Key Points
Caspase-9 is required for normal development of myeloid, lymphoid, and erythroid cells in mice.
Loss of caspase-9 results in increased DNA damage and mutation burden after exposure to alkylating agents.
Abstract
Apoptosis and the DNA damage responses have been implicated in hematopoietic development and differentiation, as well as in the pathogenesis of myelodysplastic syndromes (MDS) and leukemia. However, the importance of late-stage mediators of apoptosis in hematopoiesis and leukemogenesis has not been elucidated. Here, we examine the role of caspase-9 (Casp9), the initiator caspase of the intrinsic apoptotic cascade, in murine fetal and adult hematopoiesis. Casp9 deficiency resulted in decreased erythroid and B-cell progenitor abundance and impaired function of hematopoietic stem cells after transplantation. Mouse bone marrow chimeras lacking Casp9 or its cofactor Apaf1 developed low white blood cell counts, decreased B-cell numbers, anemia, and reduced survival. Defects in apoptosis have also been previously implicated in susceptibility to therapy-related leukemia, a disease caused by exposure to DNA-damaging chemotherapy. We found that the burden of DNA damage was increased in Casp9-deficient cells after exposure to the alkylator, N-ethyl-nitrosourea (ENU). Furthermore, exome sequencing revealed that oligoclonal hematopoiesis emerged in Casp9-deficient bone marrow chimeras after alkylator exposure. Taken together, these findings suggest that defects in apoptosis could be a key step in the pathogenesis of alkylator-associated secondary malignancies.
Introduction
The intrinsic apoptotic cascade is a process of programmed cell death triggered by stimuli within the cell, most often DNA damage. When irreparable DNA damage is sensed by components of the DNA damage response (DDR) pathway, p53 is activated and the cell is triggered to undergo mitochondrial outer membrane permeabilization (MOMP), which initiates downstream activation of the caspase cascade and ultimately leads to the orderly destruction of the cell. Because mammalian cells are regularly exposed to DNA damage during replication, an appropriate DDR and induction of apoptosis is crucial to normal homeostasis. Within the hematopoietic compartment, accumulation of DNA damage is particularly deleterious. Age-related changes in hematopoiesis, including decreased stem-cell function and a myeloid bias, are thought to be largely caused by the aberrant accumulation of DNA damage over time.1-3 Mutations in genes necessary for nucleotide excision repair (XPD), telomere maintenance (mTR and Terc), nonhomologous end joining (Ku80, Lig4), mismatch repair (Msh2), and homologous recombination repair (Rad50, Brca2) all cause increases in detectable DNA damage and are associated with impaired hematopoietic stem cell (HSC) function and premature exhaustion of stem cells.4-8 Loss of Tp53, a critical mediator of the cell cycle and apoptosis upstream of the mitochondria, promotes cell-cycle entry and self-renewal of HSCs in mice.9,10 These more proliferative Tp53-deficient HSCs have an initial advantage in competitive transplantation but are less effective than wild-type (WT) HSCs in serial transplantation.9
Exposure to genotoxins places an additional stress on the DDR and the intrinsic apoptotic pathways. Most chemotherapeutic agents act by inducing DNA damage, leading to both beneficial (tumor-cell killing) and deleterious consequences (myelosuppression and secondary malignancy). Therapy-related acute myeloid leukemia (tAML) is a lethal, late complication of exposure to DNA-damaging drugs used for the treatment of cancer or autoimmune disease.11 Current evidence suggests that changes in DDR and apoptosis may contribute to the pathogenesis of this disease.12 Somatic mutations of TP53 are more common in tAML than in de novo AML, and tAML samples typically have lower expression of TP53.13,14 Hypermethylation of the BRCA1 promoter is also more common in tAML than in de novo AML.15 There is also evidence of increased DNA damage in tAML cells, including microsatellite instability, a sign of DNA damage that is rare in de novo AML.15 tAML is also associated with accelerated telomere shortening, which is linked to genomic instability.16,17 In addition, peripheral blood CD34+ cells from patients before the development of tAML have elevated production of mitochondrial reactive oxygen species (ROS), sustained ROS elevation, and increased γH2AX foci after treatment with nitrogen mustard, suggesting that HSCs from patients at risk for tAML can have preexisting mitochondrial defects and abnormal accrual of DNA damage.18
The direct mediators of apoptosis are neither recurrently mutated nor significantly dysregulated in patients with AML.19 Nevertheless, analysis of genetically-engineered mouse models has defined the role of some of these factors in hematopoietic development and may shed light on human disease in the future. Transgenic overexpression of BCL2 results in a proliferative phenotype in mice, with an enlarged HSC compartment.20 HSCs overexpressing BCL2 have a competitive advantage in transplantation and in colony-replating assays, though they cycle less frequently than WT cells. Effectors of apoptosis downstream of the mitochondria have not been well studied in hematopoiesis or leukemogenesis, but given the importance of upstream mediators, it is likely that such effectors would play important roles in hematopoiesis, and defects in these effectors might increase the risk of tAML. Therefore, we chose to examine the role of Casp9 (encoding the critical downstream apoptosis effector, caspase-9 [Casp9]) in hematopoiesis. To validate our findings, we also studied the effects of the loss of Apaf1, which is essential for activating Casp9. We found that loss of these key effectors caused a stem-cell defect in fetal hematopoiesis, which led to dysplastic hematopoiesis and reduced survival in adult bone marrow chimeras. Loss of Casp9 also altered the response to alkylator exposure, resulting in increased DNA damage acutely, and outgrowth of clonal hematopoiesis, an early hallmark of myeloid malignancies.21,22
Methods
Harvest and culture of fetal liver cells
Casp9+/− mice23 (backcrossed >10 generations to C57BL/6J) or Apaf1+/− mice24 (backcrossed 6 generations to C57BL/6J) were intercrossed, and fetuses were obtained at embryonic day 15.5. Tail DNA was prepared (DNeasy, Qiagen) and used for genotyping (see supplemental Methods, available on the Blood Web site). Fetal livers were homogenized to yield a single-cell suspension and viably cryopreserved. All experiments performed were approved by the Animal Studies Committee at Washington University.
Flow cytometric analysis
Fetal liver cells (FLCs) were thawed and cultured overnight in complete media (RPMI containing 1% l-glutamine, 20% fetal bovine serum) supplemented with recombinant hematopoietic cytokines (100 ng/mL stem-cell factor, 6 ng/mL IL-3, 50 ng/mL Fms-related tyrosine kinase 3 ligand, 10 ng/mL thrombopoietin, and 10 ng/mL IL-6; all from Peprotech, Rocky Hill, NJ). Kit+, lineage–, Sca+ (KLS) cells were identified by expression of c-Kit and Sca-1 and lack of expression of CD41 and lineage markers (B220, CD3, Gr1, and Ter119). Long-term stem cells (LT-HSCs) were defined as KLS cells that expressed CD150 and lacked expression of CD48. All antibodies were obtained from eBioscience (San Diego, CA).
Hematopoietic progenitor assays
FLCs were cultured in methylcellulose media (MethoCult m3434 or m3630, Stemcell Technologies, Vancouver, BC, Canada) at 37°C for 7 days. Progenitors were counted and, in the case of myeloid colonies, subclassified (colony-forming unit [CFU]-G, CFU-GM, or CFU-M). For erythroid progenitors (burst-forming unit [BFU]-E or CFU-E), plates were stained to identify hemoglobin (0.43% benzidine in 13% acetic acid and 3% H2O2) and then counted.
Induction and analysis of apoptosis and DNA damage
N-ethyl-nitrosourea (ENU; Sigma/Merck) was resuspended and serially diluted in dimethyl sulfoxide (DMSO). For in vitro assays, FLCs were treated with ENU 24 hours after thawing. Cells were incubated with ENU (or DMSO at an identical concentration) for 1 hour at 37°C. The cells were then washed in phosphate-buffered saline and incubated overnight in fresh cytokine-enriched media. Twenty-four hours after ENU treatment, cells were stained with a 1:40 dilution of Annexin V (BD Pharmingen, San Jose, CA) in Annexin V binding buffer (140 mM NaCl, 4 mM KCl, 0.75 mM MgCl2, and 10 mM HEPES) and analyzed by flow cytometry. Cells were collected at 24-hour intervals and resuspended in phosphate-buffered saline at a concentration of 5 × 105 per mL, then embedded in agarose and subjected to the alkaline comet assay using the CometAssay Kit (Trevigen, Gaithersburg, MD). Comets were photographed under 10× magnification and analyzed using CometScore software (TriTek Corp, Sumerduck, VA). For in vivo DNA damage assays, bone marrow chimeras at 12 weeks posttransplant were injected IP with 200 mg/kg ENU or DMSO and diluted 1:10 in phosphate-citrate buffer, as previously described.25 Mice were sacrificed at 72 hours posttreatment and bone marrow was harvested by centrifugation. Comet assays were performed, as described previously. For exome sequencing experiments, bone marrow chimeras at 6 and 7 weeks posttransplant were injected IP with 100 mg/kg ENU or DMSO diluted 1:10 in phosphate-citrate buffer, as previously described,25 and sacrificed at 12 weeks posttransplant.
Generation of bone marrow chimeras
Recipient mice (Ly5.1+ or Ly5.1+Ly5.2+ in a C57BL/6J background, 9-12 weeks of age) were irradiated (950 cGy) and transplanted 24 hours later (via retro-orbital injection) with 2 × 106 FLCs or bone marrow cells. Mice received trimethoprim-sulfamethoxazole (40-200 mg/5 mL; HiTech Pharmacal, Amityville, NY) in drinking water for 2 weeks after transplant, and peripheral blood was sampled at 1 month posttransplant for analysis of donor chimerism by flow cytometry. For single-genotype transplants, FLCs were thawed and cultured for 24 hours before transplantation. For competitive transplants, a 1:1 mixture of Casp9+/+ or Casp9−/− (Ly-5.2+) and WT competitor (congenic Ly-5.1+) FLCs was made immediately after thawing, with no interim culture, and the exact ratio determined by flow cytometry before transplantation. For sequential transplants, primary transplant recipients were sacrificed at 12 weeks posttransplant and whole bone marrow was harvested by centrifugation before transplantation. 2 × 106 cells were transplanted into lethally-irradiated (950 cGy) Ly5.1+Ly5.2+ secondary recipients.
Analysis of bone marrow chimeras
Peripheral blood was collected from recipients at monthly intervals after transplant. Peripheral blood counts were determined using a veterinary Coulter counter (HemaVet 950, Drew Scientific Group, Dallas, TX). Chimerism and lineage composition were determined by staining with antibodies against congenic markers (Ly5.1 and Ly5.2) and lineage markers (B220, CD3, and Gr1) and analyzed by flow cytometry. Mice were sacrificed when moribund or at 12 or 18 months posttransplant. At sacrifice, spleen weight was measured, blood counts were determined, and bone marrow and spleen cells were analyzed by flow cytometry to determine chimerism and lineage composition. Slides of bone marrow and peripheral blood were stained with Wright-Giemsa and examined.
Exome sequencing
Bone marrow cells were harvested from transplanted mice at 12 weeks after ENU or DMSO treatment. Donor-derived cells (Ly-5.2+Ly-5.1–) were sorted (MoFlo, DAKO Cytomation) into cell-culture media, and genomic DNA was prepared (DNeasy kit, Qiagen) and quantified by fluorimetry (Qubit, Life Technologies, Carlsbad, CA). Multiplexed paired-end Illumina libraries were prepared from bone marrow and normal nonhematopoietic fetal tissue matched to each donor according to the manufacturer’s recommendations (Illumina Inc., San Diego, CA). Three libraries were generated from each bone marrow sample (“tumor”) and a single library was generated from each donor (“normal”). Sequencing libraries were hybridized in solution to capture oligonucleotide probes targeting the C57BL/6J mouse exome (54.3 Mb of target sequence), according to the manufacturer’s protocol (SeqCap EZ, Roche NimbleGen, Madison, WI). Quantitative polymerase chain reaction was used to calibrate flow cell–loading concentration and cluster density. Libraries were pooled and run on a single lane of an Illumina HiSeq2000 according to the manufacturer’s recommendations (Illumina, Inc). Reads were demultiplexed and aligned to the NCBI 37/mm9 reference sequence using BWA v0.5.9.26 Binary alignment/map (BAM) files were merged and duplicates marked using Picard v1.46 (http://picard.sourceforge.net). Read pileups were generated with the SAMtools v0.1.18 mpileup command using default settings. Read counts were extracted from pileup files and analyzed with VarScan v2.3.527 using default parameters. Single-nucleotide variants (SNVs) were retained if the variant allele fraction (VAF) was ≥5% in the tumor and 0% in the matched normal sample, and at least 30 reads were obtained at the site in both tumor and normal.
Results
Loss of Casp9 alters hematopoietic stem/progenitor compartment
Casp9−/− mice die perinatally because of forebrain overgrowth.23 Therefore, to determine how defects in the postmitochondrial apoptotic pathway—specifically loss of the initiator caspase, Casp9—would affect the abundance of hematopoietic stem progenitor cells, we examined hematopoiesis using FLCs generated by intercrossing Casp+/− mice. B cell and erythroid colonies were reduced in Casp9−/− FLCs, but myeloid colony frequencies were similar (Figure 1A). To assess the stem-cell population in fetal liver, FLCs were examined by flow cytometry. Casp9−/− FLCs had a roughly threefold increase in the frequency of c-Kit+, lineage–, Sca-1+ (KLS) cells. This heterogeneous cell compartment was further fractionated using “SLAM” markers (CD48, CD150) to identify long-term HSCs. The frequency of SLAM cells within the KLS compartment was markedly reduced in Casp9−/− mice, resulting in no change overall in the frequency of long-term HSCs (Figure 1B).
Loss of Casp9 alters hematopoietic progenitor cell frequency. (A) The frequency of B-cell and erythroid progenitors measured in methylcellulose assays was decreased in Casp9−/− FLCs compared with Casp9+/+ FLCs. The frequency and type of myeloid progenitors was not significantly different. (B) The frequency of Kit+ lineage– Sca+ (KLS) cells was increased ∼threefold in Casp9−/− FLCs compared with WT or heterozygous littermates. However, the frequency of long-term stem cells (Kit+lineage–Sca+CD48–CD150+; “SLAM”) within the KLS compartment was significantly lower in Casp9−/− FLCs, resulting in no net alteration in long-term HSC (LT-HSC) abundance. (C) Similar to these in vitro findings, bone marrow harvested from lethally-irradiated WT congenic mice 12 weeks after transplantation with Casp9−/− FLCs had an increase in KLS and a decrease in SLAM cells within the KLS compartment compared with recipients of Casp9+/+ FLCs, resulting in a modest decrease in the frequency of LT-HSCs overall. *P < .05, **P < .005, ***P < .0005; ns, not significant.
Loss of Casp9 alters hematopoietic progenitor cell frequency. (A) The frequency of B-cell and erythroid progenitors measured in methylcellulose assays was decreased in Casp9−/− FLCs compared with Casp9+/+ FLCs. The frequency and type of myeloid progenitors was not significantly different. (B) The frequency of Kit+ lineage– Sca+ (KLS) cells was increased ∼threefold in Casp9−/− FLCs compared with WT or heterozygous littermates. However, the frequency of long-term stem cells (Kit+lineage–Sca+CD48–CD150+; “SLAM”) within the KLS compartment was significantly lower in Casp9−/− FLCs, resulting in no net alteration in long-term HSC (LT-HSC) abundance. (C) Similar to these in vitro findings, bone marrow harvested from lethally-irradiated WT congenic mice 12 weeks after transplantation with Casp9−/− FLCs had an increase in KLS and a decrease in SLAM cells within the KLS compartment compared with recipients of Casp9+/+ FLCs, resulting in a modest decrease in the frequency of LT-HSCs overall. *P < .05, **P < .005, ***P < .0005; ns, not significant.
We next assessed HSC frequency in adult bone marrow chimeras. Equal numbers of Casp9−/− or Casp9+/+ FLCs were transplanted into lethally-irradiated recipients, which were then characterized by flow cytometry at 3 months posttransplant. As with FLCs, the frequency of KLS cells was increased approximately threefold in recipients of Casp9−/− FLCs, but there was no overall change in the abundance of LT-HSCs (KLS-SLAM cells; Figure 1C).
Casp9-deficient stem cells are functionally impaired
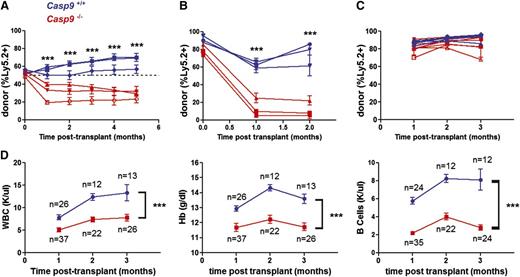
Next, we assessed the function of Casp9-deficient HSPCs using a competitive repopulation assay. Unfractionated Casp9+/+ or Casp9−/− FLCs were transplanted in a 1:1 ratio with competitor WT Ly5.1+ FLCs. The contribution of Casp9−/− cells was reduced in transplant recipients (Figure 2A), suggesting that loss of Casp9 confers a competitive disadvantage in the setting of primary transplantation. Stem-cell function was further assayed by assessing the ability of Casp9−/− cells to sequentially repopulate the hematopoietic compartment of serial recipients. Casp9−/− or Casp9+/+ FLCs were first transplanted into lethally irradiated primary recipients. Twelve weeks after this initial transplant, bone marrow was harvested and transplanted into secondary recipients. Casp9+/+ cells were able to reconstitute the hematopoietic compartment of these secondary recipients, but Casp9−/− cells failed in secondary transplants (Figure 2B). Although Casp9−/− stem cells were capable of generating a functional hematopoietic compartment in primary transplants (Figure 2C), they exhaust upon serial transplantation, providing further evidence that the immunophenotypic changes observed previously correlate with a functional defect in Casp9−/− stem cells.
Casp9−/− HSCs are functionally impaired. (A) FLCs from Casp9−/− or Casp9+/+ pups (Ly-5.2+) were transplanted 1:1 in competition with WT congenic (Ly-5.1+) FLCs into lethally-irradiated WT congenic (Ly-5.1+Ly-5.2+) recipients. Peripheral blood was analyzed for donor chimerism at monthly intervals. Casp9−/− cells demonstrated a competitive disadvantage at 1 to 5 months, suggesting that their short-term repopulating cells are functionally impaired. Each curve represents the mean ± SEM of 3 to 4 recipients from separate FLC donors (n = 3 donors per genotype, 21 recipients total). The dotted line indicates no difference in competitive advantage. (B) In secondary transplants, bone marrow cells obtained from primary recipients of Casp9−/− donor cells failed to reconstitute the hematopoietic compartment of lethally-irradiated secondary recipients, although secondary transplants from donors engrafted with Casp9+/+ cells were successful. Each curve represents the mean ± SEM of 4 recipients from separate donors (n = 3 donors of each genotype, 24 recipients total). (C) Noncompetitive transplants with Casp9−/− or Casp9+/+ FLCs had similar engraftment in lethally irradiated WT congenic (Ly-5.1+) recipients (mean % donor cells at 1 month of 87.0% and 81.2% for Casp9+/+ and Casp9−/−, respectively; P = .086). Each curve represents the mean ± SEM of 1 to 5 recipients from Casp9−/− (n = 7) or Casp9+/+ (n = 5) donors. (D) Recipients of Casp9−/− FLCs developed leukopenia, anemia, and B-cell lymphopenia compared with recipients of Casp9+/+ FLCs. ***P < .0005.
Casp9−/− HSCs are functionally impaired. (A) FLCs from Casp9−/− or Casp9+/+ pups (Ly-5.2+) were transplanted 1:1 in competition with WT congenic (Ly-5.1+) FLCs into lethally-irradiated WT congenic (Ly-5.1+Ly-5.2+) recipients. Peripheral blood was analyzed for donor chimerism at monthly intervals. Casp9−/− cells demonstrated a competitive disadvantage at 1 to 5 months, suggesting that their short-term repopulating cells are functionally impaired. Each curve represents the mean ± SEM of 3 to 4 recipients from separate FLC donors (n = 3 donors per genotype, 21 recipients total). The dotted line indicates no difference in competitive advantage. (B) In secondary transplants, bone marrow cells obtained from primary recipients of Casp9−/− donor cells failed to reconstitute the hematopoietic compartment of lethally-irradiated secondary recipients, although secondary transplants from donors engrafted with Casp9+/+ cells were successful. Each curve represents the mean ± SEM of 4 recipients from separate donors (n = 3 donors of each genotype, 24 recipients total). (C) Noncompetitive transplants with Casp9−/− or Casp9+/+ FLCs had similar engraftment in lethally irradiated WT congenic (Ly-5.1+) recipients (mean % donor cells at 1 month of 87.0% and 81.2% for Casp9+/+ and Casp9−/−, respectively; P = .086). Each curve represents the mean ± SEM of 1 to 5 recipients from Casp9−/− (n = 7) or Casp9+/+ (n = 5) donors. (D) Recipients of Casp9−/− FLCs developed leukopenia, anemia, and B-cell lymphopenia compared with recipients of Casp9+/+ FLCs. ***P < .0005.
We next assessed the hematopoietic output of Casp9−/− FLCs in primary recipients. Casp9+/+ or Casp9−/− FLCs were transplanted into lethally-irradiated recipients, and peripheral blood from these recipients was examined at monthly intervals for blood counts, chimerism, and lineage markers. Casp9−/− transplant recipients had lower white blood cell counts, lower hemoglobin levels, and a reduced frequency of B cells in peripheral blood compared with recipients of Casp9+/+ cells (Figure 2D). These data are consistent with the progenitor cell analysis in fetal liver, which showed decreased B-cell and erythroid progenitor numbers. These data suggest that loss of Casp9 biases hematopoiesis away from the B-cell and erythroid lineages while preserving normal myeloid output.
To ensure that this phenotype was the result of the loss of apoptosis associated with loss of Casp9 and not a defect unique to this particular strain, we performed similar experiments using Apaf1-deficient FLCs (because Apaf1 deficiency also causes perinatal lethality24 ). Apaf1 is the key component of the apoptosome downstream of the mitochondria and is essential for activation of Casp9 in response to apoptotic stimuli, and loss of Apaf1 causes decreased apoptosis, similar to loss of Casp9. Mice receiving Apaf1−/− FLC transplants had similar defects in hematopoiesis as observed in the Casp9−/− model: leukopenia, anemia, B lymphopenia, and preserved myeloid cell numbers (supplemental Figure 1). In summary, the impact of Apaf1 loss on hematopoiesis largely phenocopies the impact of Casp9 loss, supporting the conclusion that defects in apoptosis are responsible for the observed changes in hematopoiesis.
Loss of Casp9 or Apaf1 leads to dysplastic hematopoiesis and reduced survival
To assess the long-term consequences of Casp9 or Apaf1 deficiency on hematopoiesis, transplant recipients were followed longitudinally. Mice were sacrificed when moribund or at 12 to 18 months posttransplant. Mice receiving Casp9−/− or Apaf−/− transplants had significantly decreased survival relative to recipients of WT cells (Figure 3A-B). Evaluable mice from both cohorts demonstrated a similar phenotype, including variable leukopenia and mild anemia (supplemental Table 1). Bone marrow histology revealed a decrease in erythropoiesis and an increase in myeloid-committed cells. Within the myeloid lineage, there was also a shift to less differentiated forms. Interestingly, there was also evidence of dysplasia in the erythroid and myeloid lineages (Figure 3C). These changes were quantified and shown to be unique to Casp9−/− and Apaf1−/− transplant recipients (Figure 3D-E). One mouse transplanted with Casp9−/− cells died of recipient-derived T-cell lymphoma, likely a consequence of pretransplant radiation conditioning. In summary, loss of Casp9 or Apaf1 causes impaired hematopoiesis with dysplastic features, leading to increased mortality.
Loss of Casp9 or Apaf1 leads to dysplastic hematopoiesis and reduced survival after transplantation. (A) FLCs from Casp9+/+ or Casp9−/− pups were transplanted into lethally-irradiated WT C57BL/6J recipients. Recipients of Casp9−/− FLCs had significantly decreased survival compared with mice transplanted with Casp9+/+ FLCs. (B) Similarly, recipients of Apaf1−/− FLCs had significantly reduced survival compared with mice transplanted with Apaf1+/+ FLCs. (C) Examination of bone marrow in moribund Casp9−/− or Apaf1−/− transplant recipients revealed signs of erythroid dysplasia (arrows = nuclear blebbing), and features of abnormal myeloid differentiation, including hyperlobulated neutrophils and a shift to less differentiated forms (D). (E) The proportion of cells showing dysplastic changes in the erythroid and myeloid lineages was increased in recipients of Casp9−/− or Apaf1−/− FLCs. *P < .05, **P < .005, ***P < .0005.
Loss of Casp9 or Apaf1 leads to dysplastic hematopoiesis and reduced survival after transplantation. (A) FLCs from Casp9+/+ or Casp9−/− pups were transplanted into lethally-irradiated WT C57BL/6J recipients. Recipients of Casp9−/− FLCs had significantly decreased survival compared with mice transplanted with Casp9+/+ FLCs. (B) Similarly, recipients of Apaf1−/− FLCs had significantly reduced survival compared with mice transplanted with Apaf1+/+ FLCs. (C) Examination of bone marrow in moribund Casp9−/− or Apaf1−/− transplant recipients revealed signs of erythroid dysplasia (arrows = nuclear blebbing), and features of abnormal myeloid differentiation, including hyperlobulated neutrophils and a shift to less differentiated forms (D). (E) The proportion of cells showing dysplastic changes in the erythroid and myeloid lineages was increased in recipients of Casp9−/− or Apaf1−/− FLCs. *P < .05, **P < .005, ***P < .0005.
Loss of apoptosis results in increased DNA damage after alkylator treatment
Defects in DDR and apoptosis have also been implicated in predisposition to tAML in humans, a late complication of exposure to DNA-damaging chemotherapy agents. We hypothesize that loss of apoptosis could allow cells to inappropriately survive exposure to DNA-damaging agents, thereby giving rise to a population of cells carrying significant DNA damage.
To determine whether Casp9 deficiency allows hematopoietic cells to escape death while carrying DNA damage, we used ENU, a prototypical alkylator similar to drugs used clinically, to induce DNA damage. In vitro, we treated FLCs with varying doses of ENU and measured apoptosis by Annexin V staining at 24 hours posttreatment. DNA damage was assessed at various time points after treatment using the comet assay system, which detects single- and double-strand breaks, as well as abasic sites. FLCs deficient in Casp9 or Apaf1 had significantly lower levels of apoptosis in response to ENU, as previously reported (Figure 4A). Apoptosis peaked between 12 and 24 hours posttreatment (supplemental Figure 2A). The level of apoptosis was inversely correlated with the level of DNA damage, which was dose dependent, apparent at 48 hours posttreatment, and persistent beyond 120 hours posttreatment (Figure 4B-C).
Loss of Casp9 or Apaf1 results in decreased apoptosis and increased DNA damage after ENU treatment. (A) FLCs from Casp9−/− or Apaf1−/− pups (or WT littermates) were treated with ENU or an equivalent concentration of vehicle only (0.1% DMSO) for 1 hour and then allowed to recover for 24 hours. Cells were then stained with Annexin V to measure apoptosis. Casp9−/− and Apaf1−/− FLCs had reduced apoptosis in response to ENU. Each bar represents the mean ± SEM for 3 to 9 mice. (B) To determine whether the reduction in apoptosis detected in Casp9−/− and Apaf1−/− FLCs was associated with an increase in DNA damage in the surviving cells, the alkaline comet assay was used to measure DNA damage in cells exposed to varying doses of ENU with varying lengths of recovery time after exposure. Casp9−/− cells showed increased DNA damage at all ENU doses tested at 72 hours post-ENU treatment. DNA damage decreased over time in ENU-treated Casp9+/+ FLCs as apoptotic cells were cleared, but was sustained at high levels for up to 120 hours after ENU exposure (300 μg/mL) in Casp9−/− FLCs. Each bar represents the mean ± SEM of at least 140 nuclei from 2 to 3 (Casp9+/−) or 3 to 5 (Casp9−/− and Casp9+/+) FLC samples. (C) Comets from Casp9−/− and Apaf1−/− nuclei (300 μg/mL ENU, 72 hours after treatment) had significantly higher levels of DNA damage (% of DNA in tail) compared with other genotypes. Bars show the mean of at least 200 nuclei from at least 3 mice, with vertical lines extending to the first and third quartiles. (D) Lethally-irradiated WT congenic mice were transplanted with Casp9+/+, Casp9−/−, Apaf1+/+, or Apaf1−/− FLCs and treated with ENU (200 mg/kg IP) or vehicle only (10% DMSO) at 12 weeks posttransplant. Seventy-two hours after injection, mice were sacrificed and bone marrow nuclei were analyzed for DNA damage by comet assay. Consistent with in vitro results, Casp9−/− and Apaf1−/− bone marrow nuclei showed increased levels of DNA damage compared with WT nuclei after ENU treatment (n = 3-4 mice, at least 270 nuclei from each). *P < .05, ***P < .0005.
Loss of Casp9 or Apaf1 results in decreased apoptosis and increased DNA damage after ENU treatment. (A) FLCs from Casp9−/− or Apaf1−/− pups (or WT littermates) were treated with ENU or an equivalent concentration of vehicle only (0.1% DMSO) for 1 hour and then allowed to recover for 24 hours. Cells were then stained with Annexin V to measure apoptosis. Casp9−/− and Apaf1−/− FLCs had reduced apoptosis in response to ENU. Each bar represents the mean ± SEM for 3 to 9 mice. (B) To determine whether the reduction in apoptosis detected in Casp9−/− and Apaf1−/− FLCs was associated with an increase in DNA damage in the surviving cells, the alkaline comet assay was used to measure DNA damage in cells exposed to varying doses of ENU with varying lengths of recovery time after exposure. Casp9−/− cells showed increased DNA damage at all ENU doses tested at 72 hours post-ENU treatment. DNA damage decreased over time in ENU-treated Casp9+/+ FLCs as apoptotic cells were cleared, but was sustained at high levels for up to 120 hours after ENU exposure (300 μg/mL) in Casp9−/− FLCs. Each bar represents the mean ± SEM of at least 140 nuclei from 2 to 3 (Casp9+/−) or 3 to 5 (Casp9−/− and Casp9+/+) FLC samples. (C) Comets from Casp9−/− and Apaf1−/− nuclei (300 μg/mL ENU, 72 hours after treatment) had significantly higher levels of DNA damage (% of DNA in tail) compared with other genotypes. Bars show the mean of at least 200 nuclei from at least 3 mice, with vertical lines extending to the first and third quartiles. (D) Lethally-irradiated WT congenic mice were transplanted with Casp9+/+, Casp9−/−, Apaf1+/+, or Apaf1−/− FLCs and treated with ENU (200 mg/kg IP) or vehicle only (10% DMSO) at 12 weeks posttransplant. Seventy-two hours after injection, mice were sacrificed and bone marrow nuclei were analyzed for DNA damage by comet assay. Consistent with in vitro results, Casp9−/− and Apaf1−/− bone marrow nuclei showed increased levels of DNA damage compared with WT nuclei after ENU treatment (n = 3-4 mice, at least 270 nuclei from each). *P < .05, ***P < .0005.
To determine whether Casp9 or Apaf1 deficiency resulted in excessive DNA damage that could be detected in vivo, we generated chimeras using FLCs from both mutant strains, injected transplant recipients with 200 mg/kg ENU or equivalent volume of DMSO, and sacrificed mice at 72 hours posttreatment. Bone marrow cells from these mice were then analyzed by comet assay. Similar to the in vitro results, Casp9- or Apaf1-deficient bone marrow cells showed increased levels of DNA damage after ENU treatment (Figure 4D). This DNA damage is not simply a marker of late-stage apoptosis: the proportion of cells carrying a subdiploid complement of DNA (indicative of DNA fragmentation in late-stage apoptosis) was very low, regardless of genotype (supplemental Figure 2B). Thus, loss of apoptosis in combination with a genotoxic insult results in the accumulation of cells with high levels of DNA damage in vivo.
Loss of Casp9 leads to clonal hematopoiesis after alkylator treatment
To determine whether hematopoietic cells with genotoxin-induced DNA damage persist in vivo, an exome-sequencing approach was used. Bone marrow chimeras were generated by FLC transplantation, treated with ENU (100 mg/kg) or an equivalent amount of DMSO at 6 and 7 weeks posttransplant, and sacrificed at 12 weeks posttransplant. Six biological replicates for each genotype were included, half treated with ENU and half with DMSO. Bone marrow was harvested and sorted for donor cells. DNA was prepared from the sorted cells (“tumor”) and also from nonhematopoietic tissue taken from the original donor pup (“normal”). Libraries (from the 12 bone marrow samples and 6 paired-donor samples) were prepared and subjected to whole-exome sequencing (supplemental Figure 3). At least 40× coverage was obtained for >80% of the exome target for the tumor samples, on average, and at least 10× coverage for the normal samples (supplemental Figure 4). SNVs present in the tumor samples and absent in the matched normal samples were identified (supplemental Table 2).
Mutations corrected by DNA repair, cleared by apoptosis, or persisting in a normal background of polyclonal hematopoiesis would not be detectable in this analysis. In Casp9+/+ FLC recipients treated with DMSO, a small number of SNVs (1-11 per exome) were detectable, and this number was not significantly increased in Casp9+/+ FLC recipients treated with ENU (2-8 per exome) (Figure 5). In contrast, the frequency of SNVs was significantly higher in Casp9−/− FLC recipients treated with ENU vs DMSO (349-586 per exome vs 12-325 per exome; P < .005), with higher VAFs, indicative of oligoclonal rather than polyclonal hematopoiesis, an early hallmark of myeloid neoplasia. There were no recurrent mutations with predicted translational consequences in ENU-treated Casp9−/− samples, except for missense mutations of unknown significance in Cfh (V268I) and Trove2 (D458G) detected in 2 samples each (supplemental Table 2). The mutation spectrum in the ENU-treated Casp9−/− samples was marked by an increase in the transversion:transition ratio (supplemental Figure 5), a known signature of alkylator-induced DNA damage.28
Loss of Casp9 leads to oligoclonal hematopoiesis after alkylator treatment. FLCs from three Casp9−/− or Casp9+/+ FLCs were each transplanted into 4 lethally-irradiated WT congenic recipients. At 6 and 7 weeks posttransplant, recipient mice were injected with ENU (100 mg/kg) or an equivalent volume of 10% DMSO. At 12 weeks after treatment, donor cells were sorted from the bone marrow and exome sequencing was performed. Somatic single nucleotide variants (SNVs; present in the bone marrow and absent in matched-donor fetal tissue) were identified. SNVs were more frequent and had higher VAFs in ENU-treated vs DMSO-treated Casp9−/− cells. Within each plot, the dot represents the median VAF and the bar indicates the 95% confidence interval. The total number of somatic SNVs detected in each sample is shown below.
Loss of Casp9 leads to oligoclonal hematopoiesis after alkylator treatment. FLCs from three Casp9−/− or Casp9+/+ FLCs were each transplanted into 4 lethally-irradiated WT congenic recipients. At 6 and 7 weeks posttransplant, recipient mice were injected with ENU (100 mg/kg) or an equivalent volume of 10% DMSO. At 12 weeks after treatment, donor cells were sorted from the bone marrow and exome sequencing was performed. Somatic single nucleotide variants (SNVs; present in the bone marrow and absent in matched-donor fetal tissue) were identified. SNVs were more frequent and had higher VAFs in ENU-treated vs DMSO-treated Casp9−/− cells. Within each plot, the dot represents the median VAF and the bar indicates the 95% confidence interval. The total number of somatic SNVs detected in each sample is shown below.
Discussion
Previous work has shown that inhibition of apoptosis upstream of the mitochondrion leads to a proliferative phenotype in hematopoietic cells and increased HSC frequency. In contrast, we found that loss of Casp9 or Apaf1 in the hematopoietic compartment causes impaired hematopoiesis and reduced survival. This is likely caused by qualitative defects in HSCs from these mice. Similar to defects in DNA repair, defects in postmitochondrial apoptosis decrease stem-cell fitness and cause cells to exhaust, although LT-HSC numbers are not affected and KLS cells are increased.
The development of the HSC pool early in fetal development requires rapid proliferation and DNA replication, which exposes cells to DNA damage. HSCs are thought to be particularly susceptible to DNA damage because they go through fewer cell-cycle checkpoints when cellular machinery can detect and repair DNA damage.1 HSCs can accumulate DNA damage more readily than more differentiated cells, and the HSCs of aged mice show greater numbers of DNA breaks than age-matched progenitor cells.5 HSCs also respond differently to DNA-damaging and apoptosis-inducing stimuli. When compared with progenitor cells, HSCs show less efficient or delayed repair of DNA lesions after irradiation or alkylator treatment, maintaining DNA lesions longer than more differentiated cells.5,7,8,29,30 In Casp9-deficient mice, these damaged cells cannot complete apoptosis and a population of dysfunctional cells may accumulate within the HSC compartment. These surviving cells would have higher levels of DNA damage from the initial apoptotic stimulus and probably also as a result of the elevated levels of ROS released by the mitochondria during attempted apoptosis. We provide experimental evidence supporting this prediction, including elevated levels of DNA damage in DMSO-treated Casp9−/− mice. It has been shown previously that accumulated DNA damage results in decreased HSC self-renewal, possibly as a protective mechanism to prevent the propagation of the damage throughout the hematopoietic hierarchy.1,30,31 This decreased self-renewal could explain the qualitative defects in HSC function that we observed in competitive and sequential transplant studies and likely contributed to late-onset lethal bone marrow failure in these mice.
The phenotype we observed in Casp9-deficient mice is similar to that of mice with mutations in the DDR pathway. As noted before, HSCs from these mice accumulate DNA damage, leading to impaired function and premature exhaustion.4-8 DDR-deficient HSCs are prone to undergo apoptosis spontaneously and also generally show lower rates of proliferation, although the absolute numbers of stem cells are not altered.4-8
The phenotypes of Casp9-deficient mice and mice with defective DDR also recapitulate features of HSC aging in humans. As humans age, the number of HSCs in the bone marrow increases, but the function of each individual stem cell decreases.1 Lymphopoiesis also declines over time, possibly because of the exhaustion of lymphoid-competent stem cells, resulting in a myeloid-biased hematopoietic compartment, similar to what was observed in our mice.1-3 The aging hematopoietic system is also more prone to developing myeloid leukemias.2
We hypothesized that cells with defective apoptosis would inappropriately survive exposure to DNA-damaging agents, giving rise to a population of cells with excess DNA damage. These cells would then be prone to dysplastic differentiation and/or malignant transformation. Indeed, we observed increased DNA damage in Casp9- and Apaf1-deficient cells in vitro and in vivo after ENU treatment. With exome sequencing of Casp9−/− transplant recipients, we demonstrated that this increased DNA damage corresponds to a significantly increased mutation load. These mutations were detectable above a conservative threshold (VAF ≥5%), suggesting that they were present in clonal populations of cells within the bone marrow. Although it is possible that this clonal hematopoiesis is the result of a smaller number of input stem cells in the original transplant (“pseudoclonality”), this is unlikely for several reasons. The very slight decrease in LT-HSCs observed in transplant recipients at 12 weeks posttransplant is not sufficient to account for the clonality observed via sequencing. Furthermore, if pseudoclonality accounted for these results, we would expect to see the same in DMSO-treated Casp9−/− mice as well, which we did not. Therefore, the observed oligoclonal hematopoiesis is likely a result of the combined effects of alkylator exposure and the defective apoptosis conferred by the loss of Casp9, resulting in accumulation of mutations, some of which provide a clonal advantage. There is a paucity of sensitive tools to detect emerging oligoclonality, an early hallmark of many malignancies. Exome sequencing may prove to be useful in this context for a variety of genetically-engineered mouse models of human cancer.
This work suggests that loss of function in the apoptotic cascade has significant consequences for hematopoiesis and could play a role in cancer susceptibility. A CASP9 haplotype that alters gene expression has been associated with increased risk of lung cancer.32 Single-nucleotide polymorphisms in CASP9 and APAF1 have also been associated with a risk of developing non-Hodgkin lymphoma and chronic lymphocytic leukemia.33,34 We found that Casp9 haploinsufficiency did not significantly perturb hematopoiesis in mice, suggesting that only highly deleterious alleles (which would likely be removed by purifying selection) could plausibly affect AML susceptibility in humans. As mentioned previously, CASP9 and APAF1 are neither recurrently mutated nor differentially expressed in de novo AML,19 but further study is warranted to determine whether acquired defects in the apoptotic pathway play a role in the development of both de novo and therapy-related AML in humans.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Richard Flavell (Yale University) for providing Casp9+/− embryos, Dr Jeffery Klco for assistance with interpretation of bone marrow morphology, Dr Anthony Rongvaux for assistance with genotyping protocols, Mieke Hoock and Deborah Leflamme for their assistance in maintaining our mouse colonies, and Masayo Izumi and David Lu for technical assistance.
This work was supported by the National Institutes of Health, National Cancer Institute grant P01CA101937 and National Heart, Lung, and Blood Institute grant F30HL114316.
Authorship
Contribution: E.P.L. and T.A.G. designed the study; E.P.L. performed mouse hematopoiesis, transplants, and functional assays; R.F. performed sequence production; L.D., M.M., and C.A.M. performed bioinformatics analysis; and E.P.L., T.A.G., M.J.W., D.C.L., P.W., J.F.D., E.R.M., T.J.L., and R.K.W. edited and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for T.A.G. is Massachusetts General Hospital, Boston, MA.
Correspondence: Timothy A. Graubert, Massachusetts General Hospital Cancer Center, 10 North Grove St, Lawrence House 204, Boston, MA 02114; e-mail: tgraubert@partners.org.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal