Key Points
Haploidentical transplantation from KIR ligand–mismatched donors with activating KIRs reduces nonrelapse mortality and improves survival.
Activating KIR genetics should be considered when selecting donors for T cell–depleted haploidentical hematopoietic transplantation.
Abstract
Because activating killer cell immunoglobulinlike receptors (KIRs) are heterogeneously expressed in the population, we investigated the role of donor activating KIRs in haploidentical hematopoietic transplants for acute leukemia. Transplants were grouped according to presence vs absence of KIR-ligand mismatches in the graft-vs-host direction (ie, of donor-vs-recipient natural killer [NK]-cell alloreactivity). In the absence of donor-vs-recipient NK-cell alloreactivity, donor activating KIRs had no effects on outcomes. In the 69 transplant pairs with donor-vs-recipient NK-cell alloreactivity, transplantation from donors with KIR2DS1 and/or KIR3DS1 was associated with reduced risk of nonrelapse mortality, largely infection related (KIR2DS1 present vs absent: hazard ratio [HR], 0.25; P = .01; KIR3DS1 present vs absent: HR, 0.18; P = .006), and better event-free survival (KIR2DS1 present vs absent: HR, 0.31; P = .011; KIR3DS1 present vs absent: HR, 0.30; P = .008). Transplantation from donors with KIR2DS1 and/or KIR3DS1 was also associated with a 50% reduction in infection rate (P = .003). In vitro analyses showed that KIR2DS1 binding to its HLA-C2 ligand upregulated inflammatory cytokine production by alloreactive NK cells in response to infectious challenges. Because ∼40% of donors able to exert donor-vs-recipient NK-cell alloreactivity carry KIR2DS1 and/or KIR3DS1, searching for them may become a feasible, additional criterion in donor selection.
Introduction
Hematopoietic transplantation from human leukocyte antigen (HLA) haplotype–mismatched (haploidentical) donors is an established treatment of patients with high-risk acute leukemia who do not have a matched donor.1-12
In haploidentical, T cell–depleted peripheral blood stem cell transplants without posttransplant graft-versus-host disease (GVHD) prophylaxis,3-5 donor natural killer (NK) cells play a beneficial role in outcomes.13-17 Human NK cells possess clonally distributed, inhibitory receptors termed “killer cell immunoglobulinlike receptors” (KIRs), which recognize epitopes shared by groups of HLA-class I alleles (KIR ligands).18-20 KIR2DL1 recognizes HLA-C group 2 alleles, KIR2DL2 and KIR2DL3 recognize HLA-C group 1 alleles, and KIR3DL1 is the receptor for HLA-Bw4 alleles. NK cells become fully functional (“licensed/educated”) upon recognition of self-HLA.18 KIRs are randomly expressed in the NK-cell repertoire. NK cells that express, as their only inhibitory receptor for self, a KIR for an HLA-class I group that is absent on allogeneic targets, sense the missing expression of the self-HLA-class I KIR ligand and mediate alloreactions (“missing self” recognition).21-24 In clinical hematopoietic stem cell transplantation, KIR-ligand mismatches in the graft-vs-host (GVH) direction trigger donor-vs-recipient NK-cell alloreactivity.13-17 This occurs when the donor possesses an HLA-class I KIR ligand (either HLA-C groups 1 or 2 and/or the HLA-Bw4 group) that is absent in the recipient. Under such mismatch conditions, engrafted stem cells give rise to an NK-cell repertoire that is licensed by HLA KIR ligands on donor hematopoietic cells and, thus, enabled to recognize and react to missing self on recipient targets.17,25 Donor-vs-recipient NK-cell alloreactions control leukemia relapse and improve survival in adult and pediatric patients.13-17,26,27
Homologs of the inhibitory KIRs, with shorter cytoplasmic tails, activating KIRs transduce signals that activate NK cells.28-31 Activating KIR genes are variably present in the population. Approximately 25% of whites are homozygous for group A KIR haplotypes that contain the main inhibitory KIR genes and the activating KIR2DS4 gene (however, a KIR2DS4 deletion variant allele encodes for a nonfunctional receptor in as much as ∼80% of individuals).32 The others are either heterozygous or homozygous for group B KIR haplotypes that carry not only inhibitory KIR genes but also various combinations of activating KIR genes (KIR2DS1, 2, 3, 5, and 3DS1). Little is known about their ligands except that KIR2DS1 binds HLA-C group 2 molecules.33,34 In vitro studies showed KIR2DS1 plays a role in NK cell killing of allogeneic targets that express HLA-C2.27,34-37 Moreover, in clinical studies, activating KIRs were found to be associated with less leukemia relapse in matched unrelated and haploidentical transplants38-43 and with protection against AIDS progression and hepatitis C virus infection.44,45
In the present study, we investigated the role of donor-activating KIRs in haploidentical transplants for acute leukemia grouped according to presence or absence of donor-vs-recipient NK-cell alloreactivity. We found that transplantation from donors who exerted donor-vs-recipient NK-cell alloreactivity and carried KIR2DS1 and/or KIR3DS1 genes was associated with markedly reduced nonrelapse mortality (NRM), which was largely infection related, and with significantly better event-free survival (EFS). Interestingly, our in vitro analyses showed that KIR2DS1 binding to HLA-C2 upregulated inflammatory cytokine production by alloreactive NK cells in response to infectious challenges.
Patients and methods
Patients
The role of the donor KIR genotype was retrospectively analyzed in 161 haploidentical hematopoietic transplants, 121 for acute myeloid leukemia (AML) and 40 for acute lymphoblastic leukemia (ALL), performed between 1993 and 2012. One-hundred fifty-nine patients had been included in previous studies.3-7,13,14,16 Patients received haploidentical, T cell–depleted granulocyte colony-stimulating factor (G-CSF)–mobilized peripheral blood CD34+ cell grafts.3-7 Forty-five patients also received an infusion of donor regulatory and conventional T cells (Treg/Tcon).6,7 No posttransplant GVHD prophylaxis was given. Approval was obtained from the Institutional Ethics Committee, and participants gave written informed consent in accordance with the Declaration of Helsinki.
GVHD was assessed by consensus criteria.46 NRM was defined as death preceding leukemia relapse, and EFS was defined as the interval from transplantation to leukemia relapse, to death from all causes, or to last follow-up.
Donor selection
Candidate transplant donors were assessed for HLA compatibility by serology (HLA-A, HLA-B) and by high-resolution molecular analysis (HLA-C, HLA-DR, HLA-DP, HLA-DQ). The donor selection algorithm was: donor/recipient cytomegalovirus (CMV) serostatus, donor-vs-recipient NK-cell alloreactivity,13,14,16 and donor/recipient relationship (maternal donors),47 in this order.
Donors were considered able to exert donor-vs-recipient NK-cell alloreactivity (termed “NK alloreactive donors” in the paper) when: (1) HLA-C and HLA-B typing showed KIR-ligand mismatches in the GVH direction—that is, the donor possessed an HLA-class I KIR ligand (either HLA-C groups 1 or 2 and/or HLA-B group Bw4) that was absent in the recipient; (2) donors possessed the relevant inhibitory KIR gene(s) for missing self-recognition of recipient targets (KIR2DL2 and/or KIR2DL3 were present in everyone, KIR2DL1 was absent in ∼3% of individuals, and KIR3DL1 was absent in ∼10%)31 ; (3) donors possessed alloreactive NK-cell clones against recipient targets (see NK-cell isolation, cloning, and allocytotoxicity assay). Because our previous analyses in large numbers of individuals had shown they possessed alloreactive NK clones against HLA-C group–mismatched targets,16 in the present study, NK-cell cloning and allocytotoxicity assays were not considered necessary and were therefore not routinely performed in HLA-C group–mismatched transplant donors (however, this information was available for 29/60 HLA-C group–mismatched transplant donors included in the present study). Thus, HLA typing and donor KIR genotyping were considered sufficient to define HLA-C group–mismatched transplant donors as “NK alloreactive.” Because previous analyses had shown that alloreactive NK clones against HLA-Bw4–mismatched targets are undetectable in approximately one-third of KIR3DL1+ individuals,16 NK-cell cloning and allocytotoxicity assays were considered necessary and were therefore routinely performed.
KIR genotyping
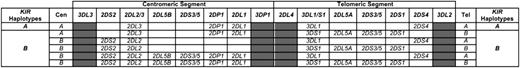
Genomic DNA was prepared from donor peripheral blood mononuclear cells (PBMCs) using the Nucleospin Tissue Kit (Macherey-Nagel, Düren, Germany). KIR genes were typed as described elsewhere16 or with the Olerup SSP KIR Genotyping kit (Olerup SSP AB, Stockholm, Sweden). KIR genes of the centromeric and telomeric segments of A and B haplotypes48-50 were analyzed as described elsewhere.39 Figure 1 shows the genomic organization of the KIR locus. Donor KIR genotypes are indicated as A/A when they did not contain B haplotypes; the centromeric segment is termed “Cen-A/A” and the telomeric “Tel-A/A.” Donor KIR genotypes are indicated as B/x when they contained at least 1 B haplotype; the centromeric segments are termed “Cen-B/x” and the telomeric “Tel-B/x.”
Genomic organization of the KIR locus.KIR genes segregate into groups A and B haplotypes. Framework genes located at the ends and in the central part of the locus (gray boxes) define 2 haplotype segments: the centromeric (Cen) and the telomeric (Tel). Common centromeric and telomeric segments are shown. In the A haplotype, KIR2DL1 and KIR2DL3 are found in the centromeric segment and KIR3DL1 and KIR2DS4 in the telomeric. Combinations of KIR2DL1/L2/L5/S2/S3/S5 are found in B-haplotype centromeric segments and combinations of KIR2DL5/S1/S3/S5 and KIR3DS1 in B-haplotype telomeric segments.
Genomic organization of the KIR locus.KIR genes segregate into groups A and B haplotypes. Framework genes located at the ends and in the central part of the locus (gray boxes) define 2 haplotype segments: the centromeric (Cen) and the telomeric (Tel). Common centromeric and telomeric segments are shown. In the A haplotype, KIR2DL1 and KIR2DL3 are found in the centromeric segment and KIR3DL1 and KIR2DS4 in the telomeric. Combinations of KIR2DL1/L2/L5/S2/S3/S5 are found in B-haplotype centromeric segments and combinations of KIR2DL5/S1/S3/S5 and KIR3DS1 in B-haplotype telomeric segments.
NK-cell isolation, cloning, and allocytotoxicity assay
NK cells were isolated from PBMCs using the Human NK Isolation Kit (Miltenyi Biotech, Bergisch Gladbach, Germany). Limiting dilution cloning and cytotoxicity assays determined alloreactive NK-cell repertoires in 110 HLA-C1+ healthy individuals (that included 10 HLA-C1–mismatched transplant donors) against allogeneic HLA-C1–missing cell targets. NK cells were plated under limiting-dilution conditions, activated with phytohemagglutinin (PHA) (Biochrom AG, Berlin, Germany), and cultured with interleukin-2 (Novartis Farma S.p.A., Origgio, Italy) and irradiated feeder cells. NK clones were screened for alloreactivity by 51Cr (PerkinElmer Inc., Waltham, MA) release assay against PHA T-cell blasts or cryopreserved primary AML cells at an effector-to-target (E:T) ratio of 10:1. Clones exhibiting >30% lysis were scored as alloreactive.
Flow cytometry
FACSCanto flow cytometer with FACSDiva software (BD Biosciences, San José, CA) and FlowJo software (Tree Star Inc., Ashland, OR) were used for multicolor immunofluorescence analyses and intracellular staining.
NK clones that were alloreactive against HLA-C1–missing cell targets were phenotyped with the following monoclonal antibodies (mAbs): anti-CD56-APC (clone NKH-1, IgG1), anti-CD3-PE-Cy7 (clone UCHT1, IgG1) (Beckman Coulter Inc., Fullerton, CA), and anti-KIR2DL2/L3/S2-FITC (clone CH-L, IgG2b) (BD Biosciences) in combination with either anti-KIR2DL1/S1-PE (clone EB6B, IgG1), anti-KIR3DL1/S1-PE (clone Z27, IgG1), anti-NKG2A-PE (clone Z199, IgG2b) (Beckman Coulter Inc.), or purified anti-KIR2DL3 (clone ECM-41, IgM, courtesy of Daniela Pende, Istituto Nazionale per la Ricerca sul Cancro, Genoa, Italy) conjugated with anti-mouse IgM-PE (Southern Biotech, Birmingham, AL). Anti-KIR2DL1/S1-PE (clone EB6B) was used in combination with anti-KIR2DL1-FITC (clone #143211, IgG1) (R&D Systems Inc., Minneapolis, MN) to distinguish between KIR2DL1 and KIR2DS1 expression. Anti-KIR3DL1/S1-PE (clone Z27) combined with anti-KIR3DL1-FITC (clone DX9, IgG1) (Miltenyi Biotech) discriminated between KIR3DL1 and KIR3DS1 expression. Alloreactive NK clones that coexpressed KIR3DS1 were not considered when investigating killing of HLA-C1–missing targets.
KIR gene expression
Gene expression confirmed KIR phenotypes in randomly selected NK clones. RNA was extracted from NK-cell clones using RNAqueous-4PCR kit (Life Technologies, Carlsbad, CA) and reverse transcribed with iScript Reverse Transcription Supermix (Bio-Rad Laboratories, Hercules, CA). cDNA was subjected to real-time polymerase chain reaction analysis using the specific primer set for the KIR genes published elsewhere51 and SsoAdvanced Universal SYBR Green Supermix (Bio-Rad Laboratories). Amplification and data acquisition were carried out on a Chromo4 PTC-200 Thermal Cycler (Bio-Rad Laboratories). The 2−ΔΔCT method was used for relative quantification to reference gene expression.
Dendritic cell generation and phenotyping
Monocytes were purified from healthy individual PBMCs by positive anti-CD14 immuno-magnetic selection (Miltenyi Biotech) and plated at a 1 × 106 cells/mL. To generate immature myeloid CD11c+ CD14– CD1a+ dendritic cells (DCs), monocytes were cultured for 6 days in the presence of 1000 UI/mL recombinant granulocyte-macrophage colony-stimulating factor and 500 UI/mL rIL-4 (Miltenyi Biotech). To generate CD1a+ CD83+ CD86+ mature DCs, immature DCs were stimulated with 2 μg/mL lipopolysaccharide (LPS) (Sigma-Aldrich/Merck, St. Louis, MO) for 24 hours or were exposed to live conidia of A fumigatus at a DC/fungi ratio of 1:1 for 24 hours, as described elsewhere.52
Myeloid DCs were phenotyped with the following mAbs: anti-CD14-FITC (clone TUk4, IgG2a) (Miltenyi Biotech); and anti-CD11c-FITC (clone 3.9, IgG1), anti-CD1a-PE (clone HI149, IgG1), anti-CD83-APC (clone HB15e, IgG1), anti-CD86-PE (clone IT2.2, IgG2b) (eBiosciences, San Diego, CA).
NK cell cytokine production
Freshly isolated NK cells were incubated with DCs at an E:T ratio of 1:2 for 6 hours in the presence of Golgi Stop (BD Biosciences) according to kit instructions. Interferon (IFN)-γ production was analyzed by intracellular staining. Because available antibodies do not discriminate among KIR2DL2, KIR2DL3, and KIR2DS2, KIR2DL3-homozygous individuals were used as NK-cell donors. Individuals with known anti-KIR2DS1/DL1 mAb (clone EB6) crossreactivity with a KIR2DL3 allele were excluded.53 NK cells that were potentially alloreactive against HLA-C1–missing cell targets were identified with the following combination of mAbs: anti-CD56-PE-Cy7 (clone NCAM16.02, IgG1) (BD Biosciences), anti-KIR2DL2/L3/S2-FITC (clone CH-L, IgG2b), anti-KIR3DL1/S1-PE (clone Z27), anti-NKG2A-PE (clone Z199), anti-KIR2DL1-PE (clone #143211, IgG1), anti-KIR3DL1/L2-PE (clone 5.133, IgG1), and anti-KIR2DS4-PE (clone JJC11.6, IgG1) (Miltenyi Biotech). Subsequently, anti-KIR2DL1/S1-APC mAb (clone EB6B, IgG1) (Beckman Coulter Inc.) was added to discriminate between KIR2DL1 and KIR2DS1, as described elsewhere.34 Cells were fixed and permeabilized with BD Cytofix/Cytoperm Plus Fixation/Permeabilization kit (BD Biosciences) and stained with anti-IFN-γ-PerCp-Cy5.5 (clone B27, IgG1) (BD Biosciences). Fluorescence compensation for spectral overlap was optimized with BD anti-mouse Ig, κ compensation beads set (BD Biosciences).
For IFN-γ production–blocking experiments, an average of 3 alloreactive NK clones per individual were pooled and incubated with DCs at an E:T ratio of 1:1 for 4 hours, in the presence or absence of anti-KIR2DL1/S1 (clone EB6B) F(ab′)2 (courtesy of Francois Romagné, Innate-Pharma, Marseille, France).54 Human IFN-γ concentrations were measured in cell-culture supernatants by an enzyme-linked immunosorbent assay kit (Life Technologies).
Statistical analyses
Demographics and prognostic variables were compared using the Fisher exact test, the χ2 test for categorical variables, and the Student t test or Mann-Whitney U test for continuous variables. The Kaplan-Meier method evaluated EFS. A log-rank test assessed the impact of donor KIR genotype on EFS. Cumulative incidence estimates were used for relapse, NRM, because they are competing risks. The cumulative incidences of acute GVHD and infectious mortality were calculated using death from any cause as a competing risk. The Gray test compared univariate competing risk outcomes.55
Multivariate analyses assessed the impact of donor-vs-recipient NK-cell alloreactivity and donor KIR genotype. They included the following variables: disease, Treg/Tcon infusions, disease status at transplant (remission vs relapse), patient age, patient and donor gender, conditioning regimens, CD34+ and CD3+ cell-graft content, donor/recipient CMV serostatus, and donor/recipient relationship (maternal donor vs others). Variables were included in a Cox regression model in a conditional forward stepwise fashion to identify factors with a significant impact on outcomes. Donor-vs-recipient NK-cell alloreactivity and disease status at transplant were included in the final model for relapse. Donor-vs-recipient NK-cell alloreactivity, disease status at transplant, and patient age per increasing year were included in the final model for EFS. When evaluating outcomes of transplants from NK-alloreactive donors, donor KIR genotype and donor/recipient relationship were included in the final model for NRM and infectious mortality; donor KIR genotype, disease status at transplant, donor/recipient relationship, and donor/recipient CMV status were included in the final model for EFS.
All P values are 2-sided and considered significant at P < .05.
Results
Overall clinical outcomes
Transplant characteristics are shown in supplemental Table 1, available on the Blood Web site. Primary engraftment was obtained in 155 of 161 patients. Rejection in 6 patients was reversed with a second transplant from the same or another haploidentical donor. Incidence of relapse was 23%. Incidence of NRM was 38.5%, owing to the fact that 62 patients died in remission, 51 of infections. The incidence of NRM was stable over time: 39% in transplants performed from 1993 to 1999, 36% from 2000 to 2006, and 43% from 2007 to 2012. Grade II-IV acute GVHD developed in 19 patients (12%), with 6 progressing to chronic GVHD. De novo chronic GVHD developed in 2 patients. At a mean follow-up of 4.07 years (range, 0.03-21.32), the probability of EFS was 38.5%.
Sixty-nine patients received transplants from NK alloreactive donors and 92 patients from non–NK-alloreactive donors. In agreement with previous studies,13,14,16 multivariate analyses showed that donor-vs-recipient NK-cell alloreactivity was associated with reduced relapse (transplantation from NK-alloreactive donors vs non–NK-alloreactive donors—HR, 0.42; 95% confidence interval [CI], 0.18-0.96; P = .04) and improved EFS in patients with AML (but not ALL) (transplantation from NK-alloreactive donors vs non–NK-alloreactive donors—HR, 0.60; 95% CI, 0.37-0.96; P = .034).
Transplantation from NK-alloreactive donors with activating KIR genes reduces NRM and improves EFS
We first evaluated the role of donor B vs A KIR haplotype centromeric segments (termed Cen-B/x and Cen-A/A, respectively) and telomeric segments (termed Tel-B/x and Tel-A/A, respectively) on transplantation outcomes in the whole series of 161 transplants and found no effects on EFS. We therefore analyzed transplants grouped according to the presence or absence of donor-vs-recipient NK-cell alloreactivity. We found no effects on EFS in the 92 transplants from non–NK-alloreactive donors (Cen-B/x vs Cen-A/A: 24.6% vs 41.9%, P = NS; Tel-B/x vs Tel-A/A: 32.5% vs 28.8%, P = NS).
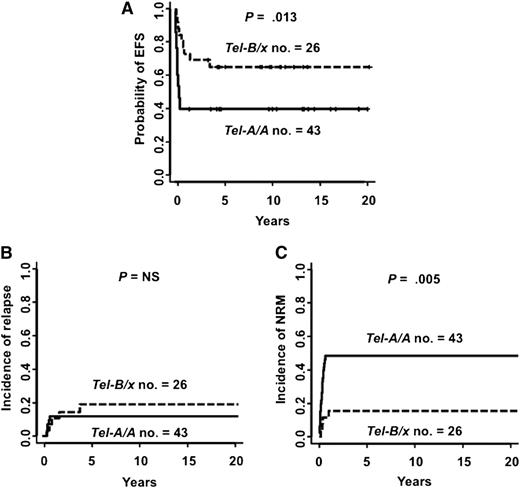
In contrast, donor activating KIR genotypes favorably affected outcomes of the 69 transplants from NK-alloreactive donors. Characteristics of patients transplanted from NK-alloreactive donors are shown in Table 1. Compared with transplantation from NK-alloreactive donors carrying Tel-A/A genes, transplantation from NK-alloreactive donors with Tel-B/x genes improved EFS (at 5 years after transplant, Tel-B/x vs Tel-A/A, 65.4% vs 39.6%; P = .013) (Figure 2A). Cox models incorporating interaction terms demonstrated that the effect of Tel-B/x genes depended on donor-vs-recipient NK-cell alloreactivity (P = .03). Transplantation from NK-alloreactive donors with Tel-B/x genes did not affect leukemia relapse (Tel-B/x vs Tel-A/A, 19.2% vs 11.6%; P = NS) (Figure 2B) or GVHD (Tel-B/x vs Tel-A/A, 11.5% vs 9.5%, P = NS). In fact, it reduced NRM (at 5 years after transplant, Tel-B/x vs Tel-A/A, 15.4% vs 48.8%; P = .005) (Figure 2C). B vs A haplotype centromeric KIR genes did not affect EFS (Cen-B/x vs Cen-A/A, 48.8% vs 50%; P = NS). Multivariate analyses showed that transplantation from NK-alloreactive donors with Tel-B/x genes was an independent factor predicting lower NRM (Tel-B/x vs Tel-A/A: HR, 0.23; 95% CI, 0.08-0.66; P = .007) and better EFS (Tel-B/x vs Tel-A/A: HR, 0.33; 95% CI, 0.14-0.76; P = .009). The other independent variables that improved EFS in NK-alloreactive transplants were maternal donors (HR, 0.25; 95% CI, 0.07-0.86; P = .028) and disease remission at transplant (HR, 0.45; 95% CI, 0.21-0.95; P = .037), with maternal donors affecting NRM (HR, 0.13; 95% CI, 0.02-0.99; P = .05) and disease remission affecting relapse (HR, 0.17; 95% CI, 0.04-0.75; P = .019). Multivariate analyses also showed that transplantation from donors with KIR2DS1 and/or KIR3DS1 (present only in B-haplotype telomeric segments) was associated with less NRM (KIR2DS1 present vs absent: HR, 0.25; 95% CI, 0.08-0.72; P = .01; and KIR3DS1 present vs absent: HR, 0.18; 95% CI, 0.05-0.62, P = .006) and better EFS (KIR2DS1 present vs absent: HR, 0.31; 95% CI, 0.13-0.76; P = .011; and KIR3DS1 present vs absent: HR, 0.30; 95% CI, 0.12-0.73; P = .008).
Characteristics of transplants with donor-vs-recipient NK-cell alloreactivity
| . | Donor KIR genotype* . | |||
|---|---|---|---|---|
| . | Cen-A/A, n = 28 . | Cen-B/x, n = 41 . | Tel-A/A, n = 43 . | Tel-B/x, n = 26 . |
| Patient sex, n (%) | ||||
| Male | 12 (43) | 20 (49) | 19 (44) | 13 (50) |
| Female | 16 (57) | 21 (51) | 24 (56) | 13 (50) |
| Median age (range), years | 42 (14-58) | 35 (14-62) | 39 (16-62) | 30.5 (14-59) |
| Disease, n (%) | ||||
| AML | 23 (82) | 37 (90) | 36 (84) | 24 (92) |
| ALL | 5 (18) | 4 (10) | 7 (16) | 2 (8) |
| Disease status at transplant, n (%) | ||||
| High-risk† complete remission | 21 (75) | 28 (68) | 28 (65) | 21 (81) |
| Chemoresistant relapse‡ | 7 (25) | 13 (32) | 15 (35) | 5 (19) |
| CMV serostatus (donor/recipient), n (%) | ||||
| Positive/positive | 26 (93) | 36 (88) | 39 (91) | 23 (88) |
| Positive/negative | 1 (3.5) | 2 (5) | 2 (5) | 1 (4) |
| Negative/positive | 0 (0) | 2 (5) | 1 (2) | 1 (4) |
| Negative/negative | 1 (3.5) | 1 (2) | 1 (2) | 1 (4) |
| Donor/recipient relationship, n (%) | ||||
| Maternal donor | 6 (21) | 7 (17) | 7 (16) | 6 (23) |
| Other donor | 22 (79) | 34 (83) | 36 (84) | 20 (77) |
| Transplant protocol, n (%) | ||||
| T cell–depleted§ | 20 (71) | 33 (80) | 33 (77) | 20 (77) |
| Treg/Tcon¶ | 8 (29) | 8 (20) | 10 (23) | 6 (23) |
| Conditioning regimen#, n (%) | ||||
| T cell–depleted protocol | ||||
| TBI, thiotepa, fludarabine, antithymocyte globulin** | 20 (71) | 33 (80.5) | 33 (77) | 20 (77) |
| Treg/Tcon protocol | ||||
| TBI, thiotepa, fludarabine, cyclophosphamide | 5 (18) | 7 (17) | 7 (16) | 5 (19) |
| TBI, thiotepa, fludarabine, alemtuzumab, or thymoglobulin | 3 (11) | 1 (2.5) | 3 (7) | 1 (4) |
| Graft composition (mean ± SD) | ||||
| CD34+ × 106/kg | 10.2 ± 3.3 | 11.9 ± 4.4 | 11.1 ± 4.0 | 11.4 ± 4.2 |
| CD3+ × 104/kg | 2.3 ± 2.2 | 3.2 ± 3.9 | 2.5 ± 2.8 | 3.3 ± 4.0 |
| . | Donor KIR genotype* . | |||
|---|---|---|---|---|
| . | Cen-A/A, n = 28 . | Cen-B/x, n = 41 . | Tel-A/A, n = 43 . | Tel-B/x, n = 26 . |
| Patient sex, n (%) | ||||
| Male | 12 (43) | 20 (49) | 19 (44) | 13 (50) |
| Female | 16 (57) | 21 (51) | 24 (56) | 13 (50) |
| Median age (range), years | 42 (14-58) | 35 (14-62) | 39 (16-62) | 30.5 (14-59) |
| Disease, n (%) | ||||
| AML | 23 (82) | 37 (90) | 36 (84) | 24 (92) |
| ALL | 5 (18) | 4 (10) | 7 (16) | 2 (8) |
| Disease status at transplant, n (%) | ||||
| High-risk† complete remission | 21 (75) | 28 (68) | 28 (65) | 21 (81) |
| Chemoresistant relapse‡ | 7 (25) | 13 (32) | 15 (35) | 5 (19) |
| CMV serostatus (donor/recipient), n (%) | ||||
| Positive/positive | 26 (93) | 36 (88) | 39 (91) | 23 (88) |
| Positive/negative | 1 (3.5) | 2 (5) | 2 (5) | 1 (4) |
| Negative/positive | 0 (0) | 2 (5) | 1 (2) | 1 (4) |
| Negative/negative | 1 (3.5) | 1 (2) | 1 (2) | 1 (4) |
| Donor/recipient relationship, n (%) | ||||
| Maternal donor | 6 (21) | 7 (17) | 7 (16) | 6 (23) |
| Other donor | 22 (79) | 34 (83) | 36 (84) | 20 (77) |
| Transplant protocol, n (%) | ||||
| T cell–depleted§ | 20 (71) | 33 (80) | 33 (77) | 20 (77) |
| Treg/Tcon¶ | 8 (29) | 8 (20) | 10 (23) | 6 (23) |
| Conditioning regimen#, n (%) | ||||
| T cell–depleted protocol | ||||
| TBI, thiotepa, fludarabine, antithymocyte globulin** | 20 (71) | 33 (80.5) | 33 (77) | 20 (77) |
| Treg/Tcon protocol | ||||
| TBI, thiotepa, fludarabine, cyclophosphamide | 5 (18) | 7 (17) | 7 (16) | 5 (19) |
| TBI, thiotepa, fludarabine, alemtuzumab, or thymoglobulin | 3 (11) | 1 (2.5) | 3 (7) | 1 (4) |
| Graft composition (mean ± SD) | ||||
| CD34+ × 106/kg | 10.2 ± 3.3 | 11.9 ± 4.4 | 11.1 ± 4.0 | 11.4 ± 4.2 |
| CD3+ × 104/kg | 2.3 ± 2.2 | 3.2 ± 3.9 | 2.5 ± 2.8 | 3.3 ± 4.0 |
TBI, total body irradiation.
Donor KIR genotypes are indicated as A/A when they did not contain B haplotypes; the centromeric segment is termed “Cen-A/A” and the telomeric “Tel-A/A.” Donor KIR genotypes are indicated as B/x when they contained at least 1 B haplotype; the centromeric segments are termed “Cen-B/x” and the telomeric “Tel-B/x.” Distribution of variables did not differ significantly in transplants from Cen-A/A vs Cen-B/x donors and Tel-A/A vs Tel-B/x donors (P > .05).
Myelodysplasia or unfavorable cytogenetics or first-line induction therapy failure in first to third complete remission.
Morphologic evidence of leukemic cells in the bone marrow and/or blood after treatment.
T cell–depleted granulocyte colony-stimulating factor (G-CSF)–mobilized peripheral blood CD34+ hematopoietic progenitor cell graft.3-5
T cell–depleted G-CSF–mobilized peripheral blood CD34+ hematopoietic progenitor cell graft and Treg/Tcon infusion.6,7
See supplemental Table 1 for details on conditioning regimens.
Four patients received cyclophosphamide instead of fludarabine.3
Transplantation from NK-alloreactive donors with B-haplotype telomeric KIR genes reduced NRM and improved EFS. (A) Probability of EFS; (B) cumulative incidence of leukemia relapse; and (C) cumulative incidence of NRM. Tel-B/x, 26 patients transplanted from NK-alloreactive donors with B-haplotype telomeric KIR genes; Tel-A/A, 43 patients transplanted from donors with A-haplotype telomeric KIR genes.
Transplantation from NK-alloreactive donors with B-haplotype telomeric KIR genes reduced NRM and improved EFS. (A) Probability of EFS; (B) cumulative incidence of leukemia relapse; and (C) cumulative incidence of NRM. Tel-B/x, 26 patients transplanted from NK-alloreactive donors with B-haplotype telomeric KIR genes; Tel-A/A, 43 patients transplanted from donors with A-haplotype telomeric KIR genes.
Infectious mortality accounted for 80% of NRM. In fact, transplantation from NK-alloreactive donors with Tel-B/x genes decreased infectious mortality (Tel-B/x vs Tel-A/A: 7.7% vs 41.9%, P = .0027) (Figure 3A). In multivariate analysis, it was an independent factor predicting lower infectious mortality (Tel-B/x vs Tel-A/A: HR, 0.13; 95% CI, 0.03-0.58; P = .007). Moreover, transplantation from NK-alloreactive donors with Tel-B/x genes was associated with a 50% reduction in the infection rate per month (Tel-B/x vs Tel-A/A: 0.91 ± 0.81 vs 0.45 ± 0.38; P = .003) (Figure 3B). The protective effect was mainly exerted on viral infection (Tel-B/x vs Tel-A/A: 0.45 ± 0.51 vs 0.20 ± 0.22; P = .0014) and, to a lesser extent, on fungal infections (Tel-B/x vs Tel-A/A: 0.22 ± 0.42 vs 0.07 ± 0.11; P = .09). Analysis of the ratio between the infection rate in patients transplanted from Tel-B/x donors and patients transplanted from Tel-A/A donors showed the protective effect was evident from the second to the fifth month after transplant (Figure 3C). Donor B vs A centromeric genotypes did not affect the infection rate (0.72 vs 0.73).
Transplantation from NK-alloreactive donors with B-haplotype telomeric KIR genes reduced infectious mortality and morbidity. (A) Cumulative incidence of infectious mortality in the 26 patients transplanted from NK-alloreactive donors with Tel-B/x genes and in the 43 patients transplanted from NK-alloreactive donors with Tel-A/A genes. Eighteen of 43 patients transplanted from donors with Tel-A/A genes died of infection (1 bacterial, 1 Toxoplasma gondii, 4 invasive fungal, 4 CMV, 2 human herpesvirus 6, 1 Epstein-Barr virus, 1 parainfluenza virus, 4 pneumonia and acute respiratory distress syndrome of unknown etiology). Only 2 of 26 patients transplanted from donors with Tel-B/x genes died of infection (1 CMV and 1 concomitant viral and fungal infection). (B) Data on infectious episodes were available for 63 of 69 patients transplanted from NK-alloreactive donors. Number of infectious episodes per month (mean ± standard deviation [SD]) in the first 6 months after transplant in patients transplanted from NK-alloreactive donors with Tel-A/A (open bar) and Tel-B/x genes (closed bar) are shown. Difference between the groups was tested by unpaired Student t test. (C) Ratios (mean with 95% CI) between the infection rates of patients transplanted from NK-alloreactive donors with Tel-B/x (diamonds) and infection rates of patients transplanted from NK-alloreactive donors Tel-A/A genes (dotted line) as evaluated monthly during the first 6 months after transplant.
Transplantation from NK-alloreactive donors with B-haplotype telomeric KIR genes reduced infectious mortality and morbidity. (A) Cumulative incidence of infectious mortality in the 26 patients transplanted from NK-alloreactive donors with Tel-B/x genes and in the 43 patients transplanted from NK-alloreactive donors with Tel-A/A genes. Eighteen of 43 patients transplanted from donors with Tel-A/A genes died of infection (1 bacterial, 1 Toxoplasma gondii, 4 invasive fungal, 4 CMV, 2 human herpesvirus 6, 1 Epstein-Barr virus, 1 parainfluenza virus, 4 pneumonia and acute respiratory distress syndrome of unknown etiology). Only 2 of 26 patients transplanted from donors with Tel-B/x genes died of infection (1 CMV and 1 concomitant viral and fungal infection). (B) Data on infectious episodes were available for 63 of 69 patients transplanted from NK-alloreactive donors. Number of infectious episodes per month (mean ± standard deviation [SD]) in the first 6 months after transplant in patients transplanted from NK-alloreactive donors with Tel-A/A (open bar) and Tel-B/x genes (closed bar) are shown. Difference between the groups was tested by unpaired Student t test. (C) Ratios (mean with 95% CI) between the infection rates of patients transplanted from NK-alloreactive donors with Tel-B/x (diamonds) and infection rates of patients transplanted from NK-alloreactive donors Tel-A/A genes (dotted line) as evaluated monthly during the first 6 months after transplant.
KIR2DS1 binding to HLA-C2 triggers cytokine production in alloreactive NK cells
Because KIR2DS1 is the only activating KIR for which binding to an HLA ligand (HLA-C2) is well documented,33,34 cytotoxicity and cytokine production were evaluated in alloreactive NK cells from HLA-C1+ healthy individuals upon triggering by HLA-C1–missing, HLA-C2/C2 targets.
Our present data show that frequencies of alloreactive NK-cell clones against HLA-C1–missing, HLA-C2/C2 PHA T-cell blasts in 45 individuals with the KIR2DS1 gene did not differ significantly from those in 65 individuals without the KIR2DS1 gene (Figure 4A). Next, such analysis was performed separately in individuals possessing the KIR2DL2 and/or KIR2DL3 inhibitory receptor genes. In fact, in HLA-C1+ individuals, NK cells that are alloreactive against HLA-C1–missing targets express KIR2DL2 and/or KIR2DL3 as their only inhibitory receptor(s) for self–HLA-C1. KIR2DL2, and to a lesser extent KIR2DL3, crossreact with HLA-C2 molecules, thereby possibly hampering allorecognition of HLA-C1–missing, HLA-C2C2 targets.27,56 Figure 4B shows that KIR2DS1 did not significantly affect the frequencies of alloreactive NK-cell clones in KIR2DL2 or KIR2DL3 homozygous or KIR2DL2/3 heterozygous individuals. Clones that expressed KIR2DS1 killed targets with equal efficacy than clones that did not express KIR2DS1 (Figure 4C). Finally, clones that expressed KIR2DS1 killed primary AML cells as efficiently as those that did not express KIR2DS1 (Figure 5).
KIR2DS1 does not affect alloreactive NK-cell repertoires. (A) A total of 11 999 NK-cell clones were generated from 110 HLA-C1+ individuals (mean 109 clones/individual [30-302]) and were screened for cytotoxicity against HLA-C1–missing, HLA-C2/C2 PHA T-cell blasts. Frequencies (mean ± SD) of alloreactive NK-cell clones in the 65 individuals without the KIR2DS1 gene (open bar) and in the 45 individuals with the KIR2DS1 gene (closed bar) are shown (P = NS by unpaired Student t test). (B) The KIR2DS1– and KIR2DS1+ individuals in (A) were grouped according to their KIR2DL2 or KIR2DL3 homozygous or KIR2DL2/3 heterozygous inhibitory genotypes. Frequencies of alloreactive NK-cell clones in KIR2DS1– (open bar) and KIR2DS1+ (closed bar) individuals are shown (P = NS by unpaired Student t test). (C) A total of 2281 clones obtained from 20 HLA-C1+ individuals (randomly selected from the 110 in [A]) were screened for allocytotoxicity against HLA-C1–missing, HLA-C2C2 PHA T-cell blasts. The contribution of KIR2DS1 to lysis was evaluated in 127 allocytotoxic NK-cell clones that expressed KIR2DL2/L3 (as evaluated by phenotypic analyses). Lysis (mean ± SD) by the 92 clones that did not express KIR2DS1 (open bar) and the 35 clones that expressed KIR2DS1 (closed bar) is shown.
KIR2DS1 does not affect alloreactive NK-cell repertoires. (A) A total of 11 999 NK-cell clones were generated from 110 HLA-C1+ individuals (mean 109 clones/individual [30-302]) and were screened for cytotoxicity against HLA-C1–missing, HLA-C2/C2 PHA T-cell blasts. Frequencies (mean ± SD) of alloreactive NK-cell clones in the 65 individuals without the KIR2DS1 gene (open bar) and in the 45 individuals with the KIR2DS1 gene (closed bar) are shown (P = NS by unpaired Student t test). (B) The KIR2DS1– and KIR2DS1+ individuals in (A) were grouped according to their KIR2DL2 or KIR2DL3 homozygous or KIR2DL2/3 heterozygous inhibitory genotypes. Frequencies of alloreactive NK-cell clones in KIR2DS1– (open bar) and KIR2DS1+ (closed bar) individuals are shown (P = NS by unpaired Student t test). (C) A total of 2281 clones obtained from 20 HLA-C1+ individuals (randomly selected from the 110 in [A]) were screened for allocytotoxicity against HLA-C1–missing, HLA-C2C2 PHA T-cell blasts. The contribution of KIR2DS1 to lysis was evaluated in 127 allocytotoxic NK-cell clones that expressed KIR2DL2/L3 (as evaluated by phenotypic analyses). Lysis (mean ± SD) by the 92 clones that did not express KIR2DS1 (open bar) and the 35 clones that expressed KIR2DS1 (closed bar) is shown.
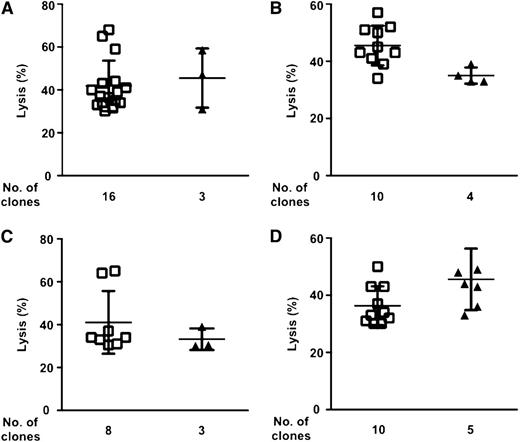
KIR2DS1 does not affect NK-cell alloreactivity against AML cells. 1265 NK-cell clones from 10 HLA-C1+ individuals were screened for cytotoxicity against a panel of 4 HLA-C1–missing, HLA-C2/C2 AML (A-D). The contribution of KIR2DS1 to lysis was evaluated in 59 allocytotoxic NK-cell clones that expressed KIR2DL2/L3 (as evaluated by phenotypic analyses). Lysis (mean ± SD) exerted by KIR2DS1– (squares) and KIR2DS1+ alloreactive NK-cell clones (triangles) is shown. (A) Lysis by 19 alloreactive NK-cell clones of 270 clones obtained from 2 individuals. (B) Lysis by 14 alloreactive NK-cell clones of 224 clones obtained from 2 individuals. (C) Lysis by 11 alloreactive NK-cell clones of 258 clones obtained from 3 individuals. (D) Lysis by 15 alloreactive NK-cell clones of 479 clones obtained from 3 individuals.
KIR2DS1 does not affect NK-cell alloreactivity against AML cells. 1265 NK-cell clones from 10 HLA-C1+ individuals were screened for cytotoxicity against a panel of 4 HLA-C1–missing, HLA-C2/C2 AML (A-D). The contribution of KIR2DS1 to lysis was evaluated in 59 allocytotoxic NK-cell clones that expressed KIR2DL2/L3 (as evaluated by phenotypic analyses). Lysis (mean ± SD) exerted by KIR2DS1– (squares) and KIR2DS1+ alloreactive NK-cell clones (triangles) is shown. (A) Lysis by 19 alloreactive NK-cell clones of 270 clones obtained from 2 individuals. (B) Lysis by 14 alloreactive NK-cell clones of 224 clones obtained from 2 individuals. (C) Lysis by 11 alloreactive NK-cell clones of 258 clones obtained from 3 individuals. (D) Lysis by 15 alloreactive NK-cell clones of 479 clones obtained from 3 individuals.
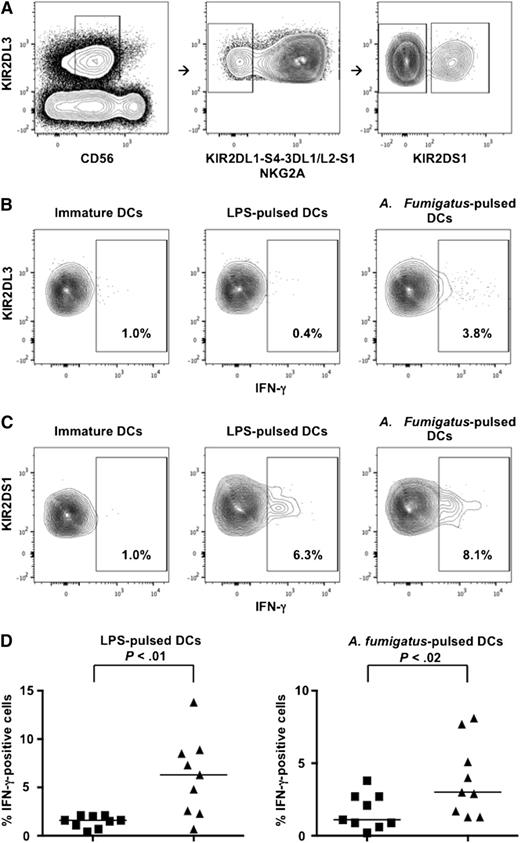
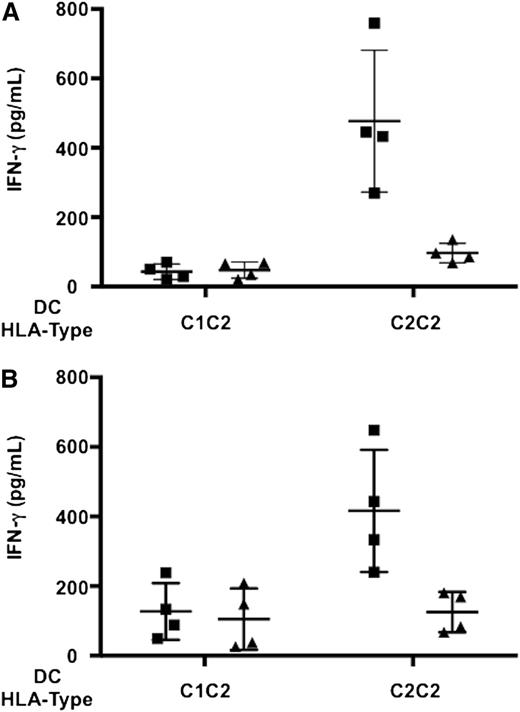
IFN-γ production analyses were performed in resting NK cells that expressed KIR2DL3 as their only inhibitory receptor for self and were therefore potentially capable of allorecognition of HLA-C1–missing targets (Figure 6A). Both KIR2DS1– and KIR2DS1+ alloreactive NK cells produced negligible IFN-γ levels upon interaction with immature DCs. KIR2DS1+ alloreactive NK cells produced significantly more IFN-γ than KIR2DS1– alloreactive NK cells upon activation with LPS- or A fumigatus–pulsed DCs (Figure 6B-D). Furthermore, IFN-γ release by alloreactive KIR2DL2/3+ NK clones that coexpressed KIR2DS1 was markedly reduced upon antibody blocking of KIR2DS1–HLA-C2 interaction (Figure 7).
KIR2DS1+ alloreactive NK cells produce more IFN-γ than KIR2DS1– alloreactive NK cells. Resting NK cells were incubated with immature or LPS- or A fumigatus–pulsed HLA-C1–missing, HLA-C2/C2 DCs. Cells were stained with the following mAb combination: anti-CD56-PE-Cy7, anti-KIR2DL2/L3/S2-FITC, anti-KIR2DL1-PE, anti-KIR3DL1/S1-PE, anti-KIR3DL1/L2-PE, anti-KIR2DS4-PE, anti-NKG2A-PE, anti-KIR2DL1/S1-APC, and anti-IFN-γ-PerCp-Cy5.5. (A) Gating strategy that identified potentially alloreactive NK cells against HLA-C1–missing, HLA-C2/C2 cell targets. Left panel: identification of KIR2DL3+ NK cells within CD56dim lymphocytes. Middle panel: identification of potentially alloreactive NK cells, which expressed KIR2DL3 and did not express KIR2DL1, KIR3DL1/S1, KIR3DL2, KIR2DS4, and NKG2A receptors. Right panel: identification of KIR2DL3+ potentially alloreactive NK cells that expressed KIR2DS1 (right quadrant) or did not express KIR2DS1 (left quadrant) (a minimum of 300 events were analyzed). (B) IFN-γ production by KIR2DS1– alloreactive NK cells. (C) IFN-γ production by KIR2DS1+ alloreactive NK cells. (D) Frequencies of IFN-γ+ cells in KIR2DS1– (squares) and KIR2DS1+ (triangles) alloreactive NK cells in 9 HLA-C1+ individuals. Each symbol represents NK cells from 1 individual. Bars represent median frequency of IFN-γ+ cells. Differences between the groups were tested by Wilcoxon matched-pairs test.
KIR2DS1+ alloreactive NK cells produce more IFN-γ than KIR2DS1– alloreactive NK cells. Resting NK cells were incubated with immature or LPS- or A fumigatus–pulsed HLA-C1–missing, HLA-C2/C2 DCs. Cells were stained with the following mAb combination: anti-CD56-PE-Cy7, anti-KIR2DL2/L3/S2-FITC, anti-KIR2DL1-PE, anti-KIR3DL1/S1-PE, anti-KIR3DL1/L2-PE, anti-KIR2DS4-PE, anti-NKG2A-PE, anti-KIR2DL1/S1-APC, and anti-IFN-γ-PerCp-Cy5.5. (A) Gating strategy that identified potentially alloreactive NK cells against HLA-C1–missing, HLA-C2/C2 cell targets. Left panel: identification of KIR2DL3+ NK cells within CD56dim lymphocytes. Middle panel: identification of potentially alloreactive NK cells, which expressed KIR2DL3 and did not express KIR2DL1, KIR3DL1/S1, KIR3DL2, KIR2DS4, and NKG2A receptors. Right panel: identification of KIR2DL3+ potentially alloreactive NK cells that expressed KIR2DS1 (right quadrant) or did not express KIR2DS1 (left quadrant) (a minimum of 300 events were analyzed). (B) IFN-γ production by KIR2DS1– alloreactive NK cells. (C) IFN-γ production by KIR2DS1+ alloreactive NK cells. (D) Frequencies of IFN-γ+ cells in KIR2DS1– (squares) and KIR2DS1+ (triangles) alloreactive NK cells in 9 HLA-C1+ individuals. Each symbol represents NK cells from 1 individual. Bars represent median frequency of IFN-γ+ cells. Differences between the groups were tested by Wilcoxon matched-pairs test.
KIR2DS1 binding to HLA-C2 triggers IFN-γ production in alloreactive NK cells. NK cell clones from 4 HLA-C1+ individuals were screened for alloreactivity against HLA-C1–missing, HLA-C2/C2 PHA T-cell blasts. Alloreactive KIR2DL2/3+ clones that coexpressed KIR2DS1 obtained from each individual were pooled and incubated with NK-resistant HLA-C1/C2 DCs or NK-susceptible HLA-C1–missing, HLA-C2/C2 DCs, pulsed with LPS (A) or A fumigatus (B). IFN-γ release (mean ± SD) in the absence (squares) or presence (triangles) of an anti-KIR2DS1 F(ab′)2 is shown. Each symbol represents IFN-γ release by NK cells from 1 individual.
KIR2DS1 binding to HLA-C2 triggers IFN-γ production in alloreactive NK cells. NK cell clones from 4 HLA-C1+ individuals were screened for alloreactivity against HLA-C1–missing, HLA-C2/C2 PHA T-cell blasts. Alloreactive KIR2DL2/3+ clones that coexpressed KIR2DS1 obtained from each individual were pooled and incubated with NK-resistant HLA-C1/C2 DCs or NK-susceptible HLA-C1–missing, HLA-C2/C2 DCs, pulsed with LPS (A) or A fumigatus (B). IFN-γ release (mean ± SD) in the absence (squares) or presence (triangles) of an anti-KIR2DS1 F(ab′)2 is shown. Each symbol represents IFN-γ release by NK cells from 1 individual.
Discussion
In T cell–depleted haploidentical hematopoietic transplantation, donor-vs-recipient NK-cell alloreactivity controls high-risk leukemia relapse.13-17,26,27 The present study identifies a further advantage of donor-vs-recipient NK-cell alloreactivity. It demonstrates that transplantation from NK-alloreactive donors who possessed KIR2DS1 and/or KIR3DS1 activating genes was associated with a markedly reduced infection rate and infectious mortality, and significantly better EFS.
The study included patients transplanted under 2 protocols: our standard T cell–depleted CD34+ hematopoietic progenitor cell graft protocol3-5 and the more recent protocol with Treg/Tcon addbacks.6,7 It might be questionable to include in the analysis patients who received Treg/Tcon cell addbacks because T cells in the graft were shown to antagonize NK-cell recovery and prevent clinically relevant NK-cell effects in unrelated and unmanipulated haploidentical transplants.9,57-60 However, in patients receiving Treg/Tcon addbacks, we previously demonstrated that reconstitution of donor-vs-recipient alloreactive NK cells was as efficient as in patients under the T cell–depleted transplant protocol.6 In fact, the survival advantage of transplantation from NK-alloreactive donors with activating KIR genes was more significant in the entire cohort (HR, 0.33; 95% CI, 0.14-0.76; P = .009) than in the T cell–depleted cohort (HR, 0.37; 95% CI, 0.15-0.88; P = .025).
Apparently, KIR-ligand mismatches and the consequent NK-cell release from recipient HLA blockade allowed activating KIRs to enhance NK-cell functions upon binding to their ligands on recipient cells. KIR2DS1 is the only activating KIR for which binding to an HLA ligand (HLA-C2) is well documented.33,34 Because KIR2DS1 is in strong linkage disequilibrium with KIR3DS148-50 and the present transplant series is relatively small, it was not possible to analyze the effect of donor KIR2DS1 interaction with recipient HLA-C2 independently of KIR3DS1. Likewise, it was not possible to analyze the effect of the KIR2DS4 gene (located in the A-haplotype telomeric segment) because of a deletion variant allele that encodes for a nonfunctional receptor in as much as ∼80% of individuals.32
In an attempt to define the underlying mechanism in vitro, we found that KIR2DS1 triggered NK cells to release IFN-γ upon interaction with KIR-ligand–mismatched, HLA-C2/C2 LPS–, or A fumigatus–pulsed DCs and that IFN-γ release was inhibited when KIR2DS1–HLA-C2 interaction was blocked. One may speculate that enhanced IFN-γ production played a role in improving responsiveness to pathogens. In fact, IFN-γ has antiviral effects, activates macrophage microbicidal functions, enhances antigen-presenting cell functions, and promotes Th1 polarization.61-66 Protective immunity against A fumigatus depends on inflammatory cytokines and Th1 responses.67 The KIR2DS1-triggered IFN-γ release in response to A fumigatus–pulsed DCs may have clinical implications because invasive Aspergillosis is a major cause of mortality in immune-compromised hosts.
Previous studies showed that KIR2DS1 boosted allocytotoxicity against HLA-C1–missing, HLA-C2/C2 targets.27,34-37 KIR2DL2, and to a lesser extent KIR2DL3, crossreact with HLA-C2 molecules, thereby possibly hampering allorecognition of HLA-C1–missing, HLA-C2C2 targets.27,56 KIR2DS1 binding to HLA-C2 was reported to overcome such inhibition, and analyses at the bulk NK-cell level suggested NK cells from HLA-C1+ individuals with the KIR2DS1 gene were more alloreactive.27,34,35 Our analyses at the clonal level documented even low-frequency alloreactive NK cells in the repertoire and were therefore particularly suited to identify donors able to mount donor-vs-recipient NK-cell alloreactivity.16 Such analyses, to date performed with >10 000 NK clones from >100 HLA-C1+ individuals, showed that alloreactive NK cells against HLA-C1–missing targets were present in KIR2DS1– individuals in frequencies that were only slightly lower than those in KIR2DS1+ individuals. Moreover, KIR2DS1 did not significantly shape the alloreactive repertoires of HLA-C1+ individuals regardless of whether they were KIR2DL2 or KIR2DL3 homozygous or KIR2DL2/L3 heterozygous. In agreement with these findings, NK-alloreactive donors exerted a remarkable graft-vs-leukemia effect regardless of their activating KIR genetics.
Studies in matched unrelated and haploidentical transplants showed that, even in the absence of KIR-ligand mismatches, donor activating KIRs were associated with less leukemia relapse38-43 or NRM.68 Under these circumstances, one may speculate that activating KIRs overrode NK-cell blockade by recipient HLA, or that T cells coexpressing activating KIRs were responsible because they expand after transplant69 and their effector functions are costimulated by activating KIRs.70-72
A previous study in adult and pediatric acute leukemia patients demonstrated that transplantation from maternal donors favorably affected survival.47 Interestingly, the present analysis in adult leukemia patients shows that such beneficial effect is evident only in transplants from NK-alloreactive donors. Studies in larger series of transplants are underway to confirm this finding.
The present investigation shows that transplantation from NK-alloreactive donors who possess KIR2DS1 and/or KIR3DS1 activating genes is associated with a remarkable reduction in infectious morbidity and mortality, and it consequently improves EFS. Because ∼40% of NK-alloreactive donors carry KIR2DS1 and/or KIR3DS1, searching for them may become a feasible, additional criterion in donor selection.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Geraldine Anne Boyd for editorial assistance and Dr Mara Merluzzi for data management.
This work was supported by a grant from the Italian Association for Cancer Research, a Translational Research grant from the Leukemia and Lymphoma Society, grants from the Italian Ministry of Further Education (Project no. 2010NECHBX_005), and the Italian Ministry of Health (European Research Area Net Translational Cancer Research, no. 7/PER-2011-2353844).
L. Ruggeri is a Leukemia and Lymphoma Society Scholar in Clinical Research.
Authorship
Contribution: A.M. designed the study, performed experiments, analyzed data, and wrote the paper; L. Ruggeri designed the study and analyzed data; E.U., A. Tosti, and F.T. contributed to NK cell cloning and cytotoxicity assays; A.P., M.S.M., A. Terenzi, A.C., and F.F. took care of patients and provided clinical data; S.B. provided A fumigatus–pulsed DCs; R.T. performed serologic and high-resolution molecular HLA typing; M.S. contributed to statistical analyses; L. Romani, F.A., and M.F.M. contributed to the design and interpretation of the study; and A.V. designed the study, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Antonella Mancusi, Division of Hematology and Clinical Immunology and Bone Marrow Transplant Program, Department of Medicine, University of Perugia, Perugia, Italy; e-mail: antonella.mancusi@unipg.it.

![Figure 3. Transplantation from NK-alloreactive donors with B-haplotype telomeric KIR genes reduced infectious mortality and morbidity. (A) Cumulative incidence of infectious mortality in the 26 patients transplanted from NK-alloreactive donors with Tel-B/x genes and in the 43 patients transplanted from NK-alloreactive donors with Tel-A/A genes. Eighteen of 43 patients transplanted from donors with Tel-A/A genes died of infection (1 bacterial, 1 Toxoplasma gondii, 4 invasive fungal, 4 CMV, 2 human herpesvirus 6, 1 Epstein-Barr virus, 1 parainfluenza virus, 4 pneumonia and acute respiratory distress syndrome of unknown etiology). Only 2 of 26 patients transplanted from donors with Tel-B/x genes died of infection (1 CMV and 1 concomitant viral and fungal infection). (B) Data on infectious episodes were available for 63 of 69 patients transplanted from NK-alloreactive donors. Number of infectious episodes per month (mean ± standard deviation [SD]) in the first 6 months after transplant in patients transplanted from NK-alloreactive donors with Tel-A/A (open bar) and Tel-B/x genes (closed bar) are shown. Difference between the groups was tested by unpaired Student t test. (C) Ratios (mean with 95% CI) between the infection rates of patients transplanted from NK-alloreactive donors with Tel-B/x (diamonds) and infection rates of patients transplanted from NK-alloreactive donors Tel-A/A genes (dotted line) as evaluated monthly during the first 6 months after transplant.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/20/10.1182_blood-2014-09-599993/4/m_3173f3.jpeg?Expires=1769112257&Signature=FTLex9egGpnVlWLGAwhlOYrUpyiPRRBeMFM~i2GOnW6Shrc33g6C7H-a3n4ru92lIVYQShTgwODMa9qTX78gC4MUEgzmUVGSYPaPBgpdsq68HuRvGYD0VsUzmSj~7tco0KesP7rP76RW9~qm2vDH9RuaQc5I6n~z~SGSgYfYmbGfuPd1l~WiozyxCguKypb7gfqS2MOvi3kh9MYSikR0TparvujLDpFhsfrKY4dXyvTKaqmIFihN~lIIOGS-~X97VaK9gArNPNvuvIyZ3FGwi7M0BabaMs0DmBiPU3dyzGg-qYDeSzvJucbWqVz6OQTNnFGW0yVrqWbl0etWWvRA3A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. KIR2DS1 does not affect alloreactive NK-cell repertoires. (A) A total of 11 999 NK-cell clones were generated from 110 HLA-C1+ individuals (mean 109 clones/individual [30-302]) and were screened for cytotoxicity against HLA-C1–missing, HLA-C2/C2 PHA T-cell blasts. Frequencies (mean ± SD) of alloreactive NK-cell clones in the 65 individuals without the KIR2DS1 gene (open bar) and in the 45 individuals with the KIR2DS1 gene (closed bar) are shown (P = NS by unpaired Student t test). (B) The KIR2DS1– and KIR2DS1+ individuals in (A) were grouped according to their KIR2DL2 or KIR2DL3 homozygous or KIR2DL2/3 heterozygous inhibitory genotypes. Frequencies of alloreactive NK-cell clones in KIR2DS1– (open bar) and KIR2DS1+ (closed bar) individuals are shown (P = NS by unpaired Student t test). (C) A total of 2281 clones obtained from 20 HLA-C1+ individuals (randomly selected from the 110 in [A]) were screened for allocytotoxicity against HLA-C1–missing, HLA-C2C2 PHA T-cell blasts. The contribution of KIR2DS1 to lysis was evaluated in 127 allocytotoxic NK-cell clones that expressed KIR2DL2/L3 (as evaluated by phenotypic analyses). Lysis (mean ± SD) by the 92 clones that did not express KIR2DS1 (open bar) and the 35 clones that expressed KIR2DS1 (closed bar) is shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/20/10.1182_blood-2014-09-599993/4/m_3173f4.jpeg?Expires=1769112257&Signature=u7jgNK7k4GdPPFx6leJH45PvDZ9C8Et0oXRjHETTBtN5lK9B9FZYfBv8exRcMzFijHr-OLqhEB3U0hQECiU9dqLwZ2biuRFzHXeEzoi8JqBJrUuan1LiL~vjMDV27O2I1xNBjYEa11FIdx4UWAG7Xa4wtxbUTaOiysMjDWIFEyCho~2aUSI~h8g2D3JSS4k88Hng62jNqe~FPd2hbx0VqiDkHnJQlkBAJbJ2~9IW~V6V4W0dATMYKcPIDFKJxulg9s1T2W85h8AOLPITML201DmDiWC5e7xHZl94pTKX3Aq1oV7VqUNQGIno-0hcl9VdmAtENAZigNDxw811r97SCQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal