Key Points
Two novel transducer modules consisting of BTK in combination with either FLT3-ITD or TLR9 induce distinct oncogenic signaling programs.
This study suggests subtype-specific treatment strategies, including BTK/FLT3 inhibitor combinations, and shows how TLR9 affects AML biology.
Abstract
Acute myeloid leukemia (AML) is driven by niche-derived and cell-autonomous stimuli. Although many cell-autonomous disease drivers are known, niche-dependent signaling in the context of the genetic disease heterogeneity has been difficult to investigate. Here, we analyzed the role of Bruton tyrosine kinase (BTK) in AML. BTK was frequently expressed, and its inhibition strongly impaired the proliferation and survival of AML cells also in the presence of bone marrow stroma. By interactome analysis, (phospho)proteomics, and transcriptome sequencing, we characterized BTK signaling networks. We show that BTK-dependent signaling is highly context dependent. In Fms-like tyrosine kinase 3 internal tandem duplication (FLT3-ITD)–positive AML, BTK mediates FLT3-ITD–dependent Myc and STAT5 activation, and combined targeting of FLT3-ITD and BTK showed additive effects. In Fms-like tyrosine kinase 3 internal tandem duplication (FLT3-ITD)–negative AML, BTK couples Toll-like receptor 9 (TLR9) activation to nuclear factor κΒ and STAT5. Both BTK-dependent transcriptional programs were relevant for cell cycle progression and apoptosis regulation. Thus, we identify context-dependent oncogenic driver events that may guide subtype-specific treatment strategies and, for the first time, point to a role of TLR9 in AML. Clinical evaluation of BTK inhibitors in AML seems warranted.
Introduction
Acute myeloid leukemia (AML) is a genetically heterogeneous disease, and causative mutations occur in various genes, including those encoding for signaling molecules.1 In particular, mutations in tyrosine kinases such as FLT3 and KIT can induce transformation and deregulated proliferation of the myeloid progeny.2 Such dependence of AML cells upon oncogenic signaling can be exploited therapeutically by using small-molecule inhibitors. For instance, kinase inhibitors targeting Fms-like tyrosine kinase 3 internal tandem duplication (FLT3-ITD) showed significant effects in preclinical and clinical studies.3 However, signaling events induced by the AML microenvironment can counteract FLT3 inhibition because they trigger AML cell survival.4 In accordance with this, pharmacologic targeting of these microenvironment-dependent pathways showed promising effects in preclinical AML models, as found, for example, for the integrin-SYK-STAT signaling axis.5,6
In this study, we identified Bruton tyrosine kinase (BTK) as an AML target. BTK is best known for its functions in the context of B-cell receptor signaling.7 Loss of BTK expression in the B lineage can result in immunodeficiency syndromes and also contributes to the pathogenesis of acute lymphoblastic leukemia,7 in which BTK acts as a tumor suppressor. In contrast, BTK acts as an oncogene in several B-cell lymphoma subtypes.8 Inhibition of BTK induced cellular apoptosis and growth reduction in chronic lymphocytic leukemias, diffuse large B-cell lymphomas of the activated B-cell type, and mantle cell lymphomas.9 Treatment of chronic lymphocytic leukemia patients with the BTK inhibitor ibrutinib led to US Food and Drug Administration approval of the compound for chronic lymphocytic leukemia.10 In contrast to its role in lymphoid (patho-)physiology, far less is known about BTK signaling in myelopoiesis. One reason for this may be that Btk-deficient mice have a profound B-cell–specific phenotype, whereas myelopoiesis is not affected.11 The functional relevance of BTK in myeloid leukemia cell lines was reported quite recently.12 However, neither BTK signaling networks nor their mechanisms of action in myeloid cells are understood. Here, we show that BTK is expressed in about 80% of human AML cases and adduce evidence for the manner in which it promotes AML cell proliferation and survival via activation of several signaling pathways; these pathways involve signaling from Toll-like receptor 9 (TLR9) and FLT3-ITD, which trigger various context-dependent downstream transcriptional programs.
Methods
Cell culture, lentiviral transduction, and immunohistochemistry
Peripheral blood samples from AML patients were obtained at diagnosis or relapse with the approval of the ethics committee of Goethe University Frankfurt. This study was conducted in accordance with the Declaration of Helsinki. Metabolic labeling of KG-1 and MV4-11 cells via stable isotope labeling with amino acids in cell culture (SILAC) for arginine and lysine and AML cell culture were performed as described earlier.6 Details of the ethics approval and the protocols for lentiviral transductions, cell proliferation, and immunohistochemistry analyses are provided in the supplemental Data available on the Blood Web site.
Mass spectrometric analysis, database search, and MaxQuant-based data processing
Results
BTK promotes cell survival and proliferation in AML
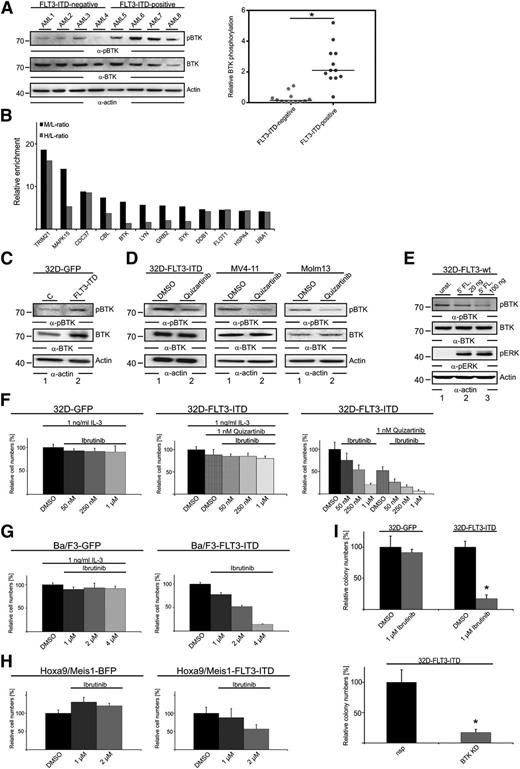
Kinase expression profiling in AML bone marrow biopsies revealed expression of BTK in 23 of the 28 AML patients analyzed (Figure 1A). In light of the oncogenic potential of BTK in lymphoid malignancies, we investigated its functional role in AML. For this purpose, we first investigated the expression and activation status of BTK in various AML cell lines and genetically well-defined AML cultures derived from AML patients in different cytogenetic risk groups (supplemental Table 1). All AML cell lines tested and all primary AML cultures expressed BTK, and most of these cell lines (with the exception of U937) and all primary AML cultures showed phosphorylation of BTK at the activation-inducing tyrosine Tyr-223, indicating constitutive kinase activity (Figure 1B).
BTK is expressed in AML and promotes cell survival and proliferation. (A) Immunohistochemical analysis of BTK expression in 28 bone marrow specimens of AML patients. For comparison, bone marrow–derived samples from healthy individuals and from diffuse large B-cell lymphoma (DLBCL) patients were stained by antibodies against BTK. (B) Cleared cellular lysates of the lymphoma cell line DG75 and various AML cell lines (left panel) or primary patient-derived AML cultures (right panel) were subjected to immunoblotting with antibodies against phosphorylated BTK (pBTK; upper panels) and BTK (middle panels). Protein loading was monitored by anti-actin immunoblotting (lower panels). (C-D) Growth curves of the AML cell lines as well as the patient-derived AML cultures FFM04, -05, -12, -21, -40, and -41 that were treated with ibrutinib at final concentrations of 10 nM (red lines), 50 nM (green lines), or 250 nM (purple lines) or with dimethylsulfoxide (DMSO) as a control (blue lines.). Data from 4 independent experiments are shown (mean ± standard deviation [SD]). *P < .05 between the different treatment groups and the control cells using Student t test. (E) Relative cell numbers of primary AML cultures that were treated with 250 nM ibrutinib for 3 cycles. Each cycle consisted of a 1-hour ibrutinib treatment followed by a 23-hour washout period. Results (mean ± SD) are from 4 independent experiments.*P < .05 between the different treatment groups and the control cells using Student t test. (F) Immunoblots of cleared cellular lysates derived from KG-1 and MV4-11 control cells (lane 1) or the respective BTK knockdown cells (lane 2) with antibodies recognizing BTK (upper panels) or actin (lower panels). (G) KG-1 and MV4-11 cells were transduced either with lentiviral vectors encoding BTK-specific shRNAs and GFP or with unspecific control shRNAs and GFP. GFP expression was subsequently monitored in the respective cell batches by flow cytometry 1 day after lentiviral transduction (day 1) and 7 days thereafter (day 7). The outlined dot plots (representative for all 3 experiments) and histograms summarizing data from 3 independent experiments (mean ± SD) show the relative abundance of GFP-expressing cells on day 1 and day 7 for the respective BTK knockdown or control cell batches, as determined by flow cytometry. *P < .05 between the BTK knockdown and the control cells (nsp) using Student t test. (H) Flow cytometric cell cycle and (I) apoptosis analysis of KG-1 and MV4-11 BTK knockdown (BTK KD) cells or control cells (nsp) at day 2 after transduction. Results (mean ± SD) are from 4 independent experiments. *P < .05 between the BTK knockdown and the control cells using Student t test.
BTK is expressed in AML and promotes cell survival and proliferation. (A) Immunohistochemical analysis of BTK expression in 28 bone marrow specimens of AML patients. For comparison, bone marrow–derived samples from healthy individuals and from diffuse large B-cell lymphoma (DLBCL) patients were stained by antibodies against BTK. (B) Cleared cellular lysates of the lymphoma cell line DG75 and various AML cell lines (left panel) or primary patient-derived AML cultures (right panel) were subjected to immunoblotting with antibodies against phosphorylated BTK (pBTK; upper panels) and BTK (middle panels). Protein loading was monitored by anti-actin immunoblotting (lower panels). (C-D) Growth curves of the AML cell lines as well as the patient-derived AML cultures FFM04, -05, -12, -21, -40, and -41 that were treated with ibrutinib at final concentrations of 10 nM (red lines), 50 nM (green lines), or 250 nM (purple lines) or with dimethylsulfoxide (DMSO) as a control (blue lines.). Data from 4 independent experiments are shown (mean ± standard deviation [SD]). *P < .05 between the different treatment groups and the control cells using Student t test. (E) Relative cell numbers of primary AML cultures that were treated with 250 nM ibrutinib for 3 cycles. Each cycle consisted of a 1-hour ibrutinib treatment followed by a 23-hour washout period. Results (mean ± SD) are from 4 independent experiments.*P < .05 between the different treatment groups and the control cells using Student t test. (F) Immunoblots of cleared cellular lysates derived from KG-1 and MV4-11 control cells (lane 1) or the respective BTK knockdown cells (lane 2) with antibodies recognizing BTK (upper panels) or actin (lower panels). (G) KG-1 and MV4-11 cells were transduced either with lentiviral vectors encoding BTK-specific shRNAs and GFP or with unspecific control shRNAs and GFP. GFP expression was subsequently monitored in the respective cell batches by flow cytometry 1 day after lentiviral transduction (day 1) and 7 days thereafter (day 7). The outlined dot plots (representative for all 3 experiments) and histograms summarizing data from 3 independent experiments (mean ± SD) show the relative abundance of GFP-expressing cells on day 1 and day 7 for the respective BTK knockdown or control cell batches, as determined by flow cytometry. *P < .05 between the BTK knockdown and the control cells (nsp) using Student t test. (H) Flow cytometric cell cycle and (I) apoptosis analysis of KG-1 and MV4-11 BTK knockdown (BTK KD) cells or control cells (nsp) at day 2 after transduction. Results (mean ± SD) are from 4 independent experiments. *P < .05 between the BTK knockdown and the control cells using Student t test.
Upon short- and long-term pharmacologic inhibition of BTK, we detected significantly decreased cell expansion in most AML cell lines and primary AML cultures that we tested (Figure 1C-E): FLT3-ITD–positive or –negative. The reduced proliferation was caused mainly by an ibrutinib-dependent cell cycle arrest in the G1/S transition and by an increase of apoptosis (supplemental Figure 1A-B). To exclude off-target effects of the BTK inhibitor ibrutinib and to confirm the proproliferative potential of BTK in AML, we silenced BTK expression in AML cells by using constructs encoding for BTK-specific short hairpin RNAs (shRNAs) and green fluorescence protein (GFP) as described previously.14 Efficient knockdown of BTK was validated by immunoblotting (Figure 1F). By flow cytometric monitoring of GFP expression, we detected a loss of (GFP-positive) BTK knockdown cells, whereas the numbers of control cells were stable in the respective control cell batches (Figure 1G). These data indicate that BTK knockdown cells have a compromised expansion potential caused by induction of apoptosis and/or G1 arrest (Figure 1H-I). Thus, BTK is expressed and activated in a large proportion of AML cases, and BTK inhibition induces cell cycle arrest and apoptosis.
FLT3-ITD induces BTK activation and sensitivity to BTK inhibition
In B cells, BTK is an important effector of B-cell receptor signaling. However, the molecular mechanisms of BTK activation and signaling in AML cells are unknown.
Because we had identified the cell lines Molm13 and MV4-11 and the primary cultures FFM40 and FFM41 (Figure 1C), all expressing FLT3-ITD in a manner sensitive to BTK inhibition, and because we detected increased BTK phosphorylation in FLT3-ITD–positive AML cells (Figures 1B and 2A), we hypothesized that BTK activation might be dependent on FLT3-ITD activity. To find out whether the two kinases interact with one another, we performed a quantitative proteomic interactome analysis. Using the quantitative immunoprecipitation combined with knockdown (QUICK) method for protein interaction screening,13 we purified the FLT3-ITD signalosomes from SILAC-labeled MV4-11 cells that had been treated either with dimethylsulfoxide (medium-labeled cells) or with quizartinib (heavy-labeled cells) and subsequently characterized them by MS. The experimental setup is outlined in supplemental Figure 1C. Among the identified FLT3-ITD interaction partners identified was BTK (Figure 2B). Furthermore, the SILAC approach allowed us to relatively quantify the interaction between each identified FLT3-ITD binding partner and FLT3-ITD in the presence and the absence of quizartinib. We found that inhibition of FLT3-ITD by quizartinib negatively affected some interactions, such as the ones with BTK and SYK, whereas others were not affected (Figure 2B and supplemental Table 2). From these experiments, we identified a previously unknown interaction between FLT3-ITD and BTK that was dependent on FLT3-ITD kinase activity. To investigate the functional relevance of this newly identified FLT3-ITD/BTK signalosome, we lentivirally expressed (1) FLT3-ITD in combination with GFP in various cell models such as the interleukin-3 (IL-3) –dependent murine hematopoietic progenitor cell lines 32D (32D-FLT3-ITD), Ba/F3 (Ba/F3-FLT3-ITD), and primary murine myeloid progenitor cells that were transformed by HOXA9 and MEIS1 (HOXA9/MEIS1-FLT3-ITD), and (2) GFP/BFP alone (32D-GFP, Ba/F3-GFP, HOXA9/MEIS1-BFP, control). As a further control, we used 32D-FLT3-ITD cells growing in the presence of the FLT3 inhibitor quizartinib and IL-3. In 32D-FLT3-ITD cells compared with 32D-GFP cells, BTK expression and phosphorylation of its activation-inducing tyrosine were strongly upregulated (Figure 2C). In line with these results, phosphorylation of BTK in 32D-FLT3-ITD, MV4-11, and Molm13 cells was strongly diminished upon quizartinib treatment (Figure 2D). Notably, stimulation of wild-type FLT3 did not affect phosphorylation of the activation-inducing tyrosine (Tyr-223) in BTK; this points to a specific functional interaction between FLT3-ITD and BTK (Figure 2E). Moreover, FLT3-ITD expressing 32D and Ba/F3 cells showed reduced cell expansion (in the absence of IL-3) upon inhibition of BTK, whereas IL-3–dependent expansion of 32D-GFP and Ba/F3-GFP cells and 32D-FLT3-ITD cells (the latter in the presence of quizartinib) was not significantly affected (Figure 2F-G and supplemental Figure 1D). Similar results were obtained when BTK expression was silenced by shRNAs in this experimental setting (data not shown). A comparable sensitizing of the cells for ibrutinib occurred upon expression of FLT3-ITD in HOXA9/MEIS1-transformed myeloid progenitor cells (Figure 2H). Their dependence on BTK was further confirmed by colony formation assays following BTK knockdown (supplemental Figure 1E). To further assess the effect of BTK inhibition on the colony formation capacity of the 32D cell batches referred to above, we performed methylcellulose-based colony formation assays in the presence or absence of ibrutinib or following knockdown of BTK. Both approaches revealed that the FLT3-ITD–dependent colony formation of 32D-FLT3-ITD cells depends on BTK activity, whereas (IL-3–dependent) 32D-GFP cells formed colonies in a BTK-independent manner (Figure 2I). These data show that BTK is part of the FLT3-ITD signalosome and is activated in an FLT3-ITD–dependent manner, suggesting that combined inhibition of both kinases might have additive antileukemic effects.
BTK acts downstream of FLT3-ITD to induce proliferation in AML cells. (A) Cleared cellular lysates of patient-derived AML cells (12 FLT3-ITD–positive and 12 FLT3-ITD–negative patients) were subjected to immunoblotting with antibodies against pBTK (upper panels) and BTK (middle panels). Protein loading was monitored by anti-actin immunoblotting (lower panels). The right panel shows the BTK phosphorylation normalized to BTK expression based on the signal intensities measured by immunoblotting. (B) The endogenous FLT3-ITD interactome was identified in MV4-11 cells using quantitative SILAC-based MS. All proteins identified that showed a medium (M) vs light (L) ratio (M/L) greater than 4 are plotted according to their M/L ratio and heavy (H) vs light (H/L) ratios of enrichment on logarithmic scales. Proteins with an M/L ratio greater than 4 were identified as interaction partners of FLT3-ITD in DMSO-treated cells. The H/L ratio represents the FLT3-ITD interactome in cells treated with 20 nM quizartinib for 1 hour. The complete list of proteins identified and quantification statistics for 2 independent experiments are provided in supplemental Table 2. (C) Cleared cellular lysates of 32D cells expressing FLT3-ITD and GFP (lane 2) and the respective GFP-expressing control cells (lane 1) were subjected to immunoblotting with antibodies against pBTK (upper panels) and BTK (middle panels). Protein loading was monitored by anti-actin immunoblotting (lower panels). (D) Cleared cellular lysates of 32D cells expressing FLT3-ITD, MV4-11 (middle panel) and Molm13 cells (right panel) that were treated with DMSO (lane 1) and 20 nM quizartinib (lane 2) were subjected to immunoblotting with antibodies against pBTK (upper panels) and BTK (middle panels). Protein loading was monitored by anti-actin immunoblotting (lower panels). (E) Cleared cellular lysates of 32D cells expressing FLT3-wild-type (wt) that were left untreated (lane 1) or were stimulated by FLT3-ligand (FL) for durations indicated (lanes 2 and 3) were subjected to immunoblotting with antibodies against pBTK, BTK, pERK, and actin. (F) Tetrazolium salt (XTT)-based proliferation analysis of 32D cells expressing FLT3-ITD and GFP, or the respective control cells expressing GFP, that were treated with either DMSO or quizartinib/ibrutinib at the final concentrations indicated. Results (from 3 independent experiments; mean ± SD) are shown for cells that had been treated for 3 days. (G-H) XTT-based proliferation analysis of (G) Ba/F3 cells expressing either GFP or FLT3-ITD in combination with GFP and (H) murine myeloid progenitor cells expressing either HOXA9 and MEIS1 in combination with BFP or HOXA9 and MEIS1 in combination with both BFP and FLT3-ITD that have been treated with ibrutinib in DMSO or with DMSO control at the final concentrations indicated. Results are representative for 3 independent experiments and are shown for cells that had been treated for 3 days; mean ± SD are shown for technical replicates. (I) Colony formation assays of 32D cells expressing FLT3-ITD and GFP or the respective control cells in the presence of DMSO or ibrutinib (upper panel) or upon BTK knockdown (lower panel). Results (mean ± SD) are from 4 independent experiments. *P < .05 between the different treatment groups (upper panel: ibrutinib compared with DMSO; lower panel: BTK KD compared with nsp) using Student t test.
BTK acts downstream of FLT3-ITD to induce proliferation in AML cells. (A) Cleared cellular lysates of patient-derived AML cells (12 FLT3-ITD–positive and 12 FLT3-ITD–negative patients) were subjected to immunoblotting with antibodies against pBTK (upper panels) and BTK (middle panels). Protein loading was monitored by anti-actin immunoblotting (lower panels). The right panel shows the BTK phosphorylation normalized to BTK expression based on the signal intensities measured by immunoblotting. (B) The endogenous FLT3-ITD interactome was identified in MV4-11 cells using quantitative SILAC-based MS. All proteins identified that showed a medium (M) vs light (L) ratio (M/L) greater than 4 are plotted according to their M/L ratio and heavy (H) vs light (H/L) ratios of enrichment on logarithmic scales. Proteins with an M/L ratio greater than 4 were identified as interaction partners of FLT3-ITD in DMSO-treated cells. The H/L ratio represents the FLT3-ITD interactome in cells treated with 20 nM quizartinib for 1 hour. The complete list of proteins identified and quantification statistics for 2 independent experiments are provided in supplemental Table 2. (C) Cleared cellular lysates of 32D cells expressing FLT3-ITD and GFP (lane 2) and the respective GFP-expressing control cells (lane 1) were subjected to immunoblotting with antibodies against pBTK (upper panels) and BTK (middle panels). Protein loading was monitored by anti-actin immunoblotting (lower panels). (D) Cleared cellular lysates of 32D cells expressing FLT3-ITD, MV4-11 (middle panel) and Molm13 cells (right panel) that were treated with DMSO (lane 1) and 20 nM quizartinib (lane 2) were subjected to immunoblotting with antibodies against pBTK (upper panels) and BTK (middle panels). Protein loading was monitored by anti-actin immunoblotting (lower panels). (E) Cleared cellular lysates of 32D cells expressing FLT3-wild-type (wt) that were left untreated (lane 1) or were stimulated by FLT3-ligand (FL) for durations indicated (lanes 2 and 3) were subjected to immunoblotting with antibodies against pBTK, BTK, pERK, and actin. (F) Tetrazolium salt (XTT)-based proliferation analysis of 32D cells expressing FLT3-ITD and GFP, or the respective control cells expressing GFP, that were treated with either DMSO or quizartinib/ibrutinib at the final concentrations indicated. Results (from 3 independent experiments; mean ± SD) are shown for cells that had been treated for 3 days. (G-H) XTT-based proliferation analysis of (G) Ba/F3 cells expressing either GFP or FLT3-ITD in combination with GFP and (H) murine myeloid progenitor cells expressing either HOXA9 and MEIS1 in combination with BFP or HOXA9 and MEIS1 in combination with both BFP and FLT3-ITD that have been treated with ibrutinib in DMSO or with DMSO control at the final concentrations indicated. Results are representative for 3 independent experiments and are shown for cells that had been treated for 3 days; mean ± SD are shown for technical replicates. (I) Colony formation assays of 32D cells expressing FLT3-ITD and GFP or the respective control cells in the presence of DMSO or ibrutinib (upper panel) or upon BTK knockdown (lower panel). Results (mean ± SD) are from 4 independent experiments. *P < .05 between the different treatment groups (upper panel: ibrutinib compared with DMSO; lower panel: BTK KD compared with nsp) using Student t test.
Additive effects of combined FLT3-BTK inhibition
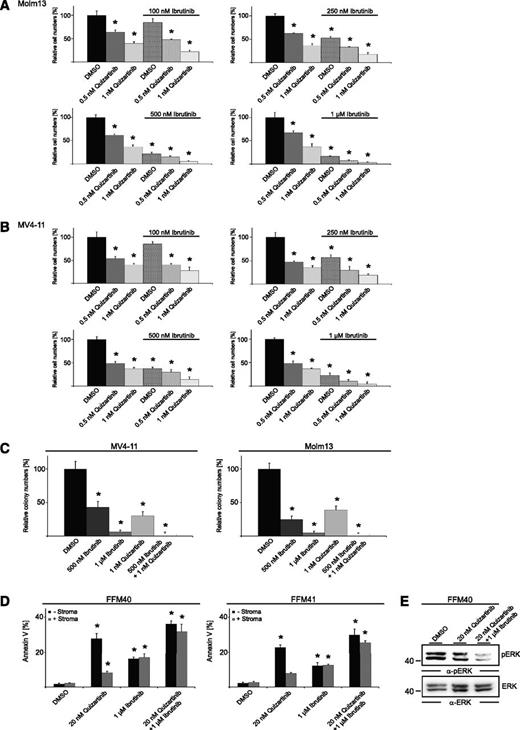
Because we identified FLT3-ITD as a functionally relevant upstream activator of BTK, we next investigated in vitro whether combined inhibition of FLT3-ITD and BTK enhances cell toxicity in comparison with treatment with these inhibitors singly. For this purpose, we treated MV4-11 and Molm13 cells, both expressing endogenous FLT3-ITD, with various concentrations of quizartinib and ibrutinib, either alone or in combination. As shown in Figure 3A-B and supplemental Figure 1F-G, we detected an additive effect of the combined treatment with regard to its antiproliferative potential. Similar effects were observed in colony formation assays (Figure 3C and supplemental Figure 1H). This suggests that this combination could be an interesting option for treatment of patients suffering from FLT3-ITD–positive AML.
FLT3/BTK inhibitor combinations show additive effects. (A-B) Proliferation analysis of Molm13 and MV4-11 cells in the presence of quizartinib, ibrutinib, or DMSO. Results (from 4 independent experiments; mean ± SD) are shown for cells that had been treated for 3 days. *P < .05 between the different treatment groups compared with DMSO using Student t test. (C) Colony formation assays of MV4-11 and Molm13 cells in the presence of DMSO, ibrutinib, quizartinib, and ibrutinib/quizartinib. Results (mean ± SD) are from 4 independent experiments. *P < .05 between the different treatment groups compared with DMSO using Student t test. (D) FFM40 and FFM41 cells were exposed to quizartinib continuously (20 nM) or ibrutinib (1 µM) for 1 hour (followed by wash-out) in suspension culture or coculture with bone marrow stroma and analyzed for annexin V binding by flow cytometry. Results from 4 independent experiments. *P < .05 between the different treatment groups compared with the respective DMSO controls using Student t test. (E) Cleared cellular lysates of FFM40 cells derived from bone marrow stroma cocultures and treated as indicated were subjected to immunoblotting with antibodies against pERK (upper panel) and ERK (lower panel).
FLT3/BTK inhibitor combinations show additive effects. (A-B) Proliferation analysis of Molm13 and MV4-11 cells in the presence of quizartinib, ibrutinib, or DMSO. Results (from 4 independent experiments; mean ± SD) are shown for cells that had been treated for 3 days. *P < .05 between the different treatment groups compared with DMSO using Student t test. (C) Colony formation assays of MV4-11 and Molm13 cells in the presence of DMSO, ibrutinib, quizartinib, and ibrutinib/quizartinib. Results (mean ± SD) are from 4 independent experiments. *P < .05 between the different treatment groups compared with DMSO using Student t test. (D) FFM40 and FFM41 cells were exposed to quizartinib continuously (20 nM) or ibrutinib (1 µM) for 1 hour (followed by wash-out) in suspension culture or coculture with bone marrow stroma and analyzed for annexin V binding by flow cytometry. Results from 4 independent experiments. *P < .05 between the different treatment groups compared with the respective DMSO controls using Student t test. (E) Cleared cellular lysates of FFM40 cells derived from bone marrow stroma cocultures and treated as indicated were subjected to immunoblotting with antibodies against pERK (upper panel) and ERK (lower panel).
However, recent studies showed that bone marrow stroma cells emit signals promoting cell survival and proliferation of AML blasts as a clinically relevant mechanism of therapy resistance. These stroma-derived signals even counteract the cytotoxic effects of quizartinib in vitro and in vivo, leading to reduced apoptosis of AML blasts in the bone marrow (compared with peripheral blood) upon quizartinib treatment.4 To test the efficacy of BTK inhibition alone or in combination with quizartinib in the presence of bone marrow stroma, we performed apoptosis assays in a coculture model consisting of primary AML cells and primary bone marrow stroma cells derived from healthy donors. Notably, ibrutinib was washed out after a 1-hour pretreatment to avoid inhibition of off-targets and to better reflect the in vivo situation in which mainly BTK remains inhibited because of the irreversible action of ibrutinib. In accordance with previous studies, we observed that quizartinib-dependent apoptosis is strongly reduced in stroma coculture compared with monoculture conditions (Figure 3D). In contrast, cytotoxic effects of BTK inhibition were not counteracted upon stroma contact, which might be explained by the detachment of AML cells from the stroma cells that we and others observed12 upon BTK inhibition. More interestingly, the combined inhibition of FLT3 and BTK showed additive effects in stroma coculture (Figure 3D) and effective suppression of ERK activation that was reported previously as a microenvironment-dependent cell survival-inducing signaling event4 (Figure 3E). Both findings support a functional relevance for FLT3/BTK inhibitor combinations in the presence or absence of bone marrow stroma cells.
However, the observed additive effect suggests that, in addition to FLT3-ITD, activators of BTK further upstream are operational, which is also reflected by the fact that AML cells devoid of FLT3-ITD responded to inhibition of BTK (Figure 1D).
A TLR9/BTK transducer module promotes AML cell expansion in FLT3-ITD–negative AML
Because sensitivity to BTK inhibition is not restricted to FLT3-ITD–expressing cells, we next investigated the BTK signalosome in FLT3-ITD–negative AML cells to identify further potential activators of BTK. For this purpose, we performed a quantitative proteomic interactome analysis (QUICK) as described above. We purified the BTK signalosome from SILAC-labeled KG-1 cells and subsequently characterized it by MS. The experimental setup is outlined in supplemental Figure 2A, and the resulting data distribution is shown in Figure 4A. Interestingly, the AML-specific BTK interaction partners differed markedly from those previously described in B cells (Table 1). Among the interactors that we identified was TLR9 which, according to previous studies that described an interaction between TLRs and BTK in other cell types,15,16 might be a potential activator of BTK in FLT3-ITD–negative AML cells. We performed co-immunoprecipitation experiments and confirmed by subsequent immunoblotting the newly identified interaction between BTK and TLR9 in further TLR9-expressing AML cell lines and the patient-derived AML cultures FFM04 and FFM12 (Figure 4B-D). Moreover, stimulation of TLR9 by CpG dinucleotides and coculture of AML cells with (apoptotic) bone marrow stroma induced tyrosine phosphorylation of BTK, further pointing toward a functional TLR9/BTK signaling axis in AML cells (Figure 4E).
An oncogenic TLR9/BTK transducer module operates in FLT3-ITD–negative AML. (A) The endogenous BTK interactome was identified in KG-1 cells using quantitative SILAC-based MS. All identified proteins (except contaminants) are plotted according to their signal intensities and their H/L ratio of enrichment on logarithmic scales. Proteins with an H/L ratio > 5 (red) were identified as interaction partners of BTK. The complete list of identified proteins and quantification statistics is provided in supplemental Table 3. (B) Cleared cellular lysates of H2228 cells (negative control, lane 1) and various AML cell lines were subjected to immunoblotting with antibodies against TLR9 (upper panel) and actin (lower panel) as loading controls. (C) KG-1, FFM04, MV4-11, and Molm13 cells were lysed and subjected to immunoprecipitation using anti-TLR9 antibodies (lane 2) or isotype-matched control antibodies (lane 1 [C]). The proteins obtained were analyzed by immunoblotting with antibodies directed against BTK (upper panels). Effective immunoprecipitation of TLR9 was confirmed by immunoblotting using TLR9-specific antibodies (lower panels). (D) FFM04 and FFM12 cells were lysed and subjected to immunoprecipitation using anti-BTK antibodies (lane 2) or isotype-matched control antibodies (lane 1 [C]). The proteins thus isolated were analyzed by immunoblotting with antibodies directed against TLR9 (upper panels). Effective immunoprecipitation of BTK was confirmed by immunoblotting using BTK-specific antibodies (lower panels). (E) Left panel: Immunoblot analyses of lysates derived from KG-1 and FFM04 cells that were left untreated (lane 1) or stimulated by CpG dinucleotides for 15, 30, or 60 minutes (lanes 2 to 4). Right panel: Immunoblot analysis of lysates derived from FFM04 cells that were either left untreated (lane 1), cocultured with bone marrow (BM) stroma cells (lane 2), or cocultured with (Annexin V-positive) apoptotic stroma cells that had been irradiated prior to coculture (lane 3). Immunoblotting was performed by using phosphosite-specific antibodies against pTyr-223 of BTK (upper panels). Protein loading was monitored by immunoblotting of BTK (lower panels). (F) XTT-based proliferation analysis of KG-1 and FFM04 cells left untreated or treated with ibrutinib and 1.5 ng/μL CpG. Results (from 4 independent experiments; mean ± SD) are shown for cells that were treated for 3 days. *P < .05 between the different treatment groups compared with the DMSO control (black bar) using Student t test. (G) KG-1, FFM04, MV4-11, and Molm13 cells were transduced either with lentiviral vectors encoding TLR9-specific shRNAs and GFP or with unspecific control shRNAs (nsp) and GFP. Subsequently, expression of TLR9 and actin was monitored by immunoblotting. (H) GFP expression was monitored in the respective transduced cell batches by flow cytometry 1 day after lentiviral transduction (day 1) and 7 days thereafter (day 7). The outlined diagram summarizes data from 4 independent experiments (mean ± SD) and shows the relative abundance of GFP-expressing cells at day 1 or day 7 for the respective TLR9 knockdown or control cell batches.
An oncogenic TLR9/BTK transducer module operates in FLT3-ITD–negative AML. (A) The endogenous BTK interactome was identified in KG-1 cells using quantitative SILAC-based MS. All identified proteins (except contaminants) are plotted according to their signal intensities and their H/L ratio of enrichment on logarithmic scales. Proteins with an H/L ratio > 5 (red) were identified as interaction partners of BTK. The complete list of identified proteins and quantification statistics is provided in supplemental Table 3. (B) Cleared cellular lysates of H2228 cells (negative control, lane 1) and various AML cell lines were subjected to immunoblotting with antibodies against TLR9 (upper panel) and actin (lower panel) as loading controls. (C) KG-1, FFM04, MV4-11, and Molm13 cells were lysed and subjected to immunoprecipitation using anti-TLR9 antibodies (lane 2) or isotype-matched control antibodies (lane 1 [C]). The proteins obtained were analyzed by immunoblotting with antibodies directed against BTK (upper panels). Effective immunoprecipitation of TLR9 was confirmed by immunoblotting using TLR9-specific antibodies (lower panels). (D) FFM04 and FFM12 cells were lysed and subjected to immunoprecipitation using anti-BTK antibodies (lane 2) or isotype-matched control antibodies (lane 1 [C]). The proteins thus isolated were analyzed by immunoblotting with antibodies directed against TLR9 (upper panels). Effective immunoprecipitation of BTK was confirmed by immunoblotting using BTK-specific antibodies (lower panels). (E) Left panel: Immunoblot analyses of lysates derived from KG-1 and FFM04 cells that were left untreated (lane 1) or stimulated by CpG dinucleotides for 15, 30, or 60 minutes (lanes 2 to 4). Right panel: Immunoblot analysis of lysates derived from FFM04 cells that were either left untreated (lane 1), cocultured with bone marrow (BM) stroma cells (lane 2), or cocultured with (Annexin V-positive) apoptotic stroma cells that had been irradiated prior to coculture (lane 3). Immunoblotting was performed by using phosphosite-specific antibodies against pTyr-223 of BTK (upper panels). Protein loading was monitored by immunoblotting of BTK (lower panels). (F) XTT-based proliferation analysis of KG-1 and FFM04 cells left untreated or treated with ibrutinib and 1.5 ng/μL CpG. Results (from 4 independent experiments; mean ± SD) are shown for cells that were treated for 3 days. *P < .05 between the different treatment groups compared with the DMSO control (black bar) using Student t test. (G) KG-1, FFM04, MV4-11, and Molm13 cells were transduced either with lentiviral vectors encoding TLR9-specific shRNAs and GFP or with unspecific control shRNAs (nsp) and GFP. Subsequently, expression of TLR9 and actin was monitored by immunoblotting. (H) GFP expression was monitored in the respective transduced cell batches by flow cytometry 1 day after lentiviral transduction (day 1) and 7 days thereafter (day 7). The outlined diagram summarizes data from 4 independent experiments (mean ± SD) and shows the relative abundance of GFP-expressing cells at day 1 or day 7 for the respective TLR9 knockdown or control cell batches.
The AML-specific BTK interactome
| Ligand . | UniProt ID . | Function . |
|---|---|---|
| TLR9 | Q9NR96 | Immune receptor |
| ALOX5 | P09917 | Lipoxygenase |
| FCGR1A | P12314 | Immune receptor |
| MAPRE1 | Q15691 | Adaptor protein |
| RYR1 | P21817 | Ryanodine receptor |
| CBX5 | P45973 | Adaptor protein |
| Ligand . | UniProt ID . | Function . |
|---|---|---|
| TLR9 | Q9NR96 | Immune receptor |
| ALOX5 | P09917 | Lipoxygenase |
| FCGR1A | P12314 | Immune receptor |
| MAPRE1 | Q15691 | Adaptor protein |
| RYR1 | P21817 | Ryanodine receptor |
| CBX5 | P45973 | Adaptor protein |
Two independent SILAC-based liquid chromatography-tandem mass spectrometry analyses were performed for identification of BTK interactors in untreated KG-1 cells. Mass-spectrometric data sets were analyzed by MaxQuant software. The proteins listed showed at least fourfold enrichment of heavy peptides compared with light ones. The protein functions listed were obtained by a literature search. Detailed statistics, including total numbers of all peptides identified and quantified, are provided in supplemental Table 3.
We next tested whether TLR9, as part of the identified BTK signalosome, is functionally relevant for AML cell proliferation and survival. To this end, we performed 2 types of experiments. First, we monitored proliferation of the AML cell line KG-1 and the patient-derived AML culture FFM04 in the presence and absence of TLR9 stimulation; second, we observed the effect of TLR9 knockdown on AML cell behavior. Upon stimulation of TLR9 with CpG, we observed significantly increased cell expansion compared with unstimulated control cells. Moreover, we found that this effect was reversible upon BTK inhibition (Figure 4F and supplemental Figure 2B). To investigate the effect of tonic TLR9 signaling on AML cells, we knocked down TLR9 in various AML cell lines by using plasmid constructs encoding TLR9-specific shRNAs in combination with GFP. TLR9 knockdown efficiency was monitored by immunoblotting (Figure 4G). When we followed GFP expression in the resulting cultures over time, we observed that the (GFP-positive) TLR9 knockdown cells became depleted from the FLT3-ITD–negative KG-1 and FFM04 cultures. In contrast, the proportion of (GFP-positive) TLR9 knockdown cells was stable over time in the FLT3-ITD–expressing MV4-11 culture, indicating that distinct BTK activation modes are operational in FLT3-ITD–positive and –negative AML cells (Figure 4H). A significant reduction of KG-1 and FFM04 TLR9 knockdown cells was observed in stroma coculture as well (supplemental Figure 2C). In summary, we identified an interaction between BTK and TLR9 that is functionally relevant in FLT3-ITD–negative AML cells. However, the signaling pathways downstream of the TLR9/BTK transducer module remain elusive.
Characterization of BTK-dependent signaling networks
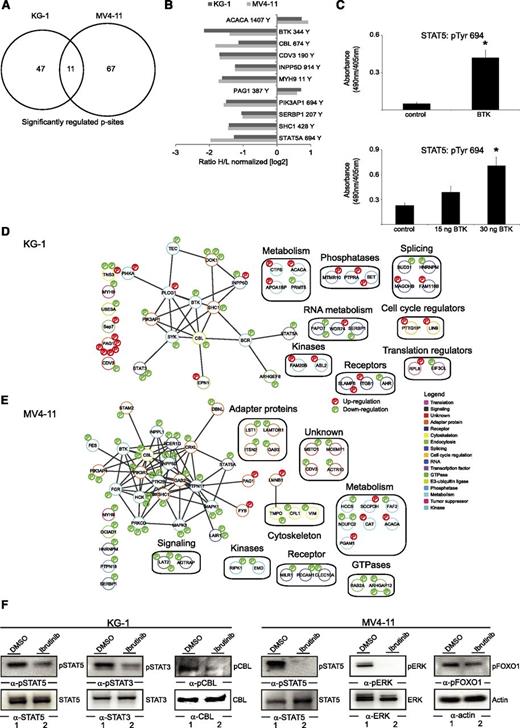
To gain insights into signaling events downstream of BTK, we performed quantitative phosphoproteomic analyses in the presence and absence of ibrutinib in KG-1 and MV4-11 cells as representatives of FLT3-ITD–negative and –positive AML cells, respectively. The experimental workflow was based on SILAC, followed by enrichment of tyrosine-phosphorylated peptides and MS analyses (supplemental Figure 2D). Upon treatment with ibrutinib, 58 of 875 phosphosites detected were identified as significantly regulated in KG-1 cells, and 78 of 571 phosphosites were similarly identified MV4-11 cells (Figure 5A, supplemental Figure 2E-F, and supplemental Table 4). However, only 11 phosphosites were concordantly regulated in both cell lines, implying that the differential BTK activation mechanisms described above involve differential downstream signaling events (Figure 5B).
BTK signaling networks in AML. (A) Venn diagram showing the number of significantly regulated pTyr sites after 1 hour of ibrutinib treatment. (B) Bar diagram outlining the concordantly regulated pTyr sites in KG-1 and MV4-11 cells. (C) Phosphorylation of Tyr 694 of STAT5 was monitored by using an in vitro kinase assay. The biotinylated peptide encompassing the amino acids TPVLAKAVDGYVKPQIK for STAT5 was used as substrate for BTK obtained from untreated KG-1 cells (upper panel) or for enzymatically active recombinant BTK (lower panel). Tyrosine phosphorylation of these peptides was monitored by anti-phosphotyrosine staining with enzyme-linked immunosorbent assay. *P < .05 between the different treatment groups compared with the controls using Student t test. (D-E) The signaling networks contain proteins that were identified as being phosphorylated (red) or dephosphorylated (green) on tyrosines in response to BTK inhibition. Proteins were grouped by Cytoscape software according to their known protein-protein interaction status listed in the STRING database. The assigned protein functions were derived by manual annotation using Uniprot, PhosphoSitePlus, and PubMed databases. (F) Cleared cellular lysates derived from KG-1 and MV4-11 cells that had either been left untreated or had been treated with 500 nM ibrutinib for 1 hour were subjected to immunoblotting using antibodies against pSTAT5, pERK, pSTAT3, pFOXO1, and phospho-c (pc)-CBL.
BTK signaling networks in AML. (A) Venn diagram showing the number of significantly regulated pTyr sites after 1 hour of ibrutinib treatment. (B) Bar diagram outlining the concordantly regulated pTyr sites in KG-1 and MV4-11 cells. (C) Phosphorylation of Tyr 694 of STAT5 was monitored by using an in vitro kinase assay. The biotinylated peptide encompassing the amino acids TPVLAKAVDGYVKPQIK for STAT5 was used as substrate for BTK obtained from untreated KG-1 cells (upper panel) or for enzymatically active recombinant BTK (lower panel). Tyrosine phosphorylation of these peptides was monitored by anti-phosphotyrosine staining with enzyme-linked immunosorbent assay. *P < .05 between the different treatment groups compared with the controls using Student t test. (D-E) The signaling networks contain proteins that were identified as being phosphorylated (red) or dephosphorylated (green) on tyrosines in response to BTK inhibition. Proteins were grouped by Cytoscape software according to their known protein-protein interaction status listed in the STRING database. The assigned protein functions were derived by manual annotation using Uniprot, PhosphoSitePlus, and PubMed databases. (F) Cleared cellular lysates derived from KG-1 and MV4-11 cells that had either been left untreated or had been treated with 500 nM ibrutinib for 1 hour were subjected to immunoblotting using antibodies against pSTAT5, pERK, pSTAT3, pFOXO1, and phospho-c (pc)-CBL.
Among the concordantly regulated phosphosites was the activation-inducing tyrosine Tyr-694 of the transcription factor STAT5, which is well known for its oncogenic potential in AML and thus constitutes a potentially relevant downstream effector of BTK. To investigate whether BTK directly phosphorylates STAT5, we performed an in vitro kinase assay by using biotinylated peptides that encompassed the activation-inducing tyrosine motif of STAT5. Affinity-purified BTK from untreated KG-1 cells was subjected to this kinase assay. STAT5-derived peptides were phosphorylated by BTK in vitro (Figure 5C). We conclude from these experiments that BTK activates STAT5 by phosphorylating Tyr-694 of STAT5, presumably through direct interaction. To exclude the possibility that a kinase potentially co-purified with BTK might contribute to phosphorylation of the STAT5 peptides, we subjected these peptides to the action of recombinant enzymatically active BTK. As shown in Figure 5C (lower panel), we again detected tyrosine phosphorylation of Tyr-694, thus confirming that BTK is able to phosphorylate STAT5 directly.
To examine how the remaining identified BTK effector proteins are integrated into signalosomes and signaling cascades, we performed a protein network analysis. We integrated information about known protein-protein interactions derived from the STRING database and information about protein functions from the KEGG, Uniprot, and PubMed databases for those proteins that we identified as differentially tyrosine phosphorylated upon BTK inhibition. This analysis revealed that the BTK effectors identified fall into a plethora of functional protein classes, including regulators of cell cycle and metabolism, which indicates a superordinate role of BTK in AML cell signaling. Within the main networks, we found STAT5, PI3-kinase, and the Map-kinase Erk to be potential mediators of BTK-induced cell survival and proliferation (Figure 5D-E). The phosphoproteomic results were confirmed by immunoblotting using phosphosite-specific antibodies directed against selected identified phosphoproteins such as pSTAT5, pSTAT3, pERK, and pc-CBL (Figure 5F). Our data show that BTK activates several oncogenic signaling pathways in AML cells, thereby inducing transcription regulation by effector proteins like STAT5.
BTK-dependent alteration of transcriptional networks depends on FLT3 mutational status
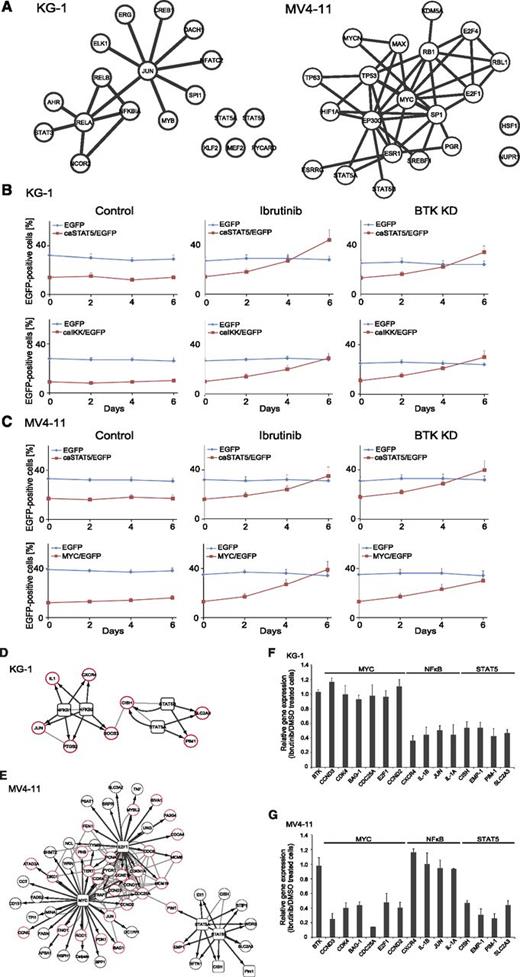
To learn more about the downstream effects of BTK activation in AML cells, we performed large-scale transcriptome sequencing in untreated and ibrutinib-treated KG-1 and MV4-11 cells (supplemental Figure 3A). In general, ibrutinib treatment induced stronger transcriptomic changes in (ITD-positive) MV4-11 cells compared with (ITD-negative) KG-1 cells (243 vs 98 significantly up- or downregulated messenger RNAs). We used Ingenuity software to determine the transcription factors that are responsible for the observed transcriptomic changes (see supplemental Data for a description of the bioinformatic work-flow) and to generate a putative network of transcription factors consistent with our observations on the basis of their known protein interaction profile (Figure 6A). Upon ibrutinib treatment, MYC and STAT5 targets were predominantly downregulated in MV4-11 cells, whereas nuclear factor kappa B (NF-κB) and STAT5 target genes were predominantly downregulated in KG-1 cells. This indicates once again that the mutational status of FLT3 influences not only the activation mode of BTK but also the molecular processes downstream of BTK (Figure 6A).
Functional dissection of BTK-dependent transcriptional programs. (A) Messenger RNAs (mRNAs) identified as significantly downregulated from the RNA sequencing analysis were submitted to Ingenuity pathway analysis for mapping of related upstream transcriptional regulators, which are shown in their respective networks. Known protein-protein interactions were included, as revealed by STRING database analysis. (B) KG-1or (C) MV4-11 cells were subjected to retroviral transduction with IRES-EGFP vectors inducing stable expression of EGFP, caSTAT5 and EGFP, caIKK and EGFP, or MYC and EGFP. The transduced cell batches were left untreated (left panel) or were treated with ibrutinib at a final concentration of 500 nM for up to 6 days (middle panels) or were treated with shRNAs specific for BTK (right panels). After confirmation of successful knockdown of BTK, cells were cultured for up to 6 days. The proportion of EGFP-positive cells was analyzed by flow cytometry. Wild-type cells were used as a negative control in all flow cytometric measurements (data not shown). (D-E) The functionally relevant and highly enriched transcription factors MYC, STAT5, E2F1, and NF-κB (square nodes) and their differentially regulated target genes (circular nodes) were used for STRING protein-protein interaction analysis (gray lines). The transcription factor to target gene relationships are indicated by black arrows. Target genes known to be involved in the regulation of the cell cycle or of apoptosis are shown with a red border. (F-G) Regulation of mRNA levels upon ibrutinib treatment were validated by quantitative polymerase chain reaction. Results are shown for 4 independent experiments (mean ± SD).
Functional dissection of BTK-dependent transcriptional programs. (A) Messenger RNAs (mRNAs) identified as significantly downregulated from the RNA sequencing analysis were submitted to Ingenuity pathway analysis for mapping of related upstream transcriptional regulators, which are shown in their respective networks. Known protein-protein interactions were included, as revealed by STRING database analysis. (B) KG-1or (C) MV4-11 cells were subjected to retroviral transduction with IRES-EGFP vectors inducing stable expression of EGFP, caSTAT5 and EGFP, caIKK and EGFP, or MYC and EGFP. The transduced cell batches were left untreated (left panel) or were treated with ibrutinib at a final concentration of 500 nM for up to 6 days (middle panels) or were treated with shRNAs specific for BTK (right panels). After confirmation of successful knockdown of BTK, cells were cultured for up to 6 days. The proportion of EGFP-positive cells was analyzed by flow cytometry. Wild-type cells were used as a negative control in all flow cytometric measurements (data not shown). (D-E) The functionally relevant and highly enriched transcription factors MYC, STAT5, E2F1, and NF-κB (square nodes) and their differentially regulated target genes (circular nodes) were used for STRING protein-protein interaction analysis (gray lines). The transcription factor to target gene relationships are indicated by black arrows. Target genes known to be involved in the regulation of the cell cycle or of apoptosis are shown with a red border. (F-G) Regulation of mRNA levels upon ibrutinib treatment were validated by quantitative polymerase chain reaction. Results are shown for 4 independent experiments (mean ± SD).
Because we identified several potentially relevant downstream effectors of BTK, we tested whether the oncogenic effect of BTK is mediated by BTK-dependent activation of the identified transcription factors MYC, NF-κB, and STAT5. For this purpose, we transduced KG-1 and MV4-11 cells retrovirally, using IRES-EGFP vectors to express EGFP either alone, as a control, or in combination with a constitutive active form of STAT5 (caSTAT5),6 a constitutively active variant of IKK (caIKK) as a well-known activator of NF-κB,17 or MYC (supplemental Figure 3B-C). Subsequently, the transduced cell batches were left untreated or were treated with ibrutinib for 6 days, and the proportion of EGFP-positive cells was monitored by flow cytometry. The initial transduction efficiency was about 30% for the empty vector controls, 14% in KG-1 (17% in MV4-11) for caSTAT5, 9% in KG-1 (14% in MV4-11) for caIKK, and 15% in KG-1 cells (12% in MV4-11 cells). It is noteworthy that the proportion of EGFP-positive cells did not change over 6 days of cell culture if the cells were left untreated (Figure 6B-C, left panel). However, after 6 days of continuous BTK inhibition or BTK knockdown, we observed a significant increase in the proportion of KG-1 cells that expressed either caSTAT5 or caIKK, but the percentage of KG-1 cells expressing MYC or EGFP alone did not change over time (Figure 6B, middle and right panels, and data not shown). In the case of MV4-11 cells, we observed an enrichment of those cells that overexpressed MYC or caSTAT5 in the presence of ibrutinib or upon BTK knockdown, but the proportion of cells expressing caIKK did not change over time (Figure 6C, middle and right panels, and data not shown). These results imply that NF-κB and STAT5 are predominantly relevant downstream of the TLR9/BTK transducer module, whereas the FLT3-ITD/BTK signaling axis involves the transcription factor MYC in addition to STAT5 to induce proliferation and cell survival. This is further supported by data derived from the FLT3-ITD–negative cell culture FFM04 and the FLT3-ITD–expressing cell line Molm13 using the same assay as described above (supplemental Figure 3D-E).
To better understand how suppression of these differential BTK-dependent transcriptional programs leads to a G1/S arrest and apoptosis in AML cells, we returned to our RNA sequencing analysis and investigated relationships of transcription factors to target genes by using Ingenuity software. The results are outlined as putative networks containing the transcription factors, their target genes, and their protein interaction profiles (Figure 6D-E). Some of those messenger RNAs (the expression of which had been identified as influenced by ibrutinib treatment) were validated by quantitative polymerase chain reaction, as shown in Figure 6F-G. It became obvious that most of the target genes identified as suppressed upon BTK inhibition are involved in the regulation of cellular proliferation and apoptosis (Figure 6D-E [marked in red] and F-G). Some genes, such as PIM1 were downregulated upon BTK inhibition in both FLT3-ITD–positive and –negative cells, whereas others were exclusively regulated in FLT3-ITD–positive cells, such as CDK4 and E2F1, an observation that was validated by immunoblotting (supplemental Figure 3F). Taken together, we observed that BTK activates distinct transcriptional programs depending on the FLT3 status: FLT3-ITD/BTK signaling converges at Myc and STAT5, whereas in FLT3-ITD-negative cells, NF-κB, and STAT5 are preferentially activated downstream of BTK.
Discussion
In this study, we have identified an unexpected functional role of BTK downstream of FLT3-ITD and TLR9. FLT3-ITD mutations occur in about 25% of AML patients and lead to hyperproliferation of myeloid progenitor cells, but differentiation responses are inhibited.18 Targeting of FLT3-ITD in AML by newer agents such as quizartinib shows significant effects in clinical trials.3 However, the responses are short-lived; this may be at least partially attributed to resistance mutations within the kinase domain of FLT3.19 Because we identified BTK as functionally relevant downstream of FLT3-ITD, BTK inhibition might be a valuable tool for the combined treatment of AML patients, because more effective inhibition of FLT3-ITD–dependent survival pathways might prevent the rapid occurrence of resistance mutations in FLT3.
The presence of FLT3-ITD in AML has proved to be an important factor for AML chemotherapy resistance, since FLT3-ITD–driven AML is much more prone to early relapse after intensive chemotherapy than is FLT3-ITD–negative disease. Inhibition of FLT3-ITD in a chemotherapeutic setting has led to ambivalent clinical results.20 In the absence of FLT3-ITD, TLR9 turned out to be a potent activator of oncogenic BTK signaling. We show here for the first time that blockade of TLR9 signaling strongly diminishes AML cell proliferation by suppressing NF-κB– and STAT5-mediated transcription. TLR9 has been described as an apoptosis sensor by recognizing damage-associated molecular patterns,21 and this process might contribute to the drug resistance of AML in the clinical setting, in which a high turnover of blasts in the bone marrow, especially under the influence of cytotoxic drugs, might stimulate the identified TLR9/BTK transducer module via damage-associated molecular pattern release. Thus, we here describe a novel microenvironment-dependent signaling axis that may represent an important mechanism of AML maintenance in various important clinical settings, including FLT3-ITD inhibition and cytotoxic therapy.
Puissant et al recently showed that the tyrosine kinase SYK is relevant for oncogenic FLT3-ITD–dependent Myc activation.22 Because we identified BTK as an additional signaling intermediate in the FLT3-ITD signalosome that activates Myc, it might well be that SYK and BTK operate within the same FLT3-ITD–dependent signaling pathway(s). Thus, their combined inhibition might be another option for improving the therapy for FLT3-ITD–positive AML.
Taken together, our results highlight the functional importance of interactions between AML cells and their microenvironment through TLRs and unravel a cooperative dependence between the mutant oncoprotein FLT3-ITD and BTK in AML pathogenesis. They also provide a rationale for the clinical evaluation of BTK inhibitors as single agents or in combination with other drugs, including FLT3 inhibitors in AML.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Monika Raabe, Anja Vogel, and Uwe Plessman for their technical support and Adelheid Cerwenka for providing reagents.
This work was supported by the Deutsche Forschungsgemeinschaft (OE 603/2-1) (T.O.) and the LOEWE Center for Cell and Gene Therapy Frankfurt (reference number: III L4-518/17.004) (T.O. and H.S.).
Authorship
Contribution: T.O. designed and supervised the study, contributed to experiments and data analysis, and wrote the manuscript; S. Mohr performed functional analysis and contributed to figure preparation; E.S. and J.B. performed transcriptome sequencing; H.U. and J.C. performed proteomic analyses; H. Braun, C.D., H. Bohnenberger, C.P., A.C., J.W., M.F.O. and S. Münch performed experiments; G.B. contributed reagents; and H.S. contributed to designing experiments involving FLT3-ITD and contributed to manuscript writing.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Thomas Oellerich, Department of Medicine II, Hematology/Oncology, Goethe University Frankfurt, Theodor-Stern-Kai 7, D-60590 Frankfurt, Germany; e-mail: thomas.oellerich@kgu.de.

![Figure 1. BTK is expressed in AML and promotes cell survival and proliferation. (A) Immunohistochemical analysis of BTK expression in 28 bone marrow specimens of AML patients. For comparison, bone marrow–derived samples from healthy individuals and from diffuse large B-cell lymphoma (DLBCL) patients were stained by antibodies against BTK. (B) Cleared cellular lysates of the lymphoma cell line DG75 and various AML cell lines (left panel) or primary patient-derived AML cultures (right panel) were subjected to immunoblotting with antibodies against phosphorylated BTK (pBTK; upper panels) and BTK (middle panels). Protein loading was monitored by anti-actin immunoblotting (lower panels). (C-D) Growth curves of the AML cell lines as well as the patient-derived AML cultures FFM04, -05, -12, -21, -40, and -41 that were treated with ibrutinib at final concentrations of 10 nM (red lines), 50 nM (green lines), or 250 nM (purple lines) or with dimethylsulfoxide (DMSO) as a control (blue lines.). Data from 4 independent experiments are shown (mean ± standard deviation [SD]). *P < .05 between the different treatment groups and the control cells using Student t test. (E) Relative cell numbers of primary AML cultures that were treated with 250 nM ibrutinib for 3 cycles. Each cycle consisted of a 1-hour ibrutinib treatment followed by a 23-hour washout period. Results (mean ± SD) are from 4 independent experiments.*P < .05 between the different treatment groups and the control cells using Student t test. (F) Immunoblots of cleared cellular lysates derived from KG-1 and MV4-11 control cells (lane 1) or the respective BTK knockdown cells (lane 2) with antibodies recognizing BTK (upper panels) or actin (lower panels). (G) KG-1 and MV4-11 cells were transduced either with lentiviral vectors encoding BTK-specific shRNAs and GFP or with unspecific control shRNAs and GFP. GFP expression was subsequently monitored in the respective cell batches by flow cytometry 1 day after lentiviral transduction (day 1) and 7 days thereafter (day 7). The outlined dot plots (representative for all 3 experiments) and histograms summarizing data from 3 independent experiments (mean ± SD) show the relative abundance of GFP-expressing cells on day 1 and day 7 for the respective BTK knockdown or control cell batches, as determined by flow cytometry. *P < .05 between the BTK knockdown and the control cells (nsp) using Student t test. (H) Flow cytometric cell cycle and (I) apoptosis analysis of KG-1 and MV4-11 BTK knockdown (BTK KD) cells or control cells (nsp) at day 2 after transduction. Results (mean ± SD) are from 4 independent experiments. *P < .05 between the BTK knockdown and the control cells using Student t test.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/12/10.1182_blood-2014-06-585216/4/m_1936f1.jpeg?Expires=1769086772&Signature=vEcM7~pAWe9cVpvCb9~3hWJJzSvikqeqy1fzouQ1Wb0mFTCzksDeiNPs5LFaO7hNjxmqTXRkg-Jm18jL4jLBFuAhb0drmQfsvMTkGSIX6wO4poIONFseDzeoBc7dknm1Dv5nydUZ49UcIKzCpWH7SqBpGv4o-jsN3P5hBYclxVMNO85ApuWKvsnHID8-jC4fvMkYIAwD4pypierxOneRadnuZjAWpL3Yhxl~u6ClbCDHqEVj0LTTqDN3aQY2JGHm0QDo1IdL8WaS7ZYNTswBKeLY8vdgJgYJx-9M1bC5lhND3O0kgtLn275zEl6K9sATX4RBuy3ablCnlNpnpyj2CQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. An oncogenic TLR9/BTK transducer module operates in FLT3-ITD–negative AML. (A) The endogenous BTK interactome was identified in KG-1 cells using quantitative SILAC-based MS. All identified proteins (except contaminants) are plotted according to their signal intensities and their H/L ratio of enrichment on logarithmic scales. Proteins with an H/L ratio > 5 (red) were identified as interaction partners of BTK. The complete list of identified proteins and quantification statistics is provided in supplemental Table 3. (B) Cleared cellular lysates of H2228 cells (negative control, lane 1) and various AML cell lines were subjected to immunoblotting with antibodies against TLR9 (upper panel) and actin (lower panel) as loading controls. (C) KG-1, FFM04, MV4-11, and Molm13 cells were lysed and subjected to immunoprecipitation using anti-TLR9 antibodies (lane 2) or isotype-matched control antibodies (lane 1 [C]). The proteins obtained were analyzed by immunoblotting with antibodies directed against BTK (upper panels). Effective immunoprecipitation of TLR9 was confirmed by immunoblotting using TLR9-specific antibodies (lower panels). (D) FFM04 and FFM12 cells were lysed and subjected to immunoprecipitation using anti-BTK antibodies (lane 2) or isotype-matched control antibodies (lane 1 [C]). The proteins thus isolated were analyzed by immunoblotting with antibodies directed against TLR9 (upper panels). Effective immunoprecipitation of BTK was confirmed by immunoblotting using BTK-specific antibodies (lower panels). (E) Left panel: Immunoblot analyses of lysates derived from KG-1 and FFM04 cells that were left untreated (lane 1) or stimulated by CpG dinucleotides for 15, 30, or 60 minutes (lanes 2 to 4). Right panel: Immunoblot analysis of lysates derived from FFM04 cells that were either left untreated (lane 1), cocultured with bone marrow (BM) stroma cells (lane 2), or cocultured with (Annexin V-positive) apoptotic stroma cells that had been irradiated prior to coculture (lane 3). Immunoblotting was performed by using phosphosite-specific antibodies against pTyr-223 of BTK (upper panels). Protein loading was monitored by immunoblotting of BTK (lower panels). (F) XTT-based proliferation analysis of KG-1 and FFM04 cells left untreated or treated with ibrutinib and 1.5 ng/μL CpG. Results (from 4 independent experiments; mean ± SD) are shown for cells that were treated for 3 days. *P < .05 between the different treatment groups compared with the DMSO control (black bar) using Student t test. (G) KG-1, FFM04, MV4-11, and Molm13 cells were transduced either with lentiviral vectors encoding TLR9-specific shRNAs and GFP or with unspecific control shRNAs (nsp) and GFP. Subsequently, expression of TLR9 and actin was monitored by immunoblotting. (H) GFP expression was monitored in the respective transduced cell batches by flow cytometry 1 day after lentiviral transduction (day 1) and 7 days thereafter (day 7). The outlined diagram summarizes data from 4 independent experiments (mean ± SD) and shows the relative abundance of GFP-expressing cells at day 1 or day 7 for the respective TLR9 knockdown or control cell batches.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/125/12/10.1182_blood-2014-06-585216/4/m_1936f4.jpeg?Expires=1769086772&Signature=3oLrUbFWrx32rZtIKvgddXC4FKGA3CFLmxmnOagaNTIDzhsPpLC2ObaQDWGuN3pLKmmF-1g-wM5XZmMJxOcl-C7qonXxzO7cqFRbt4kRvUnfopTMaapD~Vv-OuyWfgVfF24B1Xz7jeLLT59IBKOztrYAb7oml1~Lj8JF67ZJYcL7iel6QVumBpBVVGh9gMA2mQ8IFt8RuKvzipa3klqK7p5EWMGVZJGLWnXor6SHxyIUM0XP7aR8qfOf4EcCne26IZBzWAozOSAS~J5h5Q-zdwgw9-en4w3I7ulxHUpIdBlDiXe4VqCewelCt6~8LEDVW7iqDiUSG~pKpx4iOsKxfw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)