Key Points
GPI-anchor–deficient cell lines are more vulnerable to complement C5b-9 deposition and cell killing from aHUS serum.
PIGA-null reagent cell lines can be used to rapidly and reliably distinguish aHUS from other thrombotic microangiopathies.
Abstract
Atypical hemolytic uremic syndrome (aHUS) is a thrombotic microangiopathy (TMA) characterized by excessive activation of the alternative pathway of complement (APC). Atypical HUS is frequently a diagnosis of exclusion. Differentiating aHUS from other TMAs, especially thrombotic thrombocytopenic purpura (TTP), is difficult due to overlapping clinical manifestations. We sought to develop a novel assay to distinguish aHUS from other TMAs based on the hypothesis that paroxysmal nocturnal hemoglobinuria cells are more sensitive to APC-activated serum due to deficiency of glycosylphosphatidylinositol- anchored complement regulatory proteins (GPI-AP). Here, we demonstrate that phosphatidylinositol-specific phospholipase C–treated EA.hy926 cells and PIGA-mutant TF-1 cells are more susceptible to serum from aHUS patients than parental EA.hy926 and TF-1 cells. We next studied 31 samples from 25 patients with TMAs, including 9 with aHUS and 12 with TTP. Increased C5b-9 deposition was evident by confocal microscopy and flow cytometry on GPI-AP–deficient cells incubated with aHUS serum compared with heat-inactivated control, TTP, and normal serum. Differences in cell viability were observed in biochemically GPI-AP–deficient cells and were further increased in PIGA-deficient cells. Serum from patients with aHUS resulted in a significant increase of nonviable PIGA-deficient TF-1 cells compared with serum from healthy controls (P < .001) and other TMAs (P < .001). The cell viability assay showed high reproducibility, sensitivity, and specificity in detecting aHUS. In conclusion, we developed a simple, rapid, and serum-based assay that helps to differentiate aHUS from other TMAs.
Introduction
Thrombotic microangiopathies (TMAs) present with thrombocytopenia, nonimmune hemolytic anemia, peripheral blood schistocytes, and often other end-organ damage to the kidneys and central nervous system. TMAs are frequently life-threatening and have considerable clinical overlap, so prompt recognition of the underlying pathophysiology is critical.1 After exclusion of TMAs due to underlying diseases such as disseminated intravascular coagulation (DIC), drugs, malignancy, or scleroderma-associated renal crisis, the differential diagnosis is often between thrombotic thrombocytopenic purpura (TTP) and hemolytic uremic syndrome (HUS).2
TTP results from impaired postsecretion processing of ultralarge von Willebrand factor multimers due to a severe deficiency of a disintegrin and metalloprotease with thrombospondin type 1 motif, 13 (ADAMTS13). Severe ADAMTS13 deficiency (usually defined as <10%) may be inherited (Upshaw-Schulman syndrome)3,4 or acquired, resulting from immunoglobulin G (IgG) autoantibodies directed against ADAMTS13.5-7 Without treatment, TTP may lead to neurologic impairment, renal failure, and death.8 Acquired TTP is much more common than Upshaw-Schulman syndrome and is best treated with plasma exchange.1,9
HUS is classified as typical vs atypical, and most cases are associated with acute kidney injury. Typical HUS is caused by Shiga-toxin–producing organisms (most commonly Escherichia coli and Shigella dysenteriae). The mainstay of therapy is supportive care.10,11 Atypical HUS (aHUS) is most commonly caused by defects in the regulation of the alternative pathway of complement (APC). These defects are usually inherited but may also be acquired.12,13 Affected patients may have lifelong systemic complications leading to damage of multiple organ systems (renal, gastrointestinal, central nervous system, and cardiac) and death. Although plasma exchange may be effective in some cases, underlying complement-mediated damage to the kidneys and central nervous system often persists.14 Within 1 year after diagnosis, >50% of patients treated with plasma exchange or plasma infusion experience permanent renal damage, progress to end-stage renal disease, or die.15 Terminal complement inhibition with the monoclonal antibody eculizumab is highly effective for treating aHUS and is now considered the treatment of choice for this disease.16,17
In patients presenting with TMAs, it is important to obtain ADAMTS13 levels and screen for Shiga toxin before instituting definitive therapy. Plasma exchange is often initiated before the results of these assays return due to the aggressiveness of TMAs. If ADAMTS13 activity is <10%, a diagnosis of TTP is established, especially if an inhibitor of ADAMTS13 is also detected, then daily plasma exchange is continued. If ADAMTS13 activity is >10% and Shiga-toxin assay is negative, a diagnosis of aHUS must be considered.18 Unfortunately, there is no definitive test to make a diagnosis of aHUS, and given the high cost of eculizumab, definitive therapy is often delayed or not administered. Genetic testing for mutations that lead to increased activation of APC is expensive, it takes several weeks to obtain results, and it is only informative in roughly 50% to 60% of cases.12,19 Frequently, genetic variants of unknown significance are identified, providing clinicians with data of uncertain utility.20,21 More recently, biomarkers of APC such as C5a or soluble C5b-9 have been compared in aHUS and TTP, but these are not reliable in distinguishing the 2 diseases.22
Paroxysmal nocturnal hemoglobinuria (PNH) is a clonal hematopoietic stem cell disorder caused by a somatic PIGA mutation that leads to complement-mediated hemolysis. PIGA is required for the biosynthesis of glycosylphosphatidylinositol (GPI) biosynthesis; thus, PIGA-mutant cells have a deficiency or absence of GPI-anchored proteins.23 Two of the missing GPI-anchored proteins in PNH (CD55 and C59) are complement regulatory proteins; their deficiency explains the hemolytic anemia in PNH patients. CD55 regulates C3 convertases,24 and CD59 prevents the incorporation of C9 into the membrane attack complex.25 Thus, hemolysis in PNH is primarily due to activation of APC. Indeed, prior to 1990, the diagnosis of PNH was often based on the acidified serum test, also known as the Ham test. The principle of this assay is that PNH cells are more vulnerable to acidified serum, which serves to activate APC.26 Similar to aHUS, PNH is effectively treated by terminal complement inhibition.27,28
Because most aHUS patients harbor mutations that lead to activation of APC,13 we hypothesized that serum from aHUS patients would kill PNH-like cells more readily than serum from other TMAs such as acquired TTP or typical HUS (Shiga toxin). Here, we demonstrate a simple, rapid, and serum-based assay that can differentiate aHUS from other TMAs.
Methods
Study population
Patients assessed in our department from August 2014 to February 2015 with the differential diagnosis of TMA were enrolled. Routine diagnostic work-up, along with ADAMTS13 activity and inhibitor levels and Shiga-toxin screen, were obtained for all patients. Patients were clinically categorized as aHUS if their first manifestation of the syndrome met the following criteria: (1) platelet count <100 × 109/L, (2) serum creatinine >2.25 mg/dL, and (3) ADAMTS13 activity >10%. These criteria have been proposed in previous studies to clinically differentiate patients with aHUS.29,30 Patients with TMA and Shiga-toxin detection in stool culture were classified as typical HUS. TTP diagnosis was made in patients with relevant clinical features (microangiopathic hemolytic anemia and thrombocytopenia) and ADAMTS13 ≤10%.31 DIC was diagnosed in patients with thrombocytopenia, elevated d-dimers, low fibrinogen, prolonged clotting time, and underlying disorders associated with DIC, matching the International Society on Thrombosis and Haemostasis diagnostic criteria.32 Final diagnosis, course, and patient treatment were recorded. All patients gave written informed consent. This study was approved by the institutional review board and conducted in accordance with the Declaration of Helsinki.
Healthy subjects were recruited from the community as controls. Following initial experiments, normal human AB serum (H4522; Sigma-Aldrich, St Louis, MO) was used in control samples.
Blood was collected in serum separation tubes and was immediately centrifuged at 4°C. Serum was separated and stored at −80°C. Heat inactivation was performed the same day of the experiment, incubating the serum at 56°C for 30 minutes.
Cell lines
The human endothelial hybrid cell line EA.hy926 (CRL2922; ATCC, Manassas, VA) was cultured in Dulbecco’s modified Eagle medium supplemented with 1 ng/mL of endothelial growth factor, 2 mM L-glutamine, penicillin/streptomycin, and 20% fetal calf serum. The EA.hy926 cell line is a hybrid of human umbilical vein endothelial cells and human lung carcinoma cells and resembles primary cultured endothelial cells in morphologic and functional aspects.33-35 TF-1 cells (CRL2003, ATCC) were maintained in RPMI 1640 medium supplemented with 2 ng/mL of granulocyte-macrophage colony-stimulating factor, 2 mM L-glutamine, penicillin/streptomycin, and 10% fetal calf serum.
The GPI-anchored protein (GPI-AP)–deficient TF-1 cell line has been previously established in our laboratory.36 In particular, the TF-1 CD34+ cell line was treated with proaerolysin that lyses cells expressing GPI-AP. Thus, the proaerolysin-resistant clone expanded was GPI-AP deficient, having a 7-nucleotide deletion at position 291 to 297 (TTGTCAC) in exon 2 of PIGA resulting in a frameshift mutation.
Serum C5b-9 levels
Serum C5b-9 levels were determined using a commercially available enzyme-linked immunosorbent assay kit (Quidel, San Diego, CA).
C5b-9 imaging by confocal microscopy
A total of 50 000 EA.hy926 cells were plated in gelatin-coated sterile glass coverslips and cultured until 80% confluent. Adenosine 5′-diphosphate (ADP) activation was performed with 10 μM ADP (0160; Amresco, Solon, OH) for 10 minutes at room temperature. GPI-anchor cleavage of EA.hy926 cells was performed using phosphatidylinositol-specific phospholipase C (PIPLC) (P6466; Life Technologies, Carlsbad, CA). For these experiments, cells were treated with PIPLC (0.4 U/mL) for 30 minutes at 37°C and then incubated with serum for 30 minutes at 37°C. Between steps, cells were washed with Hank’s balanced salt solution (24020117, Life Technologies). Next, cells were fixed with 4% paraformaldehyde and stained with rabbit anti-complement C5b-9 (204903; Millipore, Bedford, MA) followed by Alexa Fluor 594 donkey anti-rabbit IgG secondary antibody (A21207, Life Technologies). Counterstaining with 4,6 diamidino-2-phenylindole was also performed. C5b-9 deposition was subsequently observed in a confocal inverted laser microscope (LSM 510 Meta, Zeiss).
Flow cytometry analysis of C5b-9 deposition
Flow cytometric analysis was performed on EA.hy926 cells treated with PIPLC and GPI-deficient TF1 cell line to assess C5b-9 accumulation. Because EA.hy926 cells are adherent, an extra step of trypsinization to single-cell suspension was required as compared with TF-1 suspension cells. EA.hy926 cells were treated with trypsin (10×, Life Technologies) for 3 minutes at 37°C. Of note, surface-attached C5b-9 complexes are sensitive to trypsin.37 Thus, after trypsinization, EA.hy926 cells were recovered in cell culture medium at 37°C for 30 minutes38 and then treated with PIPLC (0.4 U/mL) for 30 minutes at 37°C.
Intracellular staining was performed using the Fix & Perm Cell Fixation and Cell Permeabilization Kit (GAS 001/2, Life Technologies). Initial experiments established an optimal incubation period at 10 minutes and serum dilution at 1:4. Cells were labeled with rabbit anti-complement C5b-9 (204903, Millipore) and a secondary Alexa Fluor 488 donkey anti-rabbit IgG antibody (Jackson ImmunoResearch Laboratories, West Grove, PA). C5b-9 deposition was analyzed in a BD FACSCalibur with a total number of 10 000 events per sample.
Cell viability assay
Cells were plated in 96-well flat-bottom plates for EA.hy926 cells or U-shaped 96-well plates for TF-1 cells at a density of 4000 cells per well and cultured until confluent. EA.hy926 cells were treated with PIPLC, as described above. Then, cells were washed with phosphate-buffered saline (PBS) and incubated with serum in triplicates at a concentration of 1:4 for 30 minutes at 37°C. Serum was diluted in gelatin veronal buffer (GVB) (Sigma-Aldrich). Cells were washed again with phosphate-buffered saline and incubated with the cell proliferation reagent 4-[3-(4-lodophenyl)-2-(4-nitrophenyl)-2H-5-tetrazolio]-1.3-benzene disulfonate/WST-1 (Roche, Switzerland) for 2 hours at 37°C. WST-1 was diluted in the cell culture medium at a concentration of 1:10, and 100 μL WST-1 solution was added per well. Absorbance was measured in an iMark Microplate Absorbance Reader (Bio-Rad, Hercules, CA) at 490 nm with a reference wavelength at 595 nm, according to the manufacturer’s instructions and previous publications.39 The colorimetric assay is based on cleavage of the tetrazolium salt, WST-1, by mitochondrial dehydrogenases in viable cells.
Heat-inactivated serum was used as a negative control. Cobra venom factor (CVF) (Complement Technology, Tyler, TX), which induces activation of the alternative complement pathway, was used as a positive control according to the manufacturer’s instructions.
Reproducibility was investigated by consecutive double testing of 5 samples in 1 experiment and in 2 experiments conducted within 2 consecutive days. The assay was performed by the same investigator (E.G.)
Data analysis
Absorbance values of each sample were normalized after subtraction of the absorbance value of a blank cell. Blanks contained only 100 μL of the WST-1 solution. Percentage of viable cells was expressed as a ratio of the absorbance of each sample multiplied by 100, to the absorbance of the same sample’s heat-inactivated control. Thus, percentage of nonviable cells was calculated using the following formula: 100 − (sample absorbance × 100 / heat-inactivated sample’s absorbance).
Analysis was performed using the Statistical Package for Social Sciences (SPSS) 20.0 for Windows (SPSS, Chicago, IL). The independent-samples Student t test was used to compare differences between the mean values of 2 groups. One-way analysis of variance with Bonferroni correction or nonparametric tests were used to compare means between >2 groups. Comparisons between samples and their heat-inactivated controls were performed with a paired-samples t test. A P value ≤ .05 was considered statistically significant. Analysis of specificity, sensitivity, and the cutoff value was performed by creating a receiver-operating characteristic curve. Reproducibility was measured using Lin’s concordance correlation coefficient.
Results
Patient population
Patient characteristics at initial diagnosis and posttreatment are shown in Table 1. We studied 13 samples from 9 different patients with aHUS (2 in acute phase before treatment, 6 after initiation of eculizumab, and 5 in remission off therapy) and 14 samples from 12 different TTP patients (3 in acute phase before treatment, 3 under treatment with plasma exchange and ongoing TMA, and 8 in remission no longer on plasma exchange). We also studied serum from a patient with Shiga-toxin–associated HUS before treatment, a patient with aPIGN on prednisone, and 2 patients with DIC.
Patient clinical characteristics
| Patient no. . | Age (y), gender . | Diagnosis (trigger) . | Phase (wk) . | ADAMTS13 activity (%) . | ADAMTS13 inhibitor (%) . | PLT count (×103/μL) . | LDH (mg/dL) . | Cr (mg/dL) . | Genetic mutation . | Previous treatments . | Current treatment . | Response to treatment . | Nonviable cells (%) . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 47, F | aHUS (kidney stone) | Acute | 75 | NA | 9 | 2505 | 2.3 | CFHR3-CFHR1 homozygous deletion; CFH heterozygous c.2850G>T, p.Gln950His | — | — | — | NA |
| Remission (20) | NA | NA | 314 | 228 | 1.9 | PEx (×5) | — | No | 63 | ||||
| Eculizumab (×8) | Yes | ||||||||||||
| 2 | 52, F | aHUS (pancreatitis) | Acute | 47 | NA | 13 | 1276 | 3.0 | CFHR3-CFHR1 heterozygous deletion; ADAMTS13 heterozygous c.2195C>T, p.Ala732Val | — | Prednisone, PEx | No | NA |
| Remission (14) | NA | NA | 187 | 190 | 1.0 | Prednisone, PEx (×3) | Eculizumab (×14) | Yes | 24 | ||||
| 3 | 64, M | aHUS (unknown) | Acute | 59 | NA | 66 | 1288 | 2.5 | None | — | — | — | NA |
| Remission (1) | NA | NA | 237 | 216 | 1.3 | PEx (×14), rituximab | Eculizumab (×7) | Yes | 28 | ||||
| 4 | 38, F | aHUS (unknown) | Acute | >100 | NA | 78 | 858 | 6.8 | CFH NM_000186: heterozygous c.472G>A,p.Val158Ile; Heterozygous c.3079G>C,p.Ala1027Pro; CFHR3-CFHR1 heterozygous deletion; ADAMTS13 NM_139025: heterozygous c.2420+3G>A; heterozygous c.3677C>T,p.Thr1226Ile | — | — | — | 46 |
| Responding | >100 | <5 | 110 | 248 | 4.3 | Hemodialysis, PEx (×5), prednisone | Eculizumab (×2) | Yes | 39 | ||||
| 5 | 59, M | aHUS (Hodgkin disease) | Acute | 15 | 67 | 39 | 676 | 2.3 | None | PEx (×4) | — | No | 42 |
| Responding | 22 | 11 | 114 | 197 | 1.4 | PEx (×4) | Eculizumab (×2), brentuximab | Yes | 41 | ||||
| Remission (1) | NA | NA | 307 | 180 | 1.6 | PEx (×4) | Eculizumab (×7) | Yes | 34 | ||||
| Remission (12) | NA | NA | 330 | 170 | 1.4 | Eculizumab (×12) | — | Yes | 43 | ||||
| 6 | 65, F | aHUS (quinine) | Acute | 71 | NA | 24 | 209 | 7.8 | CFHR3-CFHR1 heterozygous deletion; CD46 NM_0022389: heterozygous c.424G>C, p.Glu142Gln | — | Hemodialysis, PEx | No | NA |
| Remission (36) | NA | NA | 445 | 168 | 1.1 | PEx (3), hemodialysis | None | No | 32 | ||||
| Eculizumab (×5) | Yes | ||||||||||||
| 7 | 35, F | aHUS (liver transplant) | Acute | NA | NA | NA | NA | NA | None | — | — | — | NA |
| Remission | NA | NA | 108 | 75 | 1.1 | — | Eculizumab (×77) | Yes | 42 | ||||
| 8 | 23, F | aHUS | Acute | 102 | NA | 62 | 2820 | 6.3 | None | — | — | — | NA |
| Remission (60) | NA | NA | 205 | 178 | 3.5 | Eculizumab (×37) | Hemodialysis | Yes | 49 | ||||
| 9 | 34, F | aHUS | Acute | 41 | NA | 17 | 1133 | 2.4 | CFHR5 heterozygous c.1357C>G,p.Pro453Ala; THBD heterozygous c.1456G>T,p.Asp486Tyr | — | — | — | — |
| Remission (105) | 101 | NA | 238 | NA | 0.8 | Prednisone, PEx, rituximab | — | Yes | 53 | ||||
| 10 | 24, F | TTP | Acute | 6 | 82 | 111 | 556 | 1.0 | CFHR3-CFHR1 homozygous deletion | — | Prednisone, PEx (×10), Rituximab (×3) | — | NA |
| Responding | 100 | 23 | 303 | 744 | 1.1 | Prednisone, PEx (×10), rituximab (×3) | Azathioprine 100 mg/d prednisone | Yes | 2 | ||||
| 11 | 42, M | TTP | Acute | <5 | 59 | 8 | 1408 | 1.4 | None | — | Prednisone | — | 8 |
| Remission (3) | <5 | 31 | 156 | 195 | 1.0 | Prednisone, PEx (×20), rituximab | — | Yes | 17 | ||||
| 12 | 23, M | TTP | Acute | 9 | NA | 26 | 732 | 1.0 | CFHR3-CFHR1 heterozygous deletion;ADAMTS13 heterozygous c.2384C>T, p.Ala795Val; heterozygous c.1368G>T, p.Gln456His | — | None | — | NA |
| Remission (1) | 9 | 33 | 209 | 113 | 1.1 | — | PEx (×11) | Yes | 1 | ||||
| Remission (8) | 8 | NA | 228 | 126 | 1.1 | PEx (×11) | Prednisone | Yes | 2 | ||||
| 13 | 50, F | TTP | Acute | <5 | 90 | 7 | NA | 0.8 | None | PEx, rituximab (×4), prednisone, azathioprine | PEx | — | NA |
| Remission (24) | NA | NA | 173 | 186 | 0.8 | PEx, rituximab (×4), prednisone, azathioprine | — | Yes | 12 | ||||
| 14 | 64, F | TTP | Acute | < 5 | 18 | 7 | 1109 | 1.1 | None | Prednisone | PEx (×7) | No | NA |
| Remission | NA | NA | 324 | NA | 1.0 | Prednisone, PEx(×12), rituximab (×4) | None | Yes | 8 | ||||
| 15 | 42, F | TTP | Acute | <5 | 57 | 61 | 331 | 0.8 | None | — | Prednisone | NA | |
| Remission | 40 | 20 | 365 | NA | 1.0 | Prednisone (×4), rituximab | — | Yes | 7 | ||||
| 16 | 35, F | TTP | Acute | <5 | 96 | 5 | 2200 | NA | NA | — | — | — | 3 |
| 17 | 45, F | TTP | Acute | <5 | 75 | 12 | 1100 | NA | NA | — | — | — | 1 |
| 18 | 48, F | TTP | Acute | <4 | NA | 25 | 521 | 3.3 | NA | Prednisone, PEx | — | — | NA |
| Remission | 52 | NA | 310 | 136 | 1.1 | Prednisone, PEx, rituximab | — | Yes | 17 | ||||
| 19 | 50, M | TTP | Acute | 6 | 26 | 28 | 574 | 0.8 | NA | Prednisone | Prednisone | — | — |
| Remission | >100 | 14 | 335 | 618 | 1.2 | Prednisone, rituximab | — | Yes | 1 | ||||
| 20 | 57, M | TTP | Acute | 4 | 10 | 7 | 1231 | 1.3 | NA | — | PEx | — | NA |
| Remission | 64 | 9 | 241 | 165 | NA | NA | PEx, rituximab | — | Yes | 14 | |||
| 21 | 19, F | TTP | Acute | 8 | NA | 56 | 750 | 1.1 | NA | — | PEx, Prednisone | — | 1 |
| 22 | 85, F | Stec-HUS | Acute | 59 | 27 | 6 | 1662 | 3.0 | None | — | None | NA | NA |
| Remission | NA | 33 | 124 | 465 | 1.6 | PEx, prednisolone | Eculizumab (×3) | Yes | 24 | ||||
| 23 | 31, M | aPIGN | Acute | 84 | NA | 234 | 138 | 2.2 | CFHR3-CFHR1 heterozygous deletion; CFH NM_000186: heterozygous c.2850G>T,p.Gln950His ADAMTS13 NM_139205: heterozygous c.2758A>G,p.Met920Val | — | Prednisone | No | 19 |
| 24 | 36, F | DIC | Acute | NA | NA | 120 | 333 | 1.9 | NA | — | None | No | 6 |
| 25 | 69, F | DIC | Acute | 53 | 6 | 44 | 360 | 1.2 | NA | — | None | No | 9 |
| Patient no. . | Age (y), gender . | Diagnosis (trigger) . | Phase (wk) . | ADAMTS13 activity (%) . | ADAMTS13 inhibitor (%) . | PLT count (×103/μL) . | LDH (mg/dL) . | Cr (mg/dL) . | Genetic mutation . | Previous treatments . | Current treatment . | Response to treatment . | Nonviable cells (%) . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 47, F | aHUS (kidney stone) | Acute | 75 | NA | 9 | 2505 | 2.3 | CFHR3-CFHR1 homozygous deletion; CFH heterozygous c.2850G>T, p.Gln950His | — | — | — | NA |
| Remission (20) | NA | NA | 314 | 228 | 1.9 | PEx (×5) | — | No | 63 | ||||
| Eculizumab (×8) | Yes | ||||||||||||
| 2 | 52, F | aHUS (pancreatitis) | Acute | 47 | NA | 13 | 1276 | 3.0 | CFHR3-CFHR1 heterozygous deletion; ADAMTS13 heterozygous c.2195C>T, p.Ala732Val | — | Prednisone, PEx | No | NA |
| Remission (14) | NA | NA | 187 | 190 | 1.0 | Prednisone, PEx (×3) | Eculizumab (×14) | Yes | 24 | ||||
| 3 | 64, M | aHUS (unknown) | Acute | 59 | NA | 66 | 1288 | 2.5 | None | — | — | — | NA |
| Remission (1) | NA | NA | 237 | 216 | 1.3 | PEx (×14), rituximab | Eculizumab (×7) | Yes | 28 | ||||
| 4 | 38, F | aHUS (unknown) | Acute | >100 | NA | 78 | 858 | 6.8 | CFH NM_000186: heterozygous c.472G>A,p.Val158Ile; Heterozygous c.3079G>C,p.Ala1027Pro; CFHR3-CFHR1 heterozygous deletion; ADAMTS13 NM_139025: heterozygous c.2420+3G>A; heterozygous c.3677C>T,p.Thr1226Ile | — | — | — | 46 |
| Responding | >100 | <5 | 110 | 248 | 4.3 | Hemodialysis, PEx (×5), prednisone | Eculizumab (×2) | Yes | 39 | ||||
| 5 | 59, M | aHUS (Hodgkin disease) | Acute | 15 | 67 | 39 | 676 | 2.3 | None | PEx (×4) | — | No | 42 |
| Responding | 22 | 11 | 114 | 197 | 1.4 | PEx (×4) | Eculizumab (×2), brentuximab | Yes | 41 | ||||
| Remission (1) | NA | NA | 307 | 180 | 1.6 | PEx (×4) | Eculizumab (×7) | Yes | 34 | ||||
| Remission (12) | NA | NA | 330 | 170 | 1.4 | Eculizumab (×12) | — | Yes | 43 | ||||
| 6 | 65, F | aHUS (quinine) | Acute | 71 | NA | 24 | 209 | 7.8 | CFHR3-CFHR1 heterozygous deletion; CD46 NM_0022389: heterozygous c.424G>C, p.Glu142Gln | — | Hemodialysis, PEx | No | NA |
| Remission (36) | NA | NA | 445 | 168 | 1.1 | PEx (3), hemodialysis | None | No | 32 | ||||
| Eculizumab (×5) | Yes | ||||||||||||
| 7 | 35, F | aHUS (liver transplant) | Acute | NA | NA | NA | NA | NA | None | — | — | — | NA |
| Remission | NA | NA | 108 | 75 | 1.1 | — | Eculizumab (×77) | Yes | 42 | ||||
| 8 | 23, F | aHUS | Acute | 102 | NA | 62 | 2820 | 6.3 | None | — | — | — | NA |
| Remission (60) | NA | NA | 205 | 178 | 3.5 | Eculizumab (×37) | Hemodialysis | Yes | 49 | ||||
| 9 | 34, F | aHUS | Acute | 41 | NA | 17 | 1133 | 2.4 | CFHR5 heterozygous c.1357C>G,p.Pro453Ala; THBD heterozygous c.1456G>T,p.Asp486Tyr | — | — | — | — |
| Remission (105) | 101 | NA | 238 | NA | 0.8 | Prednisone, PEx, rituximab | — | Yes | 53 | ||||
| 10 | 24, F | TTP | Acute | 6 | 82 | 111 | 556 | 1.0 | CFHR3-CFHR1 homozygous deletion | — | Prednisone, PEx (×10), Rituximab (×3) | — | NA |
| Responding | 100 | 23 | 303 | 744 | 1.1 | Prednisone, PEx (×10), rituximab (×3) | Azathioprine 100 mg/d prednisone | Yes | 2 | ||||
| 11 | 42, M | TTP | Acute | <5 | 59 | 8 | 1408 | 1.4 | None | — | Prednisone | — | 8 |
| Remission (3) | <5 | 31 | 156 | 195 | 1.0 | Prednisone, PEx (×20), rituximab | — | Yes | 17 | ||||
| 12 | 23, M | TTP | Acute | 9 | NA | 26 | 732 | 1.0 | CFHR3-CFHR1 heterozygous deletion;ADAMTS13 heterozygous c.2384C>T, p.Ala795Val; heterozygous c.1368G>T, p.Gln456His | — | None | — | NA |
| Remission (1) | 9 | 33 | 209 | 113 | 1.1 | — | PEx (×11) | Yes | 1 | ||||
| Remission (8) | 8 | NA | 228 | 126 | 1.1 | PEx (×11) | Prednisone | Yes | 2 | ||||
| 13 | 50, F | TTP | Acute | <5 | 90 | 7 | NA | 0.8 | None | PEx, rituximab (×4), prednisone, azathioprine | PEx | — | NA |
| Remission (24) | NA | NA | 173 | 186 | 0.8 | PEx, rituximab (×4), prednisone, azathioprine | — | Yes | 12 | ||||
| 14 | 64, F | TTP | Acute | < 5 | 18 | 7 | 1109 | 1.1 | None | Prednisone | PEx (×7) | No | NA |
| Remission | NA | NA | 324 | NA | 1.0 | Prednisone, PEx(×12), rituximab (×4) | None | Yes | 8 | ||||
| 15 | 42, F | TTP | Acute | <5 | 57 | 61 | 331 | 0.8 | None | — | Prednisone | NA | |
| Remission | 40 | 20 | 365 | NA | 1.0 | Prednisone (×4), rituximab | — | Yes | 7 | ||||
| 16 | 35, F | TTP | Acute | <5 | 96 | 5 | 2200 | NA | NA | — | — | — | 3 |
| 17 | 45, F | TTP | Acute | <5 | 75 | 12 | 1100 | NA | NA | — | — | — | 1 |
| 18 | 48, F | TTP | Acute | <4 | NA | 25 | 521 | 3.3 | NA | Prednisone, PEx | — | — | NA |
| Remission | 52 | NA | 310 | 136 | 1.1 | Prednisone, PEx, rituximab | — | Yes | 17 | ||||
| 19 | 50, M | TTP | Acute | 6 | 26 | 28 | 574 | 0.8 | NA | Prednisone | Prednisone | — | — |
| Remission | >100 | 14 | 335 | 618 | 1.2 | Prednisone, rituximab | — | Yes | 1 | ||||
| 20 | 57, M | TTP | Acute | 4 | 10 | 7 | 1231 | 1.3 | NA | — | PEx | — | NA |
| Remission | 64 | 9 | 241 | 165 | NA | NA | PEx, rituximab | — | Yes | 14 | |||
| 21 | 19, F | TTP | Acute | 8 | NA | 56 | 750 | 1.1 | NA | — | PEx, Prednisone | — | 1 |
| 22 | 85, F | Stec-HUS | Acute | 59 | 27 | 6 | 1662 | 3.0 | None | — | None | NA | NA |
| Remission | NA | 33 | 124 | 465 | 1.6 | PEx, prednisolone | Eculizumab (×3) | Yes | 24 | ||||
| 23 | 31, M | aPIGN | Acute | 84 | NA | 234 | 138 | 2.2 | CFHR3-CFHR1 heterozygous deletion; CFH NM_000186: heterozygous c.2850G>T,p.Gln950His ADAMTS13 NM_139205: heterozygous c.2758A>G,p.Met920Val | — | Prednisone | No | 19 |
| 24 | 36, F | DIC | Acute | NA | NA | 120 | 333 | 1.9 | NA | — | None | No | 6 |
| 25 | 69, F | DIC | Acute | 53 | 6 | 44 | 360 | 1.2 | NA | — | None | No | 9 |
aPIGN, atypical postinfectious glomerulonephritis; CFHR, complement factor H–related proteins; Cr, creatinine; F, female; LDH, lactate dehydrogenase; M, male; NA, not available; PAI-1, plasminogen activator inhibitor-1, plasma exchange; PLT, platelet; Stec-HUS, Shiga-toxin–associated hemolytic uremic syndrome; THBD, thrombomodulin; PEx, plasma exchange.
Serum C5b-9 levels by enzyme-linked immunosorbent assay
C5b-9 levels in pretreatment plasma samples have been suggested as useful biomarkers in differentiating aHUS from TTP.22 In our population of patients before, on, or off treatment, there was no significant difference in serum C5b-9 levels between aHUS and TTP patients (P = .640), as shown in supplemental Figure 1 (available on the Blood Web site).
C5b-9 depositions by confocal microscopy and flow cytometry
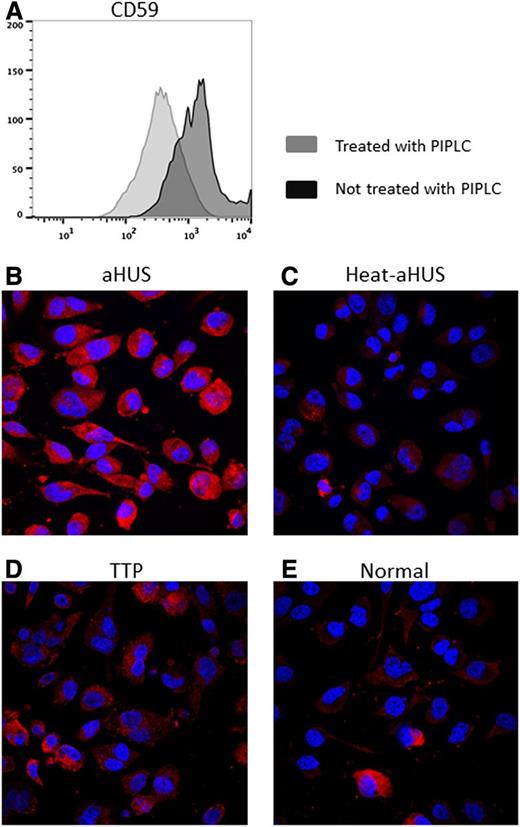
We initially measured C5b-9 deposition after exposure to patient serum on EA.hy926 cells before and after PIPLC treatment. As shown by Noris et al,40 aHUS serum showed increased C5b-9 deposition compared with normal serum in non–PIPLC-treated cells (supplemental Figure 2). In an effort to improve sensitivity and specificity of this effect, we treated EA.hy926 cells with PIPLC before exposing the cells to patient serum. PIPLC-treated EA.hy926 incubated with aHUS serum exhibited increased C5b-9 deposition as compared with normal serum, heat-inactivated aHUS serum, or TTP serum. Results from representative samples are shown in Figure 1, and representative images from all patients are shown in supplemental Figure 3.
C5b-9 deposition on PIPLC-treated human EA.hy926 cells. (A) Flow cytometric analysis of CD59 expression on ADP-activated EA.hy926 l cells treated with PIPLC vs untreated EA.hy926 cells. After trypsinization, endothelial cells were recovered in cell culture medium at 37°C for 30 minutes, treated with PIPLC, and then stained with anti-CD59 antibody. Reduced CD59 expression on PIPLC-treated cells is shown. (B-E) Confocal microscopy of ADP-activated EA.hy926 cells treated with PIPLC and stained with C5b-9 (depicted in red) and 4,6 diamidino-2-phenylindole (blue) as nuclei cell counterstaining. Magnification 40x. C5b-9 deposition is shown after incubation with aHUS serum (B) compared with heat-inactivated aHUS serum (C), TTP serum (D) and normal serum (E). PIPLC: Phosphatidylinositol-specific phospholipase C; aHUS: atypical hemolytic uremic syndrome; Heat-aHUS: heat inactivated atypical hemolytic uremic syndrome; TTP: thrombotic thrombocytopenic purpura.
C5b-9 deposition on PIPLC-treated human EA.hy926 cells. (A) Flow cytometric analysis of CD59 expression on ADP-activated EA.hy926 l cells treated with PIPLC vs untreated EA.hy926 cells. After trypsinization, endothelial cells were recovered in cell culture medium at 37°C for 30 minutes, treated with PIPLC, and then stained with anti-CD59 antibody. Reduced CD59 expression on PIPLC-treated cells is shown. (B-E) Confocal microscopy of ADP-activated EA.hy926 cells treated with PIPLC and stained with C5b-9 (depicted in red) and 4,6 diamidino-2-phenylindole (blue) as nuclei cell counterstaining. Magnification 40x. C5b-9 deposition is shown after incubation with aHUS serum (B) compared with heat-inactivated aHUS serum (C), TTP serum (D) and normal serum (E). PIPLC: Phosphatidylinositol-specific phospholipase C; aHUS: atypical hemolytic uremic syndrome; Heat-aHUS: heat inactivated atypical hemolytic uremic syndrome; TTP: thrombotic thrombocytopenic purpura.
As quantification of C5b-9 deposition by immunocytochemistry requires confocal microscopy and further analysis in specialized software, we aimed to quantify C5b-9 deposition by flow cytometry. It should be noted that initial analysis of surface C5b-9 staining showed low levels of deposition, and intracellular staining was preferred. EA.hy926 cells with normal GPI-AP expression displayed slightly increased MFI of C5b-9 staining after incubation with aHUS as compared with heat-inactivated aHUS serum. As expected, PIPLC-treated EA.hy926 cells had decreased GPI-AP expression. C5b-9 deposition on PIPLC-treated EA.hy926 l cells showed increased intensity after incubation with aHUS serum compared with the heat-inactivated control (supplemental Figure 4).
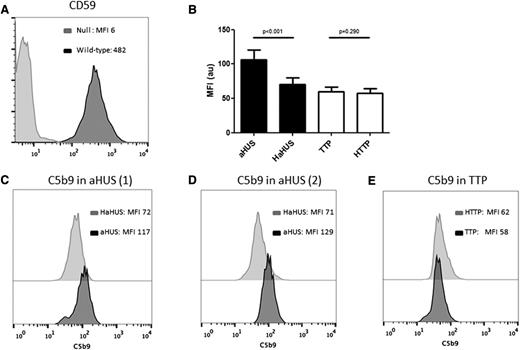
The above experiments suggested that partial biochemical removal of GPI-AP increased sensitivity to aHUS serum; thus, for future studies, we used a PIGA-null TF-1 cell line to measure C5b-9 deposition and cell viability following incubation with TMA patient serum. PIGA-mutant TF-1 cells have no surface expression of the complement regulatory proteins CD55 and CD59. As shown in Figure 2, these cells showed increased C5b-9 staining after incubation with aHUS serum as compared with heat-inactivated aHUS serum. Interestingly, similarly increased C5b-9 was observed in aHUS patients at presentation and in remission. In contrast, PIGA-null TF-1 cells incubated with TTP serum demonstrated no increase in C5b-9 staining compared with the heat-inactivated control and the normal serum. Results are summarized in Figure 2.
C5b-9 deposition by flow cytometry on PIGA-null TF-1 cells. (A) Loss of CD59 expression on PIGA null TF-1 cells vs wild type TF-1 cells. (B) Comparison of C5b-9 deposition between aHUS and TTP samples and their heat-inactivated controls (HaHUS and HTTP, respectively). (C) Representative experiment of C5b-9 deposition after incubation with acute TTP serum (black) vs heat-inactivated control (light gray). (D) C5b-9 deposition after incubation with serum from a patient with aHUS in remission (black) compared with heat-inactivated control (light gray). (E) C5b-9 deposition after incubation with serum from a second patient with acute aHUS (black) compared with heat-inactivated control (light gray). HaHUS, heat-inactivated atypical hemolytic uremic syndrome serum; HTTP, heat-inactivated thrombotic thrombocytopenic purpura; MFI, mean fluorescence intensity.
C5b-9 deposition by flow cytometry on PIGA-null TF-1 cells. (A) Loss of CD59 expression on PIGA null TF-1 cells vs wild type TF-1 cells. (B) Comparison of C5b-9 deposition between aHUS and TTP samples and their heat-inactivated controls (HaHUS and HTTP, respectively). (C) Representative experiment of C5b-9 deposition after incubation with acute TTP serum (black) vs heat-inactivated control (light gray). (D) C5b-9 deposition after incubation with serum from a patient with aHUS in remission (black) compared with heat-inactivated control (light gray). (E) C5b-9 deposition after incubation with serum from a second patient with acute aHUS (black) compared with heat-inactivated control (light gray). HaHUS, heat-inactivated atypical hemolytic uremic syndrome serum; HTTP, heat-inactivated thrombotic thrombocytopenic purpura; MFI, mean fluorescence intensity.
Cell viability
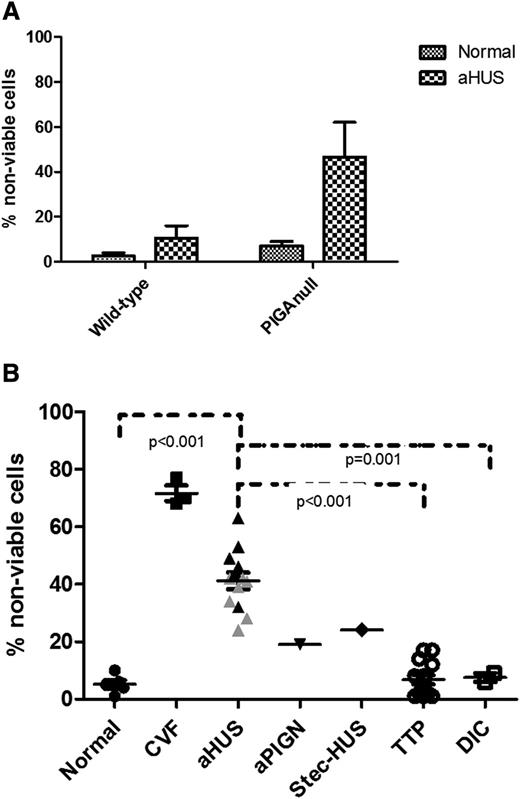
Complement induces cell killing through deposition of the membrane attack complex. In order to establish a simple, rapid, and less subjective assay, we exposed PIPLC-treated EA.hy926 cells and PIGA-null TF-1 cells to serum from our TMA patients and measured viability in a WST-1 assay. The cell viability assay was originally performed on EA.hy926 cells treated with PIPLC. Results in the first 11 patients (Figure 3) demonstrate consistently higher percentages of nonviable cells in patients with aHUS compared with other TMAs or controls. However, PIPLC-treated EA.hy926 cells display only a transient shift in CD59 expression, limiting their usefulness in a diagnostic assay.
WST-1 viability assay on PIPLC-treated EA.hy926 cells. Percentage of nonviable PIPLC-treated EA.hy926 cells among different disease entities (data are presented as mean with standard error of the mean [SEM]). aPIGN, atypical postinfectious glomerulonephritis; Ecu-aHUS, atypical hemolytic uremic syndrome treated with eculizumab; Stec-HUS: Shiga-toxin–associated hemolytic uremic syndrome.
WST-1 viability assay on PIPLC-treated EA.hy926 cells. Percentage of nonviable PIPLC-treated EA.hy926 cells among different disease entities (data are presented as mean with standard error of the mean [SEM]). aPIGN, atypical postinfectious glomerulonephritis; Ecu-aHUS, atypical hemolytic uremic syndrome treated with eculizumab; Stec-HUS: Shiga-toxin–associated hemolytic uremic syndrome.
We focused the rest of our studies on the PIGA-null TF-1 reagent cells, because genetic disruption of PIGA allowed us to eliminate the need for PIPLC treatment in our assay. Similar to our studies with C5b-9 deposition, we found the WST-1 killing assay to be more robust on the PIGA-null TF-1 cell line. As shown in Figure 4A, genetic disruption of PIGA gene augments cell killing in aHUS serum in the PIGA-null TF-1 cell line compared with the wild-type TF-1 cells. We next evaluated the sensitivity of the PIGA-null TF1 line to complement-mediated killing by serum from a variety of different patients with TMAs. aHUS serum was significantly more toxic to the PIGA-null cell line than serum from TTP (P < .001) and healthy controls (P < .001). Heat inactivation abrogated the cell killing and C5b-9 accumulation on the cells, demonstrating that the killing was associated with complement activation. As shown in Figure 4B, patients with other TMAs, including TTP (P < .001) and 1 patient with Shiga-toxin–associated HUS (P = .172), had low percentages of nonviable cells in the cell viability assay similar to levels in healthy controls. Patients with other causes of thrombocytopenia often included in the differential diagnosis of TMAs, such as DIC, also showed low levels of nonviable cells (P = .001). Intermediate levels of cell viability were found in a patient with atypical postinfectious glomerulonephritis, a disease recently shown to be associated with activation of APC.41 Because TF-1 cells do not express ABO antigens, ABO serum type had no effect on cell viability (P = .598).
Genetic disruption of PIGA augments cell killing in aHUS serum. (A) Comparison of nonviable cells between TF-1 wild-type and TF-1 PIGA-null cells (data are presented as mean with SEM) after incubation with normal and aHUS serum. (B) Percentage of nonviable PIGA-null TF1 cells among different disease entities (data are presented as mean with SEM). Serum from aHUS patients induces significantly increased percentages of nonviable cells compared with healthy controls (P < .001), TTP (P < .001), and DIC (P = .001). Gray triangles symbolize values of aHUS patients treated with eculizumab, whereas black triangles symbolize aHUS in acute phase or remission. CVF, CVF-activated serum; Stec-HUS, Shiga-toxin–associated hemolytic uremic syndrome.
Genetic disruption of PIGA augments cell killing in aHUS serum. (A) Comparison of nonviable cells between TF-1 wild-type and TF-1 PIGA-null cells (data are presented as mean with SEM) after incubation with normal and aHUS serum. (B) Percentage of nonviable PIGA-null TF1 cells among different disease entities (data are presented as mean with SEM). Serum from aHUS patients induces significantly increased percentages of nonviable cells compared with healthy controls (P < .001), TTP (P < .001), and DIC (P = .001). Gray triangles symbolize values of aHUS patients treated with eculizumab, whereas black triangles symbolize aHUS in acute phase or remission. CVF, CVF-activated serum; Stec-HUS, Shiga-toxin–associated hemolytic uremic syndrome.
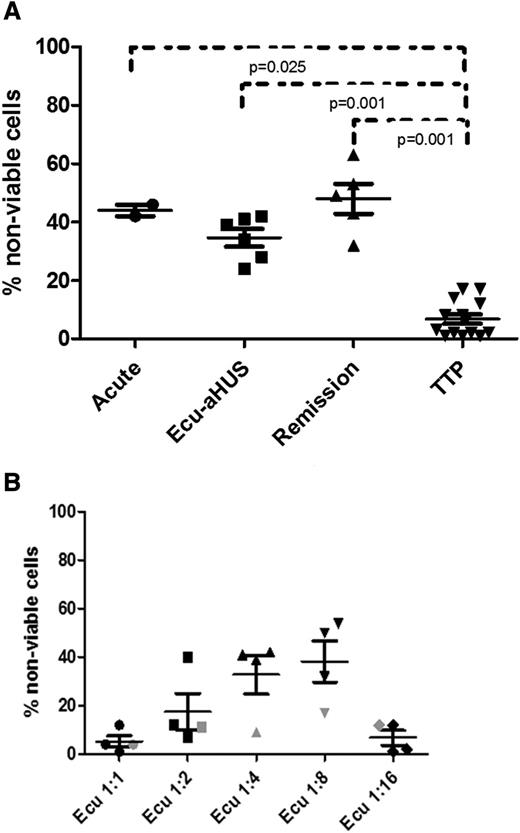
The WST-1 killing assay with PIGA-null TF-1 cells was able to distinguish aHUS from healthy controls and TTP patients regardless of whether the subjects were studied at presentation, in remission on eculizumab, or in remission off eculizumab (Figure 5A). Absorbance values of heat-inactivated aHUS and aHUS samples are shown in supplemental Table 1. We were initially surprised that the assay result was still positive in aHUS patients on eculizumab. We hypothesized that the 1:4 serum dilution was diluting the eculizumab concentration to a level that was no longer blocking terminal complement activation. To test this hypothesis, we performed our WST-1 assay on 3 aHUS patients within 7 days of their next scheduled dose of eculizumab using serial serum dilutions (1:1, 1:2, 1:4, 1:8, and 1:16), as shown in Figure 5B. Despite higher concentrations of serum as a source of complement, all 3 patients showed no increased killing compared with healthy controls at 1:1 dilutions; 2 of the 3 patients had no increase in killing at 1:2, and all 3 patients showed increased killing at 1:4 and 1:8 dilutions of their serum. There was insufficient complement in the 1:16 serum dilution in all 3 subjects to trigger complement-mediated killing in our assay. Lastly, to ensure that eculizumab was not somehow interfering with our assay, we tested serum from a PNH patient on eculizumab and saw no increased killing in the WST-1 assay.
WST-1 cell viability assay in patients with aHUS. (A) Similarly increased percentages of nonviable TF1 cells among aHUS patients in different disease states (acute phase, eculizumab treatment, and remission) compared with TTP patients. (B) Effects of serial dilution of eculizumab-treated serum on cell viability (1:1, 1:2, 1:4, 1:8, and 1:16). All aHUS patients showed no increase in killing at 1:1 and 1:16 dilutions, 2 of the 3 aHUS patients had no increase in killing at 1:2, and all 3 aHUS patients showed increased killing at 1:4 and 1:8 dilutions. Serum from a PNH patient treated with eculizumab (indicated by gray symbols) showed no increase in killing at any dilution. Ecu-aHUS, aHUS patient treated with eculizumab.
WST-1 cell viability assay in patients with aHUS. (A) Similarly increased percentages of nonviable TF1 cells among aHUS patients in different disease states (acute phase, eculizumab treatment, and remission) compared with TTP patients. (B) Effects of serial dilution of eculizumab-treated serum on cell viability (1:1, 1:2, 1:4, 1:8, and 1:16). All aHUS patients showed no increase in killing at 1:1 and 1:16 dilutions, 2 of the 3 aHUS patients had no increase in killing at 1:2, and all 3 aHUS patients showed increased killing at 1:4 and 1:8 dilutions. Serum from a PNH patient treated with eculizumab (indicated by gray symbols) showed no increase in killing at any dilution. Ecu-aHUS, aHUS patient treated with eculizumab.
We next sought to determine the cutoff value that distinguishes aHUS from TTP in our WST-1 assay on PIGA-null TF-1 cells. In the receiver-operating characteristic curve analysis, a significantly high area under the curve was found (0.996, P < .001; supplemental Figure 5). A percentage of nonviable cells higher than 21.5% was determined as a cutoff value for the diagnosis of aHUS among other TMAs with 100% sensitivity and 94.4% specificity. In terms of reproducibility, a significantly high Lin’s concordance correlation coefficient was calculated among 5 samples analyzed in the same experiment (0.923) and in 2 experiments conducted within 2 consecutive days (0.946), indicating high intra- and interexperiment reproducibility.
Discussion
The present study describes a reproducible, sensitive, and specific cell viability assay that helps to differentiate aHUS from other TMAs, irrespective of disease status (acute phase or remission, on or off therapy with terminal complement inhibition). The assay described here is analogous to the acidified serum (Ham test) for PNH (Figure 6).26 In PNH, there is a genetic mutation, PIGA, that causes a loss of complement regulatory proteins (CD55 and CD59) on PNH blood cells.23 Acidifying human serum activates APC and leads to specific lysis of PNH erythrocytes, because they are unable to protect themselves from the activated complement in acidified serum. In most aHUS cases, there are genetic mutations or antibodies that lead to activation APC in the patient’s serum. Thus, when PNH-like reagent cells (PIPLC-treated EA.hy926 cells or PIGA-null TF-1 cells) are incubated with aHUS serum, they rapidly accumulate C5b-9 and undergo cell death within 30 minutes in a WST-1 viability assay. We found the PIGA-null TF-1 cell line to be better that PIPLC-treated EA.hy926 cells in differentiating TMAs. This novel assay, in effect an “indirect Ham test,” helps to distinguish aHUS and TTP.
Model of Ham test for PNH and modified Ham test for aHUS diagnosis. The Ham test evaluates the effect of acidified serum on patient's cells (A), whereas the modified test evaluates the effect of patient's serum on GPI-AP–deficient reagent cells (B). Both tests use absorbance changes as readouts of cell viability.
Model of Ham test for PNH and modified Ham test for aHUS diagnosis. The Ham test evaluates the effect of acidified serum on patient's cells (A), whereas the modified test evaluates the effect of patient's serum on GPI-AP–deficient reagent cells (B). Both tests use absorbance changes as readouts of cell viability.
Tests that reliably, rapidly, and affordably distinguish aHUS and TTP are highly desirable given the severe morbidity and mortality associated with these diseases and the potential cost of choosing the wrong therapy (eculizumab vs plasma exchange). Currently, aHUS is predominantly a diagnosis of exclusion once typical HUS (Shiga toxin) and TTP (ADAMTS13 activity <10%) have been excluded. The most reliable assay to distinguish between these diseases is ADAMTS13 activity. Severe TTP is usually associated with <10% of plasma ADAMTS13. For that reason, ADAMTS13 activity higher than 10% in everyday clinical practice18 or even 5% in clinical trials16 is considered suggestive of aHUS diagnosis. However, partial ADAMTS13 deficiency may also be observed in aHUS patients with a heterozygous mutation in the ADAMTS13 gene.42
Because aHUS cannot be solely diagnosed based on ADAMTS13 activity and genetic testing, a number of recent studies have investigated diagnostic markers of aHUS. Complement components C5a and C5b-9 have been reported to be elevated in plasma of patients clinically characterized as aHUS before treatment, as compared with ADAMTS13-deficient TTP patients.22 However, similarly to our findings, values often overlap between the 2 groups and no cutoff value has been determined. In addition, increased plasma levels of C3b/c, convertase C3bBbP, and C5b-9 were also documented in aHUS in the acute phase as compared with remission.43 In an effort to develop an in vitro diagnostic assay, Noris et al have investigated C5b-9 deposition on ADP-activated EA.hy926 cells by confocal microscopy. In agreement with our data, increased C5b-9 deposition was found in aHUS patients in acute phase and in remission as compared with normal controls. These authors also noted increased C5b-9 deposition in unaffected mutation carriers. In contrast to our data, Noris et al were not able to detect a difference between healthy controls and aHUS patients on eculizumab therapy.40 Our assay could readily detect aHUS even in patients on eculizumab or in remission off eculizumab.
Approximately 50% to 60% of aHUS patients have known mutations in genes that either regulate or activate the APC, including complement factor H (CFH) and CFH-related proteins, complement factor I, CD46 (membrane cofactor protein), complement factor B, complement component C3, thrombomodulin, plasminogen, diacylglycerolkinase-ε (DGKE), and autoantibodies to CFH.12 Historically, penetrance of aHUS in patients with germline mutations has been estimated at 50%.44 A recent study has also revealed that penetrance is much lower in mutation-positive relatives regardless of the gene or patient age.45 Therefore, genetic modifiers and environmental factors (“triggers”) are considered crucial for aHUS manifestation (2-hit hypothesis). Our study is in agreement with the 2-hit hypothesis, showing that increased complement activation is evident not only in acute phase but also in remission when the second trigger can no longer be detected. We were also able to detect complement activation in the serum of aHUS patients on eculizumab. This observation is likely due to the fact that the concentration of eculizumab in serum diluted 1:4 is unable to block terminal complement. Indeed, killing was almost completely abrogated in serum diluted 1:1 and 1:2, suggesting that the WST-1 assay may be useful in titrating the most appropriate dose of eculizumab for aHUS patients. Our data also suggest that prompt recognition and treatment of aHUS may allow for discontinuation of eculizumab in a subset of patients as long as they are closely monitored for relapse analogous to withdrawing plasma exchange in patients with TTP who achieve remission. Indeed, 4 patients (1, 5, 6, and 8) have had their eculizumab treatment discontinued and remain in clinical remission for a median of 31 (range, 8-60) weeks. More patients and longer follow-up will be necessary to determine the wisdom of applying this approach, but given the expense of eculizumab, it will be necessary to determine the safety of this approach in future studies.
Our study has several limitations. First, due to the relatively small patient population, our assay needs to be validated in other independent TMA cohorts. In addition, our assay is based on excessive complement activation in patient sera and thus may not detect complement dysregulation due to mutations in cell-surface molecules, such as membrane cofactor protein and thrombomodulin. Similarly, novel mutations in DGKE have been described in aHUS developing with hypertension, microhematuria, proteinuria, and nephrotic syndrome.46 Because the relationship between these mutations and complement dysregulation is not fully clarified, diagnosis utilizing the present assay may not be feasible.47 Thus, our results need to be confirmed by further prospective studies with larger TMA cohorts that include patients with these mutations.
In conclusion, we have developed a modified Ham test with high reproducibility, sensitivity, and specificity in differentiating aHUS from other TMAs, based on complement-mediated apoptosis and death in GPI-AP–deficient cells. Favorable outcomes of eculizumab treatment in aHUS patients underscore the importance of this diagnostic assay in implementing treatment decisions. In an era of intense interest in precision medicine, our assay may be a promising tool to differentiate patients with increased complement activation who would benefit most from complement inhibition.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
E.G. was a Johns Hopkins-Libra fellow during the performance of these studies.
Authorship
Contribution: E.G. and R.A.B. designed and conducted the study, analyzed data, and drafted and corrected the manuscript; X.Y. and Z.Y. contributed in designing and performing research; and A.J.A., S.P.S., M.B.S., T.S.K., A.R.M., and C.J.S. contributed patients and helped in drafting and correcting the manuscript.
Conflict-of-interest disclosure: C.J.S. is Johns Hopkins principal investigator for the international aHUS registry sponsored by Alexion Pharmaceuticals (Cheshire, CT). R.A.B. is a member of the scientific advisory board of Alexion Pharmaceuticals. The remaining authors declare no competing financial interests.
Correspondence: Robert A. Brodsky, Ross Research Building, Room 1025, 720 Rutland Ave, Baltimore, MD 21205-2196; e-mail: rbrodsky@jhmi.edu.

![Figure 3. WST-1 viability assay on PIPLC-treated EA.hy926 cells. Percentage of nonviable PIPLC-treated EA.hy926 cells among different disease entities (data are presented as mean with standard error of the mean [SEM]). aPIGN, atypical postinfectious glomerulonephritis; Ecu-aHUS, atypical hemolytic uremic syndrome treated with eculizumab; Stec-HUS: Shiga-toxin–associated hemolytic uremic syndrome.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/23/10.1182_blood-2015-02-629683/4/m_3637f3.jpeg?Expires=1769799959&Signature=iHlKSU8QuaOWIFRFka4fTp5WlGuozYg9oAi8FVk-J3BmU3ug2NTJCwCgnwbW~ZyyQ857F7vPhI8Dsw7qRLxp8bbaIAse~LcXbVgwixrEryN78XnufjKKCnAFviqjVkJuisB9FBg4NdHIsS089UKjnMHna3M629-FPlQ04DqqwAZH6drevXqgb~qzgToMLXg43iQEErW-T~Mxk4kS2UB9nWcFNUf4QETHklRhqGB7YtrIpB2Jo2~Lev9QqhlQBJtS95hHcP2BGlFiV4wucP~x9rkV9zOpa5eHcN-NgfAsNUY9GDkR3gIqPNMN15bP7Y~PdRQwHT7MTmnqtx9qUYesxQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)