlying mechanisms defining this nonresponsiveness are largely unknown. Recently, Cereblon (CRBN) and Ikaros/Aiolos (IZKF1 and IZKF3) were identified as the molecular targets of IMiDs in MM3-6 via their ability to induce cytotoxicity by inhibiting interferon regulatory factor 4 (IRF4) and MYC. The action of IMiDs appears to be mediated via binding to CRBN with activation of its associated E3 ubiquitin ligase and subsequent proteasomal degradation of Ikaros.5,6 Furthermore, low CRBN and IZKF1 levels are associated with IMiD resistance,3,4,7,8 but other parallel pathways or downstream events that enhance or preclude drug responsiveness are unknown.
In the present study, a druggable genome short interfering RNA (siRNA) screen was used to identify 63 genes whose loss of expression enhances lenalidomide sensitivity in human MM cells. The strongest sensitizer was the kinase, RSK2. Using short hairpin RNA (shRNA) expression systems and small molecule inhibitors of RSK2, we validated RSK2 as the top IMiD sensitizer gene in MM cells, and demonstrated that inhibition of RSK2 is also independently cytotoxic and a broad sensitizer to MM chemotherapy through its ability to downregulate IRF4 and MYC independently of IMiDs.
Materials and methods
Cell lines, compounds, siRNA, plasmids, and reagents
Myeloma cell lines were maintained in RPMI 1640 media, supplemented with 5% to 10% fetal bovine serum and antibiotics. Lenalidomide was purchased from LC Laboratories (Woburn, MA). The Human Druggable Genome siRNA Set V3 was purchased from Qiagen (Valencia, CA). Lentiviral shRNA clones targeting RSK2 and control lentiviral constructs expressing nontargeting (NT) shRNA were purchased from The MISSION TRC shRNA libraries of Sigma-Aldrich (St. Louis, MO) as sequence-verified bacterial stocks. Human RSK2 lentiviral expression construct was purchased from GeneCopoeia (Rockville, MD). Anti-RSK2, anti-pRSK2 (Ser 227, D53A11), anti-MCL1, anti-PARP, anti-BIM, and anti-IRF4 antibodies were purchased from Cell Signaling Technology (Danvers, MA) and anti-MYC was from Epitomics (Burlingame, CA). Lipofectamine 2000 was purchased from Invitrogen (Carlsbad, CA) and MTT reagent was from Sigma-Aldrich. Phorbol myristate acetate (PMA), ionomycin, and lipopolysaccharide (LPS) were also from Sigma-Aldrich. Anti-human IL-2–APC (allophycocyanin) and the enzyme-linked immunosorbent assay (ELISA) kit for detection of tumor necrosis factor-α (TNFα) were purchased from eBioscience (San Diego, CA). TaqMan Universal PCR Master Mix and quantitative polymerase chain reaction (qPCR) probes for IRF4 and MYC were purchased from Applied Biosystems (Grand Island, NY).
High-throughput siRNA screening
SiRNAs (4 siRNA oligos per gene) were preprinted on 384-well plates alongside staggered negative (ALLStars-NT and green fluorescent protein [GFP] siRNAs) and positive control siRNAs. Primary screening experiment was conducted on KMS11 cells in the presence of various doses of lenalidomide. Briefly, the frozen plates preloaded with siRNA were thawed at room temperature and 20 μL of diluted lipofectamine 2000 solution was added to each well. After 30 minutes, 1000 cells in 20 μL of medium were added per well and then cultured at 37°C; 10 μL of medium containing various concentrations of lenalidomide was added after 24 hours and cell viability was determined at 144 hours (day 5 after lenalidomide addition) by CellTiter-Glo luminescence assay and read on a Molecular Devices Analyst GT multimode plate reader.
Hit selection and secondary screening
The raw luminescence values collected from the primary high-throughput siRNA screens were processed and annotated with their respective target gene, plate wells, and siRNA IDs. Each gene was targeted with 4 siRNA sequences and each siRNA sequence targeting a gene was treated with 5 different drug doses. A 4-parameter sigmoidal curve-fitting method was applied for hit selection as described.9 The EC50 for each siRNA was calculated and compared with the EC50 of controls (the plate median). Any significant shift (a twofold difference) in the EC50 of the sample siRNA compared with the control EC50 was determined to be a sensitizer hit. After further removing hits due to screen-related quality errors and compared with gene expression profile and our other screening data, a total of 160 potential candidate targets were selected from the primary screen.
The confirmation screening was performed with 4 original siRNAs, 4 drug doses (0,10, 37, and 200 μM), and 2 biological runs, and GFP siRNA was chosen as the negative control reference. The data collected from each siRNA at different treatments were normalized to the corresponding averaged GFP control siRNAs for each well. The output from ratio normalization was then used as an input to RNAi gene enrichment ranking algorithm (RIGER).10 RIGER, which is a java extension of the GENE-E software package (http://www.broadinstitute.org/cancer/software/GENE-E/) was applied to determine the enrichment of multiple siRNAs targeting the same gene. The signal-to-noise metric for ranking siRNAs and the Kolmogorov–Smirnov method were used to convert individual siRNA to genes. RIGER is nonparametric in its approach, and uses Gene Set Enrichment Analysis methodology11 and Kolmogorov–Smirnov to calculate gene scores from multiple siRNAs targeting a gene.
The output generated by RIGER is a list of ranked genes with normalized enrichment scores (NES).11 SiRNAs are assigned a score based on their differential phenotypic effect between 2 classes. False discovery rate, which is computed as a P value, is also assigned to each gene. The dataset was split into 2 untreated (siRNA+vehicle) and treated (siRNA+drug) classes. RIGER algorithm was applied to the dataset. A P value of <.05 was used to select the list of sensitizer hits and a P value <.01 was used to select top sensitizer hits.
Pathway/network enrichment analysis
To assess possible interactions between sensitizers after lenalidomide treatment, pathway/network analysis was performed using MetaCore software (GeneGo, Thomson Reuters). Briefly, the genes identified in the screening were overlaid onto a preliminary global molecular network developed from information contained in the MetaCore database. Analysis was set up using human data with a P value of <.01 as a cutoff. Enrichment analysis consisted of matching gene IDs of possible targets for the “common,” “similar,” and “unique” sets with gene IDs in functional ontologies in MetaCore. The probability of a random intersection between a set of IDs and the size of target list with ontology entities is estimated in the P value of hypergeometric intersection. The lower P value means a higher relevance of the entity to the dataset, which shows in a higher rating for the entity.
Validation by introduction of RSK2 shRNAs and RSK2 complementary DNA
Five lentiviral RSK2 shRNA constructs were obtained and modified by replacing the puramycin-resistant gene expression cassette with the GFP expression cassette. Lentivirus were generated and used to infect myeloma cells as described.3 The infection efficiency was measured by FACScan analysis of GFP expression at 48 hours postinfection. Cell viability assay was set up 48 hours postinfection in the absence or presence of different doses of lenalidomide, and measured by MTT assay at day 5 or 6 posttreatment. Cells were harvested at 48 or 72 hours postinfection for western blot analysis of RSK2 expression. Three of the RSK2 shRNAs were demonstrated to knockdown RSK2 efficiently. The targeting sequences for each shRNA were: 5′-GCCTGAAGATACATTCTATTT-3′, 5′-CCGGAATCTATTCGAATTTGT-3′, and 5′-GCTCAGCGGAGAGGTATTAAA-3′.
For assessment of lenalidomide sensitivity following exogenous RSK2 expression, H929 cells were infected with lentivirus expressing wild-type human RSK2 or control (empty vector) virus. After selecting the infected cells with puromycin and confirming RSK2 expression by immunoblotting, cell viability with and without lenalidomide for 5 days was measured by MTT assay.
Small molecule RSK2 inhibitor studies
To assess whether the RSK2 inhibitors BI-D1870, RMM46, and SL0101-1 synergize with lenalidomide to induce cytotoxicity of myeloma cells, various myeloma cell lines were treated with different combinations of each drug for 5 to 6 days, followed by MTT assay. The data were normalized and analyzed using CalcuSyn mathematical modeling12,13 to calculate a combination index and generate an isobologram.
For experiments with bone marrow stromal cells (BMSCs), BMSCs were generated from bone marrow samples of MM patients. Briefly, mononuclear cells were isolated from patient bone marrow samples and cultured in 10 mL RPMI 1640 supplemented with 10% fetal bovine serum. After the first week, the medium and suspension cells were removed and the media was replenished every 3 days. When an adherent cell monolayer developed, the adherent cells were trypsinized to harvest BMSCs. In MTT assay, 5000/well BMSCs were seeded in 96-well plates overnight before adding myeloma cells and the drug.
The ELISA analysis of TNFα and staining for intracellular IL-2 production
Human peripheral blood mononuclear cells (PBMCs) were isolated from buffy coats of healthy donors by Ficoll-Hypaque gradient centrifugation. For TNFα production, PMBCs were pretreated with lenalidomide (1 μM) or BI-D1870, or a combination of both drugs for 1 hour before stimulation with LPS (1 ug/mL) for 18 hours. The supernatants were harvested and tested for TNFα production by ELISA. For IL-2 production, Jurkat T cells were pretreated with lenalidomide (1 μM) or BI-D1870, or a combination of both drugs for 1 hour, and then stimulated with PMA (1 ng/mL)/ionomycin (1 μg/mL) for 40 hours. The cells were fixed and permeabilized by the addition of cytofix/cytoperm solution, stained with anti-IL-2–APC and analyzed by flow cytometry.
qPCR analysis of IRF4 and MYC expression
Total RNA was isolated using RNeasy Plus Mini Kit (QIAGEN) and reverse-transcribed using QuantiTect Reverse Transcription Kit (QIAGEN). qPCR was performed using TaqMan Universal PCR Master Mix with predesigned probes, and the comparative CT (threshold cycle) method was used for relative quantification on an ABI 7900HT Fast Real-Time PCR System (Applied Biosystems).
Western blot
Western blots were performed on whole-cell lysate extracted from various control and experimental samples. Briefly, equal amounts of protein were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels followed by transfer to polyvinylidene difluoride membranes. Membranes were probed with primary antibodies overnight. Detection was performed by the enhanced chemical luminescence method.
Results
Identification of lenalidomide sensitizer genes from high-throughput RNAi screening
To identify genes that are critical for drug response to lenalidomide, a synthetic siRNA screen with 27 968 siRNAs targeting 6992 druggable genes (one siRNA per well) was performed. An adherent KMS11 human MM line (HMCL) was reverse transfected using conditions that resulted in 98% successful transfection and <10% nonspecific lethality. A dose-response curve for lenalidomide was established against this cell line under screening conditions, and during screening, increasing doses of lenalidomide were added by robot to KMS11 cells 24 hours posttransfection. The results from primary screening were evaluated for multiple quality control metrics. All expected performance parameters passed the quality control with >95% global transfection efficiency and <0.15 cross-validation values across the dataset. A heat map of the raw data from the total 27 508 data points in the primary screen demonstrated the successful titration of cytotoxic response to increasing doses of lenalidomide (see supplemental Figure 1A, available on the Blood Web site). A waterfall plot of EC50 values for each siRNA from the primary screen that affect lenalidomide sensitivity after knockdown identified gene targets of interest (supplemental Figure 1B). Using a 2-log difference from the mean as a cutoff, we identified 449 sensitizer candidates (a 6.4% hit rate) and 96 resistance-inducing candidates (a 1.37% hit rate). The primary screen hit rate was reduced by including only plausibly expressed gene candidates and only genes with at least 2 siRNAs sensitizing MM cells to lenalidomide. Thus, a total of 160 genes were selected for secondary confirmation screening (supplemental Data 1C). Confirmatory screening was performed in duplicate using the 4 original siRNA oligos (4 siRNAs per gene) in the presence or absence of 3 concentrations of lenalidomide. Each data point for siRNA was normalized to GFP control siRNAs, followed by analysis using the RIGER algorithm. All siRNAs were ranked based on their lethal activity changes before and after lenalidomide treatment (supplemental Figure 2). In secondary screening, we confirmed 63 genes that significantly enhanced lenalidomide cytotoxicity upon silencing (P < .05) (supplemental Figure 2). Furthermore, 23 of the 63 genes were more potent lenalidomide modulators (P < .01). Ribosomal protein S6 kinase (RPS6KA3 or RSK2) was at the top of the ranking list with a P value of .0005 (Figure 1A). Figure 1B lists the top ranked genes with NES. RSK2, I-κ-B kinase-α (IKK1 or CHUK), and a phosphorylation-dependent transcription factor (CREB1) were all sensitizers, and together associate to regulate several signaling pathways such as the toll signaling pathways. Several sensitizers that encode proteins belonging to the same family were identified, including 5 vesicle trafficking GTPase RAB family proteins (RAB4A, RAB8A, RAB26, RAB30, and RAB36), 3 genes encoding potassium channel proteins (KCNE2, KCNG2, and KCNQ1), and 2 genes encoding peroxisome (PEX) family proteins (PEX1 and PEX10).
Top ranking lenalidomide sensitizers identified from druggable genome screens. The data collected from each siRNA were normalized and divided into untreated (siRNA only) and treated (siRNA+drug) classes, followed by analysis using the RIGER algorithm. When a stringent P value of <.01 was applied, 23 genes were selected as top lenalidomide modulators. (A) The 4 siRNAs targeting the top 23 sensitizer hits were ranked using a heat map plot based on their lethal activity changes after lenalidomide treatment. Relative: GENE-E converts values to heat map colors using the mean and maximum values for each row or the standard deviations from the row mean for each row (Source: https://www.broadinstitute.org/cancer/software/GENE-E/doc.html). “Untreated vs treated” are scores generated by the RIGER method for each siRNA. It is a score that is based on siRNA only (untreated) vs siRNA+lenalidomide (treated). (B) A list of top ranked genes with NES and P values are shown.
Top ranking lenalidomide sensitizers identified from druggable genome screens. The data collected from each siRNA were normalized and divided into untreated (siRNA only) and treated (siRNA+drug) classes, followed by analysis using the RIGER algorithm. When a stringent P value of <.01 was applied, 23 genes were selected as top lenalidomide modulators. (A) The 4 siRNAs targeting the top 23 sensitizer hits were ranked using a heat map plot based on their lethal activity changes after lenalidomide treatment. Relative: GENE-E converts values to heat map colors using the mean and maximum values for each row or the standard deviations from the row mean for each row (Source: https://www.broadinstitute.org/cancer/software/GENE-E/doc.html). “Untreated vs treated” are scores generated by the RIGER method for each siRNA. It is a score that is based on siRNA only (untreated) vs siRNA+lenalidomide (treated). (B) A list of top ranked genes with NES and P values are shown.
To assess for possible interactions between the 63 sensitizers, pathway/network analysis was performed using MetaCore software (supplemental Figure 3). The top pathway maps included several pathways involved in the development and immune response, such as IGF1, IL-6, and toll-mediated signaling, which are also known to associate with MM cell growth and survival.14,15
RSK2 knockdown inhibits MM growth and enhances lenalidomide-mediated MM cytotoxicity
Being the most potent lenalidomide modulator identified, RSK2 was further validated in the present study. Phosphorylated RSK2 was detected in all tested HMCLs (data not shown). The gene expression profile data indicated that most primary MM samples express RSK2, but no correlation was detected between RSK2 expression and IMiD response (data not shown). To study RSK2 function in MM cells, 5 RSK2 shRNA lentiviral constructs were obtained and 3 of these knocked down RSK2 efficiently. In 3 genetically variable MM cell lines, western blot analysis confirmed that RSK2 was knocked down at 48 hours (H929 and MM1.S) or 72 hours (JJN3) after infection (Figure 2A). Introduction of RSK2 shRNAs induced a substantial reduction of cell viability at day 6 postinfection in MM1.S and H929 cell lines, but only a 10% viability reduction was observed in the lenalidomide-resistant JJN3 cell line (Figure 2B). Furthermore, lenalidomide-induced cytotoxicity was enhanced in RSK2 shRNA lentivirus infected cells vs control cells (Figure 2C). Interestingly, despite the comparative lack of direct RSK2 knockdown-induced cytotoxicity, JJN3 cells, which are resistant to lenalidomide, became sensitive after the introduction of RSK2 shRNAs.
RSK2 knockdown inhibited myeloma growth and enhanced lenalidomide-mediated myeloma cytotoxicity. (A) JJN3 (left), MM1.S (middle), and H929 (right) cells were infected with control virus (NT) and RSK2 shRNAs (#52, #54, and #55) lentivirus, and immunoblotting was performed at 48 or 72 hours after virus infection to confirm RSK2 knockdown by shRNAs. (B) Cell viability was measured by MTT assay at day 6 after infection. The data from cells infected with RSK2 shRNAs were normalized to NT control. (C) Escalating doses of lenalidomide were added at 24 hours after virus infection and cell viability was measured at day 5 after treatment (the data from each treatment were normalized to vehicle-treated control). shRNA (#52) in H929 cells and shRNA (#55) in MM1.S cells alone were lethal to most of those cells and we were not able to evaluate drug response in them.
RSK2 knockdown inhibited myeloma growth and enhanced lenalidomide-mediated myeloma cytotoxicity. (A) JJN3 (left), MM1.S (middle), and H929 (right) cells were infected with control virus (NT) and RSK2 shRNAs (#52, #54, and #55) lentivirus, and immunoblotting was performed at 48 or 72 hours after virus infection to confirm RSK2 knockdown by shRNAs. (B) Cell viability was measured by MTT assay at day 6 after infection. The data from cells infected with RSK2 shRNAs were normalized to NT control. (C) Escalating doses of lenalidomide were added at 24 hours after virus infection and cell viability was measured at day 5 after treatment (the data from each treatment were normalized to vehicle-treated control). shRNA (#52) in H929 cells and shRNA (#55) in MM1.S cells alone were lethal to most of those cells and we were not able to evaluate drug response in them.
RSK2 small molecule inhibitors synergize with lenalidomide to induce cytotoxicity of MM cells
To further confirm the shRNA knockdown results, we next studied the effects of high potency, specific, small molecule RSK2 inhibitors alone or in combination with lenalidomide on MM cell growth. Three selective small molecule RSK2 inhibitors, BI-D1870, RMM46, and SL0101-1, were tested. BI-D1870 is a selective RSK2 inhibitor previously demonstrated to induce cytotoxicity of myeloma cells through the inhibition of RSK2 activity.16 Treatment of 4 HMCLs with BI-D1870 alone induced a dose-dependent cytotoxicity (supplemental Figure 4A). When HMCLs were treated with a combination of various doses of BI-D1870 and lenalidomide, a synergistic effect in inducing MM-cell cytotoxicity was observed (Figure 3A and supplemental Figure 4B). The small molecules, RMM46 (Figure 3B and supplemental Figure 4B) and SL0101-1 (supplemental Figure 4C), also demonstrated synergistic anti-myeloma activity with lenalidomide on all HMCLs tested. Conversely, the overexpression of wild-type RSK2 in H929 cells impaired the synergistic activity between lenalidomide and BI-D1870 (supplemental Figure 5).
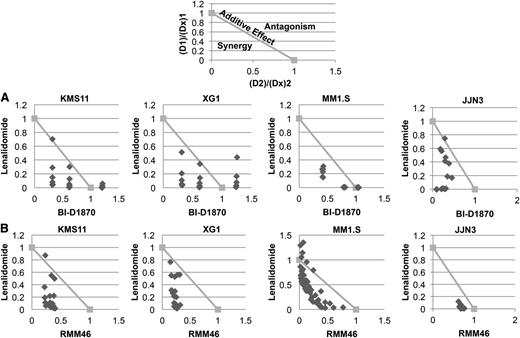
RSK2 inhibitor inhibited myeloma cell growth and synergized with lenalidomide to induce cytoxicity of myeloma cells. Four HMCLs (KMS11, XG1, MM1.S, and JJN3) were treated with various doses of lenalidomide (Len, μM) and (A) BI-D1870 (BI, μM) or (B) RMM46 (RM, μM) for 6 days, followed by MTT assay to measure cell viability. The data from each treatment were normalized to vehicle-treated control and then analyzed using CalcuSyn software to generate normalized isobolograms. An example of a normalized isobologram with the ranges indicative of synergy, additive effects, and antagonism has been provided above the experimental data. The normalized isobolograms below depict the combinations of lenalidomide (D1), BI-1870 or RMM46 (D2), and the effect of the drug observed at the corresponding dose (D × 1, D × 2). Any data points falling below the line have a combination index <1 and are synergistic. Data points on or above the line are considered additive or antagonistic, respectively. Each point on the normalized isobologram corresponds to a specific combination of lenalidomide and BI-D1870/RMM46.
RSK2 inhibitor inhibited myeloma cell growth and synergized with lenalidomide to induce cytoxicity of myeloma cells. Four HMCLs (KMS11, XG1, MM1.S, and JJN3) were treated with various doses of lenalidomide (Len, μM) and (A) BI-D1870 (BI, μM) or (B) RMM46 (RM, μM) for 6 days, followed by MTT assay to measure cell viability. The data from each treatment were normalized to vehicle-treated control and then analyzed using CalcuSyn software to generate normalized isobolograms. An example of a normalized isobologram with the ranges indicative of synergy, additive effects, and antagonism has been provided above the experimental data. The normalized isobolograms below depict the combinations of lenalidomide (D1), BI-1870 or RMM46 (D2), and the effect of the drug observed at the corresponding dose (D × 1, D × 2). Any data points falling below the line have a combination index <1 and are synergistic. Data points on or above the line are considered additive or antagonistic, respectively. Each point on the normalized isobologram corresponds to a specific combination of lenalidomide and BI-D1870/RMM46.
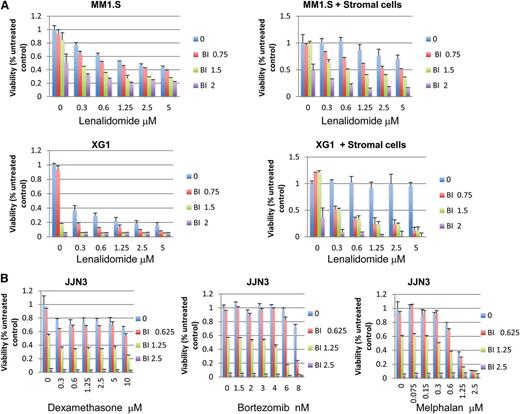
As shown in Figure 4A, BMSCs protected MM cells from either lenalidomide or BI-D1870–induced cytotoxicity but could not protect the same cells from the synergistic effects of the 2 drugs used in combination. The viability of BMSCs was not inhibited by either each drug alone (Len or BI-D1870) or in combination (supplemental Figure 6).
RSK2 inhibition synergizes with lenalidomide to overcome BMSC-mediated drug resistance and it also enhances MM cytotoxicity mediated by other anti-myeloma drugs. (A) Myeloma cells (top panel, MM1.S cells; middle panel, XG1 cells) were incubated with indicated doses of drugs in the absence (left) or presence (right) of BMSCs. BMSCs were seeded at 5000/well in 96-well plates overnight before adding myeloma cells and indicated doses of drugs. MTT assays were read at day 5 after treatment. (B) JJN3 HMCLs were treated with indicated doses of BI-D1870, bortezomib (middle), dexamethasone (left), and melphalan (right) for 3 days, followed by MTT assay to measure cell viability.
RSK2 inhibition synergizes with lenalidomide to overcome BMSC-mediated drug resistance and it also enhances MM cytotoxicity mediated by other anti-myeloma drugs. (A) Myeloma cells (top panel, MM1.S cells; middle panel, XG1 cells) were incubated with indicated doses of drugs in the absence (left) or presence (right) of BMSCs. BMSCs were seeded at 5000/well in 96-well plates overnight before adding myeloma cells and indicated doses of drugs. MTT assays were read at day 5 after treatment. (B) JJN3 HMCLs were treated with indicated doses of BI-D1870, bortezomib (middle), dexamethasone (left), and melphalan (right) for 3 days, followed by MTT assay to measure cell viability.
To determine whether RSK2 inhibitors also affect the anti-MM activity of other clinically useful drugs, we next studied the combinatorial effects of an RSK2 inhibitor with other standard anti-myeloma drugs by MTT assay. As shown in Figure 4B, BI-D1870 appears to also synergize with other anti-MM drugs. For example, treatment of JJN3 cells with 6 nM bortezomib and 1.25 μM BI-D1870 caused an 87% MM cell death, whereas treatment of the same cells with 6 nM bortezomib or 1.25 μM BI-D1870 alone resulted in only 3% and 44% cell death, respectively. This result suggests that RSK2 sensitizes MM cells by interrupting a common pathway critical for MM cell survival.
Effects of RSK2 inhibitors on the immune-modulatory activity of lenalidomide
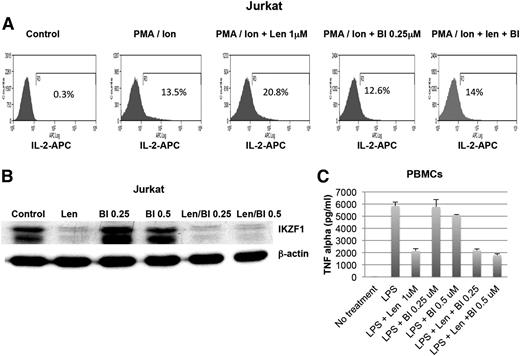
IMiDs have a wide spectrum of effects on the immune system, including augmentation of natural killer cell activity, IL-2 cytokine production, and T-cell activity and inhibition of proinflammatory cytokine production, such as TNFα, IL-6, and IL-12 from human PBMCs.2 To evaluate whether RSK2 inhibitors affect the immunomodulatory activity of IMiDs, the effects of RSK2 inhibitors alone or in combination with lenalidomide on TNFα production from monocytic cells and IL-2 production from T cells were measured. Consistent with previous studies,5,17 we demonstrated that lenalidomide increases IL-2 production in PMA/ionomycin-stimulated Jurkat T cells simultaneously, with substantial downregulation of IKZF1. RSK2 inhibition did not downregulate IKZF1, but it was able to abolish lenalidomide-enhanced production of IL-2 in Jurkat T cells (Figure 5A-B).
Effects of RSK2 inhibition on lenalidomide-mediated immune-modulatory activities. (A) Jurkat T cells (top panel) were pretreated with vehicle (control) and indicated doses of lenalidomide (Len, 1 μM) and BI-D1870 (BI, 0.25 and 0.5 μM) for 1 hour, and then stimulated with PMA (1 ng/mL)/ionomycin (Ion, 1 μg/mL) for 40 hours. The cells were fixed, stained with anti-IL-2–APC and analyzed by flow cytometry. The expression of IL-2 after different combination of treatment (from left to right) are shown. (B) The treated Jurkat cells were also harvested for analysis of IKZF1 expression by western blot. (C) Human PBMCs were pretreated with lenalidomide (1 μM) or BI-D1870 (BI), or a combination of both drugs for 1 hour and then stimulated with LPS (1 ug/mL). The supernatants were harvested and tested for TNFα production by ELISA.
Effects of RSK2 inhibition on lenalidomide-mediated immune-modulatory activities. (A) Jurkat T cells (top panel) were pretreated with vehicle (control) and indicated doses of lenalidomide (Len, 1 μM) and BI-D1870 (BI, 0.25 and 0.5 μM) for 1 hour, and then stimulated with PMA (1 ng/mL)/ionomycin (Ion, 1 μg/mL) for 40 hours. The cells were fixed, stained with anti-IL-2–APC and analyzed by flow cytometry. The expression of IL-2 after different combination of treatment (from left to right) are shown. (B) The treated Jurkat cells were also harvested for analysis of IKZF1 expression by western blot. (C) Human PBMCs were pretreated with lenalidomide (1 μM) or BI-D1870 (BI), or a combination of both drugs for 1 hour and then stimulated with LPS (1 ug/mL). The supernatants were harvested and tested for TNFα production by ELISA.
We further tested the effects of an RSK2 inhibitor on LPS-induced TNFα production from human PBMCs. As shown in Figure 5C, RSK2 inhibitors did not substantially affect either LPS-induced TNFα production or lenalidomide inhibition of LPS-induced TNFα production.
Mechanism of modulation of lenalidomide sensitivity by RSK2 inhibition
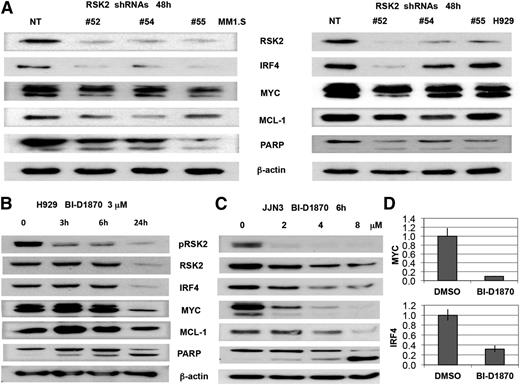
Recently, lenalidomide was shown to induce IKZF1 and IKZF3 degradation, which consequently downregulates IRF4,5,6 a survival factor for myeloma.18 RSK2 inhibition was reported previously to downregulate MYC, MCL1, and induce BIM expression.16 In order to understand whether modulation of RSK2 also enhances lenalidomide-regulated downstream proteins, western blot analysis was performed on MM cells after RSK2 knockdown or small molecule RSK2 inhibition. RSK2 knockdown and inhibition resulted in a significant reduction of IRF4 or MYC (Figure 6A), in a time- and dose-dependent manner (Figure 6B-D). To further demonstrate that the synergy of lenalidomide and BI-D1870 in inducing MM toxicity is correlated with biochemical changes in IRF4, MYC, and MCL1 expression, H929 was treated with low doses of BI-D1870 and lenalidomide, either alone or in combination, followed by MTT assay and immunoblotting. As shown in Figure 7A, at day 5 after treatment, the combination of lenalidomide and BI-D1870 cells induced synergistic cytotoxicity in H929 cells. Cell lysates collected at 48 hours after treatment indicated a synergistic reduction of IRF4 and MYC with induction of BIM and apoptosis (Figure 7B). In the IMiD resistant JJN3 MM cells, the combination of BI-D1870 and lenalidomide was also shown to induce similar synergistic effects on IRF4 and induction of apoptosis (Figure 7C-D). We demonstrated that in contrast to lenalidomide, at 6 hours posttreatment, an RSK2 inhibitor alone does not significantly affect IKZF1 and IKZF3 protein levels in JJN3 cells (Figure 7D). Apparently, both drugs therefore use noncompetitive mechanisms to deplete IRF4 together, but not when used alone, and overcome a critical IRF4 threshold for survival.
Knockdown or inhibition of RSK2 in myeloma cell lines induced apoptotic signaling and downregulation of IRF4, MYC, and MCL1. (A) Myeloma cell lines (MM1.S and H929) were infected with NT and RSK2 shRNAs lentivirus. After 48 hours, cells were harvested and used for immunoblotting assay to measure the expression of IRF4, MYC, MCL1, BIM, and PARP. (B) H929 cells were incubated with BI-D1870 (BI, 3 μM) at indicated time points, followed by immunoblotting analysis. (C) JJN3 cells were incubated with BI-D1870 at indicated doses for 6 hours, followed by immunoblotting analysis. (D) JJN3 cells were incubated with 2 μM of BI-D1870 for 3 hours, followed by qPCR analysis of MYC and IRF4 expression at transcription level.
Knockdown or inhibition of RSK2 in myeloma cell lines induced apoptotic signaling and downregulation of IRF4, MYC, and MCL1. (A) Myeloma cell lines (MM1.S and H929) were infected with NT and RSK2 shRNAs lentivirus. After 48 hours, cells were harvested and used for immunoblotting assay to measure the expression of IRF4, MYC, MCL1, BIM, and PARP. (B) H929 cells were incubated with BI-D1870 (BI, 3 μM) at indicated time points, followed by immunoblotting analysis. (C) JJN3 cells were incubated with BI-D1870 at indicated doses for 6 hours, followed by immunoblotting analysis. (D) JJN3 cells were incubated with 2 μM of BI-D1870 for 3 hours, followed by qPCR analysis of MYC and IRF4 expression at transcription level.
The combination of lenalidomide and BI-D1870 demonstrated a significant enhancement of cytotoxicity and downregulation of IRF4 and MYC. H929 (A-B) and JJN3 (C-D) were treated with the indicated doses of lenalidomide and BI-D1870 (μM) for various times, and was followed by MTT assay (A and C, day 5) and immunoblotting assay (B and D, at various time points).
The combination of lenalidomide and BI-D1870 demonstrated a significant enhancement of cytotoxicity and downregulation of IRF4 and MYC. H929 (A-B) and JJN3 (C-D) were treated with the indicated doses of lenalidomide and BI-D1870 (μM) for various times, and was followed by MTT assay (A and C, day 5) and immunoblotting assay (B and D, at various time points).
Discussion
Using a high-throughput RNAi screening in human MM cells, we identified 63 genes whose suppression enhanced lenalidomide sensitivity. We found that many of these genes were associated with immune responses or related to MM cell growth and survival. For example, 3 sensitizers identified from our screen, including RSK2, CHUK (or IKK1), and CREB1, have previously been demonstrated to interact and were associated with significance together in NF-κB, toll, and IGF1 signaling pathways. For example, RSK2 has been demonstrated to activate NF-κB activity through the phosphorylation of IκBα.19 CHUK-activated NF-κB signaling was previously shown to cooperate with CREB1.20,21 RSK2 has also been demonstrated to phosphorylate and activate CREB1.22 CREB1 regulates diverse cellular responses23-25 and is a direct target of MIR203, whose expression inhibited cellular proliferation by targeting CREB1 messenger RNA in MM.26
We also noticed that several sensitizers belonging to the RAB gene family. The RAB family of proteins constitutes the largest branch of the Ras GTPase superfamily and is engaged in the regulation of intracellular vesicle trafficking and protein transport,27 implying that these processes are important for lenalidomide response. Three potassium channel genes were also identified as sensitizer hits, which is of interest as the primary target of lenalidomide. CRBN is known to regulate the assembly and neuronal surface expression of large-conductance calcium-activated potassium channels in brain regions involved in memory and learning,28 likely via its interaction with KCNT1. Two sensitizers encode PEX biogenesis factors, suggesting that PEX function might also be important for IMiD response. PEXs are associated with energy metabolism. Interestingly, CRBN has been reported to associate with and negatively regulate AMP-activated protein kinase,29 which play a role in cellular energy homeostasis.
In the present study, we focused on the strongest kinase sensitizer hit, RSK2. RSK2 encodes a member of the 90 kDa ribosomal kinase family of serine/threonine kinases (RSK1-4), all of which are downstream substrates of ERK and play a role in various cellular processes, including gene expression, cell cycle, survival, and proliferation.30-32 This family of kinases contains two nonidentical kinase catalytic domains, located at the carboxy- and N-terminal with a linker between them. The N-terminal kinase domain is responsible for the phosphorylation of exogenous substrates, and the carboxy-terminal kinase domain and linker region regulates RSK activation.31 Activation of RSK2 involves multiple steps ending with the Ser227 phosphorylation at the NTKD.30 RSK2 in turn phosphorylates a wide range of substrates, including RSK2, CREB1, and histones.33,34 Although a previous study demonstrated that oncogenic activation of FGFR3 [via the t(4;14)] in MM phosphorylates and activates RSK2 to induce hematopoietic transformation,35 a recent study from a different group found that RSK2 is commonly activated in MM cells regardless of type of cytogenetic or upstream molecular signaling,16 suggesting that RSK2 can be activated by multiple mechanisms in MM and it may be a potential therapeutic target for all MM patients. Consistent with this, we detected phosphorylated RSK2 in all MM cell lines, and we also demonstrated that knockdown of RSK2 or inhibition of RSK2 activity with selective small molecule inhibitors induced cytotoxicity in HMCLs of variable genetic background. Our immunoblotting assay further indicated that RSK2 knockdown or inhibition induced apoptosis signaling, as well as downregulation of IRF4, MYC, and MCL1. All of these proteins are critical for the survival of MM cells.18,36,37 RSK2 was demonstrated to phosphorylate and regulate C/EBPβ,38 which was reported to control transcription factors critical for survival of MM cells, including IRF4.39 RSK2 has also been shown to directly regulate proteins involved in apoptosis, such as BAD40,41 and BIM.16,42 In JJN3 cells, which have among the lowest baseline levels of IKZF1 (and thought to explain their inherent lenalidomide resistance),8 IRF4 was not substantially downregulated by lenalidomide itself unless an RSK2 inhibitor was added. Unlike lenalidomide, treatment of MM cells with an RSK2 inhibitor alone does not appear to induce IKZF1 and IKZF3 degradation, and thus the mechanism of IRF4 depletion are complementary and nonoverlapping. This ability to target the MM-cell survival factors IRF4 and MYC most likely explains why RSK2 inhibition also enhances the activity of other anti-myeloma drugs and can overcome inherent IMiD resistance.
RSK2 knockout mice revealed normal T-cell development, but these T cells had lower production of IL-2 in response to anti-CD3 and anti-CD28 stimulation in vitro.43 Indeed, unlike lenalidomide, RSK2 inhibitors do not increase the ability of lenalidomide to upregulate IL-2 or to significantly reduce LPS-induced TNFα production in PBMCs. All results indicate that the synergistic effects of RSK2 inhibition and lenalidomide are confined to inducing myeloma cell cytotoxicity.
In conclusion, our high-throughput screening identified multiple gene targets that associate with increasing sensitivity to IMiDs in MM cells, of which, RSK2 was the most potent. RSK2 inhibition is cytotoxic, produces synergistic effects with all tested MM therapies, and overcomes IMiD resistance likely by cooperatively downregulating IRF4 and MYC. Clinical studies of RSK2 inhibition in MM would be appropriate and may overcome drug resistance.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Donald Chow and Dr James Bogenberger for their suggestions on data analysis, and Latha Pathangey for technique assistance on immunostaining of intracellular IL-2 production.
This study was funded by grants from the National Institutes of Health, National Cancer Institute (CA167511-01A1-02A) (A.K.S.) and the Leukemia and Lymphoma Society (4-02A) (A.K.S.). K.M.K. received a research grant from the Deutsche Forschungsgemeinschaft (KO-4604/1-1).
National Institutes of Health
Authorship
Contribution: A.K.S., Y.X.Z., and H.Y. contributed to the conception and design of the study; Y.X.Z., E.B., C.-X.S., L.A.B., J.E.S., K.M.K., P.J., M.A., C.S., and M.C. contributed to the acquisition and analysis of the data; Y.X.Z. and A.K.S. drafted the article; and all authors read, revised the article critically for important intellectual content, gave final approval of the version to be published, and contributed to the interpretation of the data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: A. Keith Stewart, Mayo Clinic, 13400 E. Shea Blvd, CRB Room 1-001, Scottsdale, AZ 85259; e-mail: stewart.keith@mayo.edu.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal