Key Points
Additional chromosomal changes modulate the outcome of patients with high-risk multiple myeloma.
Abstract
In multiple myeloma, cytogenetic changes are important predictors of patient outcome. In this setting, the most important changes are deletion 17p, del(17p), and translocation of chromosomes 4 and 14, t(4;14), conferring a poor outcome. However, a certain degree of heterogeneity is observed in the survival of these high-risk patients. We hypothesized that other chromosomal changes may impact the outcome. We retrospectively analyzed a large series of 242 patients displaying either t(4;14) (157 patients) or del(17p) (110 patients), 25 patients presenting both abnormalities, using single nucleotide polymorphism array. In patients with t(4;14), del(1p32), del22q, and >30 chromosomal structural changes negatively impacted progression-free survival (PFS). For overall survival (OS), del(13q14), del(1p32), and the number of chromosomal structural changes worsened the prognosis of patients. For patients with del(17p), del6q worsened the prognosis of patients, whereas trisomy 15 and monosomy 14 were found to have a protective effect on PFS. For OS, del(1p32) worsened the prognosis of patients, whereas having >8 numerical changes was found to have a protective effect on survival. This study, which is the largest series of high-risk patients analyzed with the most modern genomic technique, identified 1 main factor negatively impacting survival: del(1p32).
Introduction
Although chromosomal changes are rarely observed at conventional karyotype in multiple myeloma (MM),1-5 genomic studies based on comparative genomic hybridization or single nucleotide polymorphism (SNP) arrays did show that they are present in >90% of the patients.6,7 Some of these chromosomal abnormalities have been shown to very accurately predict outcome for patients with MM. Several studies showed that several chromosomal changes were associated with a poor outcome, including translocation of chromosomes 4 and 14, t(4;14), and deletion 17p, del(17p),8-11 with less or discordant data for t(14;16) and t(14;20).12-14 In contrast, no specific abnormality has been associated with a good outcome, apart from the absence of all high-risk changes with low β2-microglobulin (β2m) level.15
Regarding the 2 major high-risk abnormalities, t(4;14) and del(17p), studies are rather concordant in showing that the outcome of patients with del(17p) is uniformly poor, whereas a certain degree of heterogeneity is observed for t(4;14). A first step in understanding this heterogeneity was made in 2012 with a Mayo Clinic publication showing that high-risk abnormalities lose part of their prognostic value when associated with hyperdiploidy.16 Based on fluorescence in situ hybridization (FISH), they showed that 55 of 133 patients (41%) with high-risk abnormalities also displayed trisomies of the odd chromosomes. They concluded that these latter patients did not display a poor outcome, as compared with other high-risk patients.
Because FISH by definition is used to analyze only specific chromosomal changes, and not the whole genome, we designed a study based on an unbiased technique (ie, SNP array). This technique allows the identification of all copy number changes through the genome. We aimed to explore cytogenetic abnormalities that influence predictive and prognostic values among t(4;14) and del(17p) separately in MM patients. We report here the results of an analysis of 242 patients displaying either t(4;14) (157 patients) or del(17p) (110 patients), 25 patients presenting both abnormalities.
Methods
Patients
This study was approved by the institutional review board of the Institut Universitaire du Cancer of Toulouse (France). Among a series of 986 patients analyzed by SNP array with a minimal follow-up of 36 months, we identified 242 patients displaying either t(4;14) (identified by interphase FISH as previously described)11 or del(17p). Del(17p) was identified on the SNP-array profiles and was then verified by FISH in order to precisely define the percentage of plasma cells displaying the deletion. According to our previous results,11 only patients displaying del(17p) in at least 60% of the plasma cells were kept in this study. There were 157 patients with t(4;14) and 110 patients with del(17p), 25 patients presenting the 2 abnormalities.
Regarding treatment, 44 patients received a melphalan-prednisone-thalidomide or melphalan-prednisone-Velcade combination (18%), 73 patients received a vincristin-adriamycin-dexamethasone induction followed by high-dose melphalan and autologous stem cell transplant (ASCT) (30%), and 125 patients received a Velcade-dexamethasone induction followed by high-dose melphalan and ASCT (52%).
SNP-array analysis
All SNP-array analyses were performed in newly diagnosed patients on bone marrow aspirate collected prior to any therapy. After plasma cell purification using the StemCell Technology (Vancouver, BC, Canada) or Miltenyi Biotec (Paris, France) technique, DNA was extracted, labeled, and hybridized on Affymetrix chips according to Affymetrix (Santa Clara, CA) recommendations. We used either the SNP 6.0 or the CytoScan chips, depending on the date of hybridization. For all patients, we used a visual evaluation of gains and losses, focused on the more frequent aberrations: del(1p32), del(1p12), 1q gain, 6p gain, del(6q), del(8p) del(12p), del(13q14), monosomy 14, del(14q), del(16q), del(17p13), del(22q), Xq gain in males, del(Xp) in females, and nullisomy Y. For all the patients, ploidy was assessed based on the number of chromosomes, defining no hyperdiploidy (≤46 chromosomes), mild hyperdiploidy (47 to 50 chromosomes), and large hyperdiploidy (>50 chromosomes). For t(4;14), we noted the number of fused signals, the classical one with 2 fusions, and the abnormal one with only 1 fusion, as previously reported.17,18
Statistical analysis
Because of different cytogenetic profiles between t(4;14) and del(17p) patients, the following analyses were performed separately in the t(4;14) population and the del(17p) population. The groups were not mutually exclusive in order to explore if the presence of t(4;14) modifies the outcome of patients displaying del(17p) and conversely. Progression-free survival (PFS) was defined as the time interval between diagnosis and progression or death, whichever occurred earlier. Overall survival (OS) was defined as the time from diagnosis to death. PFS and OS were analyzed by the Kaplan-Meier method, log-rank test, and Cox proportional hazards model to estimate the hazard ratio (HR) along with 95% confidence intervals (95% CIs).
To test the predictive and prognostic value of each cytogenetic abnormality, the first models were adjusted on age (continuous variable), treatment (no ASCT vs high-dose therapy/ASCT + vincristin-adriamycin-dexamethasone vs high-dose therapy/ASCT + Velcade-dexamethasone), and β2m (<3.5 vs ≥3.5 mg/L), which are well-known prognostic factors. To correct for confounding between markers, cytogenetic abnormalities with P value <.15 were entered into multivariate Cox models. The model was reduced using backward elimination until only significant effects remained. First-order interactions were explored. To test the independent effect of nullisomy Y and Xq gain in males, these markers were introduced in the final model. The same strategy was applied for del(Xp) in females. Interactions involving sex chromosomes were not tested because samples were too small to yield estimates with acceptable reliability. The proportionality assumptions have been checked with Cox-Snell residuals. For the OS analysis, we observed a violation of the proportional hazards assumption for del(13q14) in the subgroup of t(4;14)-positive patients, so an interaction term with a function of time was added to model the evolvement of the HR over time. Tests were 2-sided, and P values <.05 were considered significant. All analyses were conducted using Stata Version 11.0.
Results
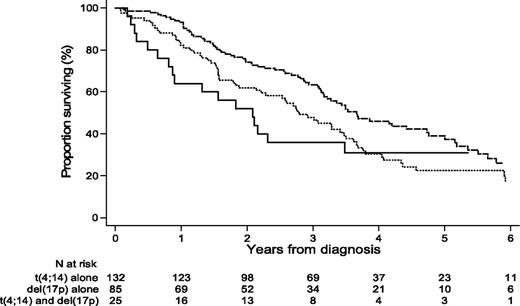
For analysis, we separated the patients into 2 groups: group 1 with t(4;14) (157 patients) and group 2 with del(17p) (110 patients). Twenty-five patients were in both groups because they presented the 2 abnormalities. Median age at diagnosis was 59.6 years (range, 33-86). The sex ratio (male/female) was 1.26 (56% were male). The median follow-up was 4.86 years. The PFS and OS of patients presenting t(4;14) alone, del(17p) alone, or both abnormalities irrespective of additional abnormalities are shown in Figure 1.
OS of patients displaying t(4;14) alone, del(17p) alone, or both t(4;14) and del(17p), irrespective of the presence of additional abnormalities.
OS of patients displaying t(4;14) alone, del(17p) alone, or both t(4;14) and del(17p), irrespective of the presence of additional abnormalities.
Group 1 = t(4;14)
In this group, 144 patients have relapsed, and 103 have died. The median PFS was 1.4 years, and the median OS was 3.5 years. The median β2m level was 4.0 mg/L (range, 1.2-38.3). The distribution of International Staging System stages I, II, and III was 35.4%, 26.4%, and 38.2%, respectively. Based on FISH results, 31.2% of the patients presented the abnormal configuration with only 1 fused signal. In all these patients, SNP-array analysis showed a loss of the telomeric part of 1 chromosome 4. In contrast to previous reports on this particularity, in which the explanation was a loss of the derivative chromosome 14,17,18 we clearly show here that this configuration results from an unbalanced translocation.
Distributions of chromosomal changes are summarized in Table 1 using mutually exclusive groups of patients. One hundred eleven patients did not have hyperdiploidy (71%), 27 patients presented mild hyperdiploidy (17.2%), and 19 patients had large hyperdiploidy (12.1%). The median number of chromosomal structural changes was 11 (range, 1-71). Del(13q14) (mostly monosomies) was present in 130 patients (83%). Del(1p32) and del(1p12) were observed in 9.6% and 40.1% of patients, respectively. Chromosome 1q gains were observed in 105 patients (66.9%). Chromosome 6p gains and del(6q) were present in 17.2% and 29.3% of patients, respectively, whereas del(8p) was seen in 26.8% of patients. Del(12p) was found in 35.7% of patients, whereas del(14q) and monosomy 14 were observed in 51% and 18.5% of patients, respectively. Del(16q) was seen in 10.8% of patients, and del(22q) was present in 47.8% of patients. Finally, regarding sex chromosomes, Xq gains and nullisomy Y were present in 34.1% and 24.2% of males, respectively, and del(Xp) was observed in 63.6% of females.
Frequency of the most important chromosomal abnormalities in the t(4;14) only (n = 132), del(17p) only (n = 85), and high-risk (n = 25) subgroups
| Chromosomal changes . | t(4;14) alone, n = 132 . | Del(17p) alone, n = 85 . | t(4;14) and del(17p) positive, n=25 . |
|---|---|---|---|
| Hyperdiploidy | |||
| No | 66.7 | 60.6 | 92.0 |
| Mild | 20.4 | 21.1 | 0.0 |
| Large | 12.9 | 18.3 | 8.0 |
| Number of chromosomes (median IQR) | 45 (44-48) | 45 (43-50) | 45 (43-45) |
| Structural chromosomal changes | 46.2 | 28.2 | 4.0 |
| <10 | |||
| 10-30 | 49.2 | 42.2 | 56.0 |
| ≥30 | 4.6 | 29.6 | 40.0 |
| Structural chromosomal changes (median IQR) | 10 (6-17) | 16 (9-34) | 23 (15-41) |
| Del(13q14) | 81.1 | 74.7 | 92.0 |
| Del(1p12) | 37.1 | 52.1 | 56.0 |
| Del(1p32) | 6.8 | 29.6 | 24.0 |
| Chromosome 1q gain | 68.9 | 31.0 | 56.0 |
| Chromosome 6p gain | 18.9 | 18.3 | 8.0 |
| Del(6q) | 24.2 | 42.3 | 56.0 |
| Del(8p) | 25.0 | 45.1 | 36.0 |
| Del(12p) | 34.9 | 31.0 | 40.0 |
| Del(14q) | 47.7 | 46.5 | 68.0 |
| Monosomy 14 | 17.4 | 19.7 | 24.0 |
| Del(16q) | 8.3 | 53.5 | 24.0 |
| Del(22q) | 47.7 | 43.7 | 48.0 |
| Nullisomy Y | 24.7 | 24.3 | 21.4 |
| Chromosomal changes . | t(4;14) alone, n = 132 . | Del(17p) alone, n = 85 . | t(4;14) and del(17p) positive, n=25 . |
|---|---|---|---|
| Hyperdiploidy | |||
| No | 66.7 | 60.6 | 92.0 |
| Mild | 20.4 | 21.1 | 0.0 |
| Large | 12.9 | 18.3 | 8.0 |
| Number of chromosomes (median IQR) | 45 (44-48) | 45 (43-50) | 45 (43-45) |
| Structural chromosomal changes | 46.2 | 28.2 | 4.0 |
| <10 | |||
| 10-30 | 49.2 | 42.2 | 56.0 |
| ≥30 | 4.6 | 29.6 | 40.0 |
| Structural chromosomal changes (median IQR) | 10 (6-17) | 16 (9-34) | 23 (15-41) |
| Del(13q14) | 81.1 | 74.7 | 92.0 |
| Del(1p12) | 37.1 | 52.1 | 56.0 |
| Del(1p32) | 6.8 | 29.6 | 24.0 |
| Chromosome 1q gain | 68.9 | 31.0 | 56.0 |
| Chromosome 6p gain | 18.9 | 18.3 | 8.0 |
| Del(6q) | 24.2 | 42.3 | 56.0 |
| Del(8p) | 25.0 | 45.1 | 36.0 |
| Del(12p) | 34.9 | 31.0 | 40.0 |
| Del(14q) | 47.7 | 46.5 | 68.0 |
| Monosomy 14 | 17.4 | 19.7 | 24.0 |
| Del(16q) | 8.3 | 53.5 | 24.0 |
| Del(22q) | 47.7 | 43.7 | 48.0 |
| Nullisomy Y | 24.7 | 24.3 | 21.4 |
IQR, interquartile range.
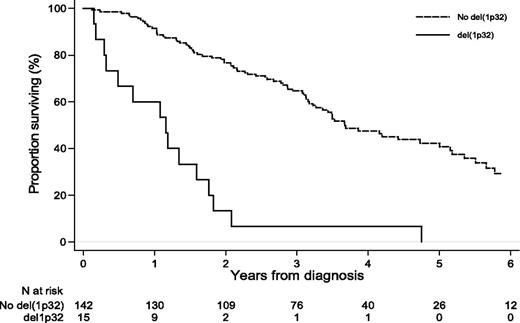
Adjusted on age, treatment type, and β2m level, the associations between chromosomal changes and PFS or OS are shown in Table 2. After controlling for confounding effects, the statistically independent significant markers for PFS were del(1p32) (adjusted HR [aHR] = 5.9; 95% CI, 2.9-12.1; P < .001), del22q (aHR = 1.5; 95% CI, 1.00-2.2; P = .048), and >30 chromosomal structural changes (aHR = 2.3; 95% CI, 1.2-4.2; P = .01). For OS, del(13q14) (aHR = 2.6; 95% CI, 1.3-5.3; P = .01), del(1p32) (aHR = 4.9; 95% CI, 2.5-9.6; P < .001; Figure 2), and the number of chromosomal structural changes (aHR = 1.7; 95% CI, 1.01-2.8; P < .05 for 10-30 structural changes and aHR = 2.5; 95% CI, 1.2-5.1; P = .01 for >30 structural changes) worsened the prognosis of patients. In this subgroup of patients, the effect of del(13q14) was found to increase over time (P < .03). No statistically significant effect on PFS or OS was found for nullisomy Y.
Proportional Cox regression analyses of chromosomal changes for PFS and OS among patients displaying t(4;14)
| Chromosomal changes . | PFS . | OS . | ||||
|---|---|---|---|---|---|---|
| aHR* . | 95% CI . | P . | aHR . | 95% CI . | P . | |
| Del(13q14) | 1.66 | 1.02-2.71 | .043 | 2.89 | 1.49-5.61 | .002 |
| Del(1p12) | 1.51 | 1.04-2.20 | .032 | 1.09 | 0.71-1.67 | .696 |
| Del(1p32) | 6.98 | 3.43-14.18 | <.001 | 5.32 | 2.77-10.26 | <.001 |
| Del(17p) | 1.53 | 0.91-2.59 | .112 | 2.01 | 1.11-3.66 | .022 |
| Trisomy 5 | 0.62 | 0.31-1.24 | .173 | 0.43 | 0.16-1.18 | .102 |
| Trisomy 9 | 0.69 | 0.43-1.11 | .126 | 0.51 | 0.27-0.96 | .036 |
| Trisomy 15 | 0.69 | 0.45-1.04 | .078 | 0.43 | 0.24-0.76 | .004 |
| Gain 1q | 1.28 | 0.85-1.90 | .234 | 1.29 | 0.81-2.06 | .289 |
| Del(6q) | 1.41 | 0.92-2.15 | .114 | 1.80 | 1.13-2.88 | .014 |
| Del(8p) | 1.09 | 0.72-1.65 | .685 | 1.52 | 0.96-2.42 | .074 |
| Del(12p) | 1.47 | 1.01-2.16 | .046 | 2.12 | 1.36-3.30 | .001 |
| Monosomy 14 | 0.64 | 0.39-1.05 | .077 | 0.88 | 0.50-1.54 | .645 |
| Del(16q) | 1.79 | 0.99-3.20 | .052 | 1.52 | 0.83-2.79 | .175 |
| Del(22q) | 1.51 | 1.04-2.20 | .031 | 1.30 | 0.84-2.02 | .235 |
| Gain Xq in males | 1.15 | 0.69-1.90 | .599 | 0.93 | 0.50-1.70 | .803 |
| Nullisomy Y | 1.51 | 0.87-2.63 | .150 | 1.68 | 0.90-3.16 | .105 |
| Hyperdiploidy | ||||||
| No | ||||||
| Mild | 0.92 | 0.56-1.51 | .738 | 0.63 | 0.34-1.15 | .131 |
| High | 0.61 | 0.35-1.08 | .090 | 0.37 | 0.16-0.87 | .022 |
| Numerical changes (ref: <4) | ||||||
| 4-8 | 0.73 | 0.49-1.08 | .118 | 0.84 | 0.52-1.35 | .473 |
| ≥8 | 0.60 | 0.31-1.20 | .148 | 0.49 | 0.19-1.26 | .137 |
| Structural changes (ref: <10) | ||||||
| 10-30 | 1.53 | 1.03-2.26 | .034 | 1.75 | 1.08-2.83 | .023 |
| ≥30 | 2.17 | 1.19-3.97 | .012 | 2.75 | 1.40-5.37 | .003 |
| Chromosomal changes . | PFS . | OS . | ||||
|---|---|---|---|---|---|---|
| aHR* . | 95% CI . | P . | aHR . | 95% CI . | P . | |
| Del(13q14) | 1.66 | 1.02-2.71 | .043 | 2.89 | 1.49-5.61 | .002 |
| Del(1p12) | 1.51 | 1.04-2.20 | .032 | 1.09 | 0.71-1.67 | .696 |
| Del(1p32) | 6.98 | 3.43-14.18 | <.001 | 5.32 | 2.77-10.26 | <.001 |
| Del(17p) | 1.53 | 0.91-2.59 | .112 | 2.01 | 1.11-3.66 | .022 |
| Trisomy 5 | 0.62 | 0.31-1.24 | .173 | 0.43 | 0.16-1.18 | .102 |
| Trisomy 9 | 0.69 | 0.43-1.11 | .126 | 0.51 | 0.27-0.96 | .036 |
| Trisomy 15 | 0.69 | 0.45-1.04 | .078 | 0.43 | 0.24-0.76 | .004 |
| Gain 1q | 1.28 | 0.85-1.90 | .234 | 1.29 | 0.81-2.06 | .289 |
| Del(6q) | 1.41 | 0.92-2.15 | .114 | 1.80 | 1.13-2.88 | .014 |
| Del(8p) | 1.09 | 0.72-1.65 | .685 | 1.52 | 0.96-2.42 | .074 |
| Del(12p) | 1.47 | 1.01-2.16 | .046 | 2.12 | 1.36-3.30 | .001 |
| Monosomy 14 | 0.64 | 0.39-1.05 | .077 | 0.88 | 0.50-1.54 | .645 |
| Del(16q) | 1.79 | 0.99-3.20 | .052 | 1.52 | 0.83-2.79 | .175 |
| Del(22q) | 1.51 | 1.04-2.20 | .031 | 1.30 | 0.84-2.02 | .235 |
| Gain Xq in males | 1.15 | 0.69-1.90 | .599 | 0.93 | 0.50-1.70 | .803 |
| Nullisomy Y | 1.51 | 0.87-2.63 | .150 | 1.68 | 0.90-3.16 | .105 |
| Hyperdiploidy | ||||||
| No | ||||||
| Mild | 0.92 | 0.56-1.51 | .738 | 0.63 | 0.34-1.15 | .131 |
| High | 0.61 | 0.35-1.08 | .090 | 0.37 | 0.16-0.87 | .022 |
| Numerical changes (ref: <4) | ||||||
| 4-8 | 0.73 | 0.49-1.08 | .118 | 0.84 | 0.52-1.35 | .473 |
| ≥8 | 0.60 | 0.31-1.20 | .148 | 0.49 | 0.19-1.26 | .137 |
| Structural changes (ref: <10) | ||||||
| 10-30 | 1.53 | 1.03-2.26 | .034 | 1.75 | 1.08-2.83 | .023 |
| ≥30 | 2.17 | 1.19-3.97 | .012 | 2.75 | 1.40-5.37 | .003 |
HR adjusted for age, treatment type, and β2m level.
Group 2 = del(17p)
In this group (110 patients), 99 patients relapsed, and 83 died. The median PFS was 1.3 years, and the median OS was 2.7 years. The median β2m level was 4.4 mg/L (range, 1.4-32.1). The distribution of International Staging System stages I, II, and III was 28.4%, 26.9%, and 44.8%, respectively.
Chromosomal changes are summarized in Table 1. Seventy-two patients did not have hyperdiploidy, 18 patients presented mild hyperdiploidy, and 20 patients had large hyperdiploidy. The median number of chromosomal structural changes was 17 (range, 0-74). Del(13q14) (mostly monosomies) was present in 87 patients (79.1%). Del(1p32) and del(1p12) were observed in 28.2% and 52.7% of patients, respectively. Chromosome 1q gains were observed in 38 patients (34.5%). Chromosome 6p gains and del(6q) were present in 15.5% and 46.4% of the patients, respectively, whereas del(8p) was seen in 41.8% of patients. Del(12p) was found in 30% of patients, whereas del(14q) and monosomy 14 were observed in 52.7% and 23.6% of patients, respectively. Del(16q) was seen in 47.3% of patients, and del(22q) was present in 46.4% of patients. Finally, regarding sex chromosomes, Xq gains and nullisomy Y were present in 41.4% and 22.4% of the males, respectively, and del(Xp) was observed in 63.5% of females.
Adjusted on age, treatment type, and β2m level, chromosomal parameters that were associated with PFS and OS are described in Table 3. After controlling for confounding effects, the statistically independent significant markers for PFS were del(6q) (aHR = 1.9; 95% CI, 1.1-3.1; P = .01), which worsened the prognosis of patients, whereas trisomy 15 (aHR = 0.5; 95% CI, 0.3-0.8; P = .01) and monosomy 14 (aHR = 0.5; 95% CI, 0.3-0.9; P = .03) were found to have a protective effect on PFS. For OS, del(1p32) (aHR = 2.2; 95% CI, 1.3-3.9; P = .01) worsened the prognosis of patients, whereas >8 numerical changes (aHR = 0.4; 95% CI, 0.2-0.7; P = .004) was found to have a protective effect on survival. In the subgroup of male patients, gain of Xq was associated with a shorter PFS (aHR = 2.0; 95% CI, 1.01-4.1; P < .05). No statistically significant effect on PFS or OS was found for nullisomy Y.
Proportional Cox regression analyses of chromosomal changes for PFS and OS among patients displaying del(17p)
| Chromosomal changes . | PFS . | OS . | ||||
|---|---|---|---|---|---|---|
| aHR* . | 95% CI . | P . | aHR . | 95% CI . | P . | |
| Del(13q14) | 1.84 | 1.01-3.34 | .045 | 1.85 | 0.96-3.56 | .064 |
| Del(1p12) | 1.27 | 0.81-1.97 | .300 | 0.87 | 0.53-1.43 | .586 |
| Del(1p32) | 1.36 | 0.78-2.38 | .281 | 1.73 | 0.98-3.05 | .058 |
| t(4;14) | 1.19 | 0.66-2.15 | .562 | 1.20 | 0.63-2.28 | .577 |
| Trisomy 5 | 0.56 | 0.31-1.03 | .064 | 0.43 | 0.21-0.87 | .019 |
| Trisomy 9 | 0.67 | 0.39-1.13 | .134 | 0.52 | 0.28-0.94 | .031 |
| Trisomy 15 | 0.50 | 0.30-0.84 | .009 | 0.53 | 0.30-0.94 | .030 |
| Gain 1q | 2.18 | 1.33-3.58 | .002 | 1.30 | 0.77-2.18 | .325 |
| Del(6q) | 1.75 | 1.08-2.86 | .024 | 1.26 | 0.75-2.11 | .381 |
| Del(8p) | 0.77 | 0.48-1.24 | .285 | 1.14 | 0.69-1.88 | .606 |
| Del(12p) | 1.01 | 0.63-1.60 | .978 | 1.21 | 0.73-1.99 | .456 |
| Monosomy 14 | 0.59 | 0.33-1.07 | .081 | 0.78 | 0.42-1.45 | .429 |
| Del(16q) | 1.08 | 0.68-1.71 | .757 | 1.46 | 0.87-2.45 | .153 |
| Del(22q) | 1.37 | 0.87-2.15 | .169 | 1.52 | 0.93-2.47 | .092 |
| Gain Xq in males | 1.86 | 0.96-3.60 | .067 | 1.18 | 0.60-2.34 | .638 |
| Nullisomy Y | 1.46 | 0.61-3.45 | .394 | 2.57 | 1.03-6.46 | .044 |
| Hyperdiploidy | ||||||
| No | ||||||
| Mild | 0.75 | 0.42-1.36 | .688 | 1.06 | 0.58-1.94 | .843 |
| High | 0.41 | 0.21-0.80 | .009 | 0.43 | 0.20-0.93 | .033 |
| Numerical changes (ref: <4) | ||||||
| 4-8 | 0.91 | 0.52-1.57 | .732 | 0.96 | 0.53-1.76 | .905 |
| ≥8 | 0.43 | 0.24-0.77 | .004 | 0.44 | 0.23-0.84 | .013 |
| Structural changes (ref: <10) | ||||||
| 10-30 | 2.04 | 1.10-3.79 | .024 | 1.33 | 0.68-2.61 | .401 |
| ≥30 | 1.51 | 0.77-2.96 | .228 | 1.28 | 0.62-2.63 | .497 |
| Chromosomal changes . | PFS . | OS . | ||||
|---|---|---|---|---|---|---|
| aHR* . | 95% CI . | P . | aHR . | 95% CI . | P . | |
| Del(13q14) | 1.84 | 1.01-3.34 | .045 | 1.85 | 0.96-3.56 | .064 |
| Del(1p12) | 1.27 | 0.81-1.97 | .300 | 0.87 | 0.53-1.43 | .586 |
| Del(1p32) | 1.36 | 0.78-2.38 | .281 | 1.73 | 0.98-3.05 | .058 |
| t(4;14) | 1.19 | 0.66-2.15 | .562 | 1.20 | 0.63-2.28 | .577 |
| Trisomy 5 | 0.56 | 0.31-1.03 | .064 | 0.43 | 0.21-0.87 | .019 |
| Trisomy 9 | 0.67 | 0.39-1.13 | .134 | 0.52 | 0.28-0.94 | .031 |
| Trisomy 15 | 0.50 | 0.30-0.84 | .009 | 0.53 | 0.30-0.94 | .030 |
| Gain 1q | 2.18 | 1.33-3.58 | .002 | 1.30 | 0.77-2.18 | .325 |
| Del(6q) | 1.75 | 1.08-2.86 | .024 | 1.26 | 0.75-2.11 | .381 |
| Del(8p) | 0.77 | 0.48-1.24 | .285 | 1.14 | 0.69-1.88 | .606 |
| Del(12p) | 1.01 | 0.63-1.60 | .978 | 1.21 | 0.73-1.99 | .456 |
| Monosomy 14 | 0.59 | 0.33-1.07 | .081 | 0.78 | 0.42-1.45 | .429 |
| Del(16q) | 1.08 | 0.68-1.71 | .757 | 1.46 | 0.87-2.45 | .153 |
| Del(22q) | 1.37 | 0.87-2.15 | .169 | 1.52 | 0.93-2.47 | .092 |
| Gain Xq in males | 1.86 | 0.96-3.60 | .067 | 1.18 | 0.60-2.34 | .638 |
| Nullisomy Y | 1.46 | 0.61-3.45 | .394 | 2.57 | 1.03-6.46 | .044 |
| Hyperdiploidy | ||||||
| No | ||||||
| Mild | 0.75 | 0.42-1.36 | .688 | 1.06 | 0.58-1.94 | .843 |
| High | 0.41 | 0.21-0.80 | .009 | 0.43 | 0.20-0.93 | .033 |
| Numerical changes (ref: <4) | ||||||
| 4-8 | 0.91 | 0.52-1.57 | .732 | 0.96 | 0.53-1.76 | .905 |
| ≥8 | 0.43 | 0.24-0.77 | .004 | 0.44 | 0.23-0.84 | .013 |
| Structural changes (ref: <10) | ||||||
| 10-30 | 2.04 | 1.10-3.79 | .024 | 1.33 | 0.68-2.61 | .401 |
| ≥30 | 1.51 | 0.77-2.96 | .228 | 1.28 | 0.62-2.63 | .497 |
HR adjusted for age, treatment type, and β2m level.
Discussion
In MM, cytogenetic changes evaluated by interphase FISH on identified plasma cells play a major role in the outcome of the patients.8-11 Among all the chromosomal abnormalities identified in MM, del(17p) and t(4;14) define a high-risk population, with significantly shorter PFS and OS. In this population, del(17p) is uniformly associated with a very poor outcome, whereas a degree of heterogeneity is observed with t(4;14). A single publication addressed the role of other chromosomal changes in the outcome of 109 high-risk patients.16 Based on interphase FISH, this study showed that the presence of trisomies (suggesting hyperdiploidy) significantly improved the prognosis of high-risk patients. The main pitfall of the interphase FISH technique is that data on chromosomal changes are restricted to the selected probes. In order to circumvent this issue, we used SNP-array technology in a series of 242 patients presenting t(4;14) and/or del(17p). The main advantage of this technology is that all copy number changes present in the genome are seen and analyzable.
We separately analyzed patients with t(4;14) and those with del(17p). Twenty-five patients presenting both abnormalities were thus analyzed in each subgroup. In the area of new agent therapies, the median OSs reported for patients displaying t(4,14) or del(17p) seem relatively low. However, the introduction of bortezomib compared with other treatments is relatively recent, and the patients included in these treatment groups were monitored for shorter time periods. Consequently, the median OS may have been underestimated. Nevertheless, those estimates are consistent with estimate reported in the study by the Mayo Clinic.16
Among the 157 patients with t(4;14), one-third presented an abnormal FISH configuration, with loss of the der(14)-fused signal. This configuration is the consequence of an unbalanced translocation, with loss of the telomeric part of chromosome 4, harboring the FGFR3 gene. This configuration is not associated with a specific outcome as compared with the classical one. These results suggest that FGFR3 is not playing any role in the disease aggressiveness, because it is not overexpressed in the unbalanced translocation, in contrast to the classical one. Hyperdiploidy was observed in only 29% of the t(4;14) patients, in contrast to 52% in the general population (966 patients analyzed by SNP array, unpublished data). Several differences were observed in this subgroup as compared with the general population, including a more complex molecular karyotype (median 11 structural changes vs 8 for the general population), more frequent del(13q14) (83% vs 51%), more frequent 1q gains (67% vs 39%), more frequent del(12p) (35.7% vs 13.5%), more frequent del(14q) and monosomy 14 (51% and 18.5% vs 28% and 11.3%, respectively), less frequent del(16q) (10.8% vs 25.4%), more frequent del(22q) (47.8% vs 26%), and more frequent nullisomy Y in males (24.2% vs 15.1%). In the multivariate analyses, PFS was shorter in patients with del(1p32), del(22q), and a large number of structural abnormalities. For OS, the significant factors were del(13q14), del(1p32), and the number of structural abnormalities. Hyperdiploidy or individual trisomies did not impact either the PFS or the OS. Thus, in this large series of patients with t(4;14), we were not able to identify “good-risk” parameters. In contrast, several chromosomal changes were associated with a worse prognosis.
In patients with del(17p) present in >60% of their plasma cells, hyperdiploidy was present in 35% of them. Regarding chromosomal changes differently expressed than in the general population, we found more frequent complex molecular karyotypes, more frequent del(13q14) (79.1% vs 51%), more frequent del(1p32) (28.2% vs 12.3%), more frequent del(6q) (46.4% vs 24.2%), more frequent del(8p) (41.8% vs 23.9%), more frequent del(12p) (30% vs 13.5%), more frequent del(14q) and monosomy 14 (52.7% and 23.6% vs 28% and 11.3%, respectively), more frequent del(16q) (47.3% vs 25.4%), more frequent del(22q) (46.4% vs 26%), and more frequent nullisomy Y in males (22.4% vs 15.1%). In the multivariate analyses, PFS was shorter in patients with del(6q), in the treatment subgroup melphalan-prednisone-thalidomide + melphalan-prednisone-Velcade, and in the subgroup of male patients with a gain of Xq, and longer for patients with monosomy 14 and trisomy 15. For OS, del(1p32) worsened the prognosis, and the number of numerical abnormalities (≥8 abnormalities) was found to have a protective effect.
Hyperdiploidy, as measured by the total number of chromosomes, was not an independent factor of progression risk. After adjustment, only trisomy 15 independently improved PFS, but not OS. However, if other chromosomal changes are not included in the statistical analysis, large hyperdiploidy did improve both PFS and OS. This point may contribute to the explanation of the discordant results obtained with the Mayo Clinic study, which did not take into account these parameters. In contrast, the number of structural chromosomal changes, which reflects the genomic complexity, impaired PFS in both groups and OS in the t(4;14) group. By counting the number of structural changes, chromosomes, or numerical abnormalities, regardless of their precise nature (protective or risk factors), we can only obtain a risk marker. In our study, only the gains and losses of the most frequent aberrations were specifically recorded. Once high-risk patients were identified on the basis of specific chromosomal aberrations, the fact that the number of anomalies (whatever type) remains independently associated with outcome suggests that cytogenetic aberrations not specifically measured, which differ from the ones retained in our analysis, can also explain difference in survival times. This finding is probably because of incomplete adjustment on specific chromosomal changes. So we argue that the number of structural changes, chromosomes, or numerical abnormalities are markers for the existence of other candidate cytogenetic factors but are not in themselves risk factors.
Del(1p32) was the more consistent specific cytogenetic abnormality found in this study. We and others have already reported that 1p32 deletions are major negative prognostic factors for PFS and OS for patients with MM and that deletion 1p32 is associated with t(4;14) (P = .001) and del(17p) (P = .002). Moreover, Boyd et al19 have published a study of deletions at 1p12, 1p22, and 1p32 where del(1p32) was present in 11.3% of patients and impaired survival; they also described putative target genes in the 1p32 region: CDKN2C, FAF1, and MTF2.
Our findings can explain why the outcome of patients with del(17p) is uniformly poor as only a few other specific cytogenetic factors are independently associated with outcomes. Conversely, the heterogeneity of survival among t(4;14) patients can be explained by several other cytogenetic abnormalities found to be independently associated with progression or death.
Our study also has some limitations. It is not possible to draw causal inferences from observational studies, and some associations in exploratory analysis may only reflect chance findings. Moreover, the complex relationships between cytogenetic abnormalities present particular modeling challenges because of frequent interrelationships among structural aberrations. Even though our series of high-risk patients was large, the number of outcome events per predictor variables was relatively small to correctly address the problem of overfitting. Moreover, the sample size was still not sufficient to ensure precise estimates. So our results should be interpreted with caution and need to be replicated in further studies.
In conclusion, this study in a large series of high-risk patients, analyzed with the most modern genomic technique, identified 1 main specific factor negatively impacting survival, del(1p32), which would be important to investigate in future studies, but also in the routine assessment of chromosomal risk in patients with MM.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by a grant from the National Institutes of Health, National Cancer Institute (Dana-Farber Cancer Institute) (PO1 CA100707-12), the CAPTOR program (French government), and a dedicated grant from Onyx.
Authorship
Contribution: F.M., N.M., J.C., S.M., and H.A.-L. designed the study; B.H., V.L.-C., and H.A.-L. wrote the manuscript; A.C., V.L.-C., and E.Y. performed statistical analyses; and M.-L.C., C.H., X.L., G.M., L.K., M.R., A.-M.S., K.B., L.V., L.G., M. Macro, D.C., M. Mohty, T.F., P.M., and M.A. provided patient samples and follow-up data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hervé Avet-Loiseau, Unité de Génomique du Myélome, IUC-T Oncopole, 1 Avenue Irène Joliot-Curie, 31059 Toulouse, France; e-mail: avet-loiseau.h@chu-toulouse.fr.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal