Key Points
The hyperactive phenotype of lymphoma-associated mutations is caused by increased oligomerization propensity of the MyD88 TIR domain.
The TIR domain of mutants interacts with wild-type MyD88, explaining why heterozygous mutation could be sufficient as a driver mutation.
Abstract
Myeloid differentiation 88 (MyD88) is the key signaling adapter of Toll-like and interleukin-1 receptors. Recurrent lymphoma-associated mutations, particularly Leu265Pro (L265P), within the MyD88 Toll/interleukin-1 receptor (TIR) domain sustain lymphoma cell survival due to constitutive nuclear factor κB signaling. We found that mutated TIR domains displayed an intrinsic propensity for augmented oligomerization and spontaneous formation of cytosolic Myddosome aggregates in lymphoma cell lines, mimicking the effect of dimerized TIR domains. Blocking of MyD88 oligomerization induced apoptosis. The L265P TIR domain can recruit the endogenous wild-type MyD88 for oligomer formation and hyperactivity. Molecular dynamics simulations and analysis of additional mutations suggest that constitutive activity is caused by allosteric oligomerization.
Introduction
Myeloid differentiation 88 (MyD88) is a pivotal signaling protein in the innate immune system, participating in Toll-like receptor (TLR) signaling pathways during a host’s response to infection.1 Patients with germline MYD88 loss-of-function mutations suffer from severe susceptibility to pyogenic bacterial infection during childhood and only survive into adulthood on a strict therapy of antibiotics.2 Conversely, somatic MyD88 gain-of-function mutations were recently discovered in diffuse large B-cell lymphoma (DBLCL), Waldenström’s macroglobulinemia (WM), and chronic lymphocytic leukemia (supplemental Tables 1 and 2 available on the Blood Web site). These data suggest that such mutations, particularly the most prominent Leu265Pro (L265P) mutation, are oncogenic driver mutations that sustain B-cell survival and thus oncogenesis. Indeed, MyD88-mutated cells (cell lines and primary cells) could be selectively killed by ablation of MyD88 downstream signaling using short hairpin RNA or pharmacologic inhibition.3,4 Although hyperactivation of nuclear factor κB (NF-κB) has been described as a key feature of MyD88-mutated B cells, the molecular mechanism of how the amino acid alterations in lymphoma-associated MyD88 mutations exert this phenotype has to be resolved.
MyD88 is composed of an N-terminal death domain (DD)5 and a highly conserved C-terminal Toll/interleukin-1 receptor (TIR) domain.6 Whereas the DD and an intermediate linker domain7 are responsible for downstream signal propagation via kinases of the interleukin-1 receptor-associated kinases (IRAKs),8 the MyD88 TIR domain integrates signals from upstream TLR and interleukin-1 receptor. Association of TLRs mediated by their agonists triggers the formation of TIR-domain dimers, which results in downstream signaling and expression of genes involved in the host defense system.1,9 TIR domains comprise 135- to 160-amino-acid residues with 20% to 30% sequence conservation between the adaptors and TLR families. Most TIR domains comprise 5 α helices (A-E) surrounding 5 central β strands (A-E). Three regions that are functionally important and highly conserved in the amino acid sequence are defined as boxes 1 to 3 located at the βA strand, BB loop, and αE helix, respectively.10 Although the hydrophobic core of TIR domains is conserved, the surface residues vary among TIR domains of different signaling proteins, modulating their specificity.11 Three-dimensional structures of TIR domains from MyD88,11,12 Mal,13 TRIF,14,15 TRAM,15 TLR1,16 TLR2,16 TLR10,17 and IL-1RAPL7 are known. Several regions within the MyD88 TIR-domain surface have been proposed to mediate homo- or heterotypic interactions during signaling, although some of these predictions are contradictory.11,12,18,19 MyD88 TIR-TIR interactions are thought to nucleate the formation of a high-molecular-weight (HMW) (>180 kDa) “Myddosome” complex containing 14 DD domains of MyD88 and IRAKs20 ; however, this crystal structure lacks the MyD88 TIR domain. Therefore, the molecular mechanism and stoichiometry of the activating signaling complex involving TIR-domain interaction between receptors and adaptors remain poorly understood. The isolated MyD88 wild-type (WT) TIR domain has a dominant-negative effect on TLR signaling,6 but forced TIR domain dimerization, eg, based on the intrinsic affinity of TIR domains of TLR421 or tandem constructs of MyD88 TIR domains (tethered TIR domains), constitutively initiate signaling, suggesting that TIR di/oligomerization may initiate nucleation of Myddosomes in the absence of an incoming TLR signaling.22 Whether and how these results relate to a physiologically relevant process has not been determined so far.
Given the importance of the MyD88 TIR domain in cell activation in the context of B-cell malignancies, our study aimed to elucidate the molecular mechanism underlying activation of MyD88. Because lymphoma-associated mutations in the MyD88 TIR domain cause the constitutive activation of MyD88, we surmised that this is primarily a consequence of an enhanced interaction at the level of the MyD88 TIR domain. This hypothesis was tested using computational methods and experimental approaches in cellular model systems and in lymphoma cell lines expressing endogenous levels of MyD88.
Methods
Full details can be found in the supplemental Methods.
Reagents and cell cultures
All reagents were from Sigma unless stated otherwise. The MyD88-blocking peptide (Pepinh-MYD)23 was from Invivogen. Concentrations and preincubation times are given in the figure legends. Human embryonic kidney 293 (HEK293) cells (European Collection of Cell Cultures), MyD88-deficient cells (HEK293-I3A; a kind gift from George R. Stark, Department of Molecular Genetics, Lerner Research Institute, Cleveland, OH), WT MyD88 (OCI-LY19, BJAB, U2392), or heterozygously MYD88-mutated (OCI-LY10, TMD8, HBL1 with L265P; HLY1 with S119C, SUDHL2 with S122R) or homozygously L265P-mutated (OCI-LY3) DLBCL cell lines (from Manfred Kögl [Boehringer Ingelheim RCV, Vienna] or Stephan Hailfinger [Department of Biochemistry, University of Tübingen]) were cultured as described in the supplemental Methods.
siRNA knockdown of endogenous MyD88
Qiagen control (#1027280) or MYD88 (Hs_MYD88_10 SI02100902) small interfering RNA (siRNA) oligonucleotides were transfected (Lipofectamine, Life Technologies) at 40 nM together with MyD88 TIR-domain plasmids (amino acids 155-294, N-terminal streptavidin-hemagglutinin tag) and NF-κB firefly luciferase and constitutive Renilla luciferase in 24-well format. Cells were lysed 48 hours later (Promega passive lysis buffer) and either luciferase activity measured by dual luciferase assay (DLA) or the same lysates analyzed by immunoblot using specific antibodies against MyD88 or hemagglutinin.
DLA
Cells were transiently transfected using JetPEI (Polyplus Transfection) with plasmids for different signaling proteins and firefly luciferase under the NF-κB or interferon-β (IFN-β) promoter and constitutive Renilla luciferase. Twenty-four hours after transfection, cells were optionally stimulated for 24 hours with lipopolysaccharide (LPS) from Escherichia coli 055:B5 (Sigma), lysed in a passive lysis buffer (Promega), and measured for luciferase assay.
SDS polyacrylamide gel electrophoresis and immunoblot
For protein expression studies, HEK293 or HEK293-I3A cells were plated in 6-well plates (Techno Plastic Products AG) and 24 hours later transfected using JetPEI with 1 µg of appropriate plasmid. Cells were lysed 48 hours later in a RIPA buffer with protease inhibitors (Sigma). MyD88 (D80F5; Cell Signaling), green fluorescent protein (Life Technologies), or α/β-tubulin antibody (#2148, Cell Signaling) were used for immunoblot (see the supplemental Methods). MyD88 and IRAK1 protein aggregation in DLBCL cell lines was analyzed by lysing 3 × 106 cells (Promega passive lysis buffer with protease and phosphatase inhibitors [Roche] and Benzonase [Sigma]). Insoluble HMW aggregates were pelleted (16 000g, 10 minutes), washed with phosphate-buffered saline, and boiled with shaking in Invitrogen 4x reducing sodium dodecyl sulfate (SDS) sample buffer. For immunoblots, Cell Signaling (MyD88 D80F5, IRAK1 D51G7) and Sigma anti-β-tubulin (clone TUB 2.1) antibodies and enhanced chemiluminescence signal quantification were used.
Luminescence-based mammalian interactome mapping (LUMIER)
MyD88 full-length, TIR, or DD domain constructs were described previously.24 The 104 cells/well in triplicate 96-wells were transfected with 20 ng Protein A and Renilla fusion plasmids. Forty-eight hours posttransfection cells were lysed, raw Renilla activity measured, and Protein A–tagged proteins and Renilla-tagged interactors copurified (Dynabeads M-280 sheep anti-mouse immunoglobulin G, Life Technologies). Bound Renilla luciferase activity was measured, and an interaction signal was processed by dividing the activity of bound Renilla luciferase by the amount of raw Renilla activity for each transfection and then subtracting the signal for a Protein A–only (no bait) condition.
Quantitative polymerase chain reaction
The messenger RNA (mRNA) was isolated using the RNeasy Mini Kit (Qiagen), transcribed to complementary DNA (High Capacity RNA-to-cDNA Kit; Life Technologies) and MyD88 mRNA expression quantified relative to TBP using TaqMan primers (Hs01573837_g1 and Hs00427621_m1, respectively; Life Technologies) on a real-time cycler (Applied Biosystems; 7500 fast).
Confocal microscopy
HEK293 cells were plated on 8-well plates (Ibidi) and transiently transfected as indicated. mCitrine-TIR in pcDNA3 was constructed using polymerase chain reaction (PCR) ligation between mCitrine and MyD88-mCerulean. ECFP-MyD88, IRAK1- and IRAK4-EGFP constructs were generated from the respective ENTR clones24 and DEST clones containing the fluorescent protein coding sequences (gift of S. Pusch, German Cancer Research Center Heidelberg, Germany). Twenty-four hours later, cells were analyzed using a Leica TCS SP6 confocal microscope (using software LAS AF) under a 63× oil-immersion objective (numerical aperture = 1.4). Excitation and emission wavelengths for all fluorophores are listed in the supplemental Methods.
Molecular dynamics simulations and analysis
All molecular dynamics simulation studies were performed using GROMACS 4.6.325 with OPLS-AA26 and CHARMM27 force fields and TIP3P26 as the water model with a MyD88 TIR nuclear magnetic resonance (NMR) structure (Protein Data Bank accession 2JS7). Structures were energy minimized by the steepest descent approach and subjected to 2 nanoseconds of equilibration at 310 K and 1 atm prior to the actual run. The 40-nanosecond studies were performed on all 20 models present in the NMR (2JS7) structure, while the 100-nanosecond studies were performed on the most representative model 7, which was obtained by structure clustering implemented in UCSF Chimera.5 Analysis of the results was performed using g_rms and g_rmsf present in the GROMACS package. The last 10 nanoseconds of the longer runs gave qualitatively the same root mean square fluctuations (RMSF) graph as the one computed for the whole 100 nanoseconds, showing sufficient equilibration. Models were visualized and manually inspected using Pymol.
Data analysis and statistics
For statistical analysis (GraphPad Prism 5), an unpaired t test was used, if Gaussian distribution was fulfilled (D’Agostino-Pearson, Shapiro-Wilk, and Kolmogorov-Smirnov tests). Otherwise, the Mann-Whitney U t test of unpaired samples was used to account for non-Gaussian distribution. Throughout, an asterisk (*) indicates statistical significance at P < .05.
Results
Lymphoma-associated TIR-domain MyD88 mutants exhibit increased activation of the proinflammatory signaling
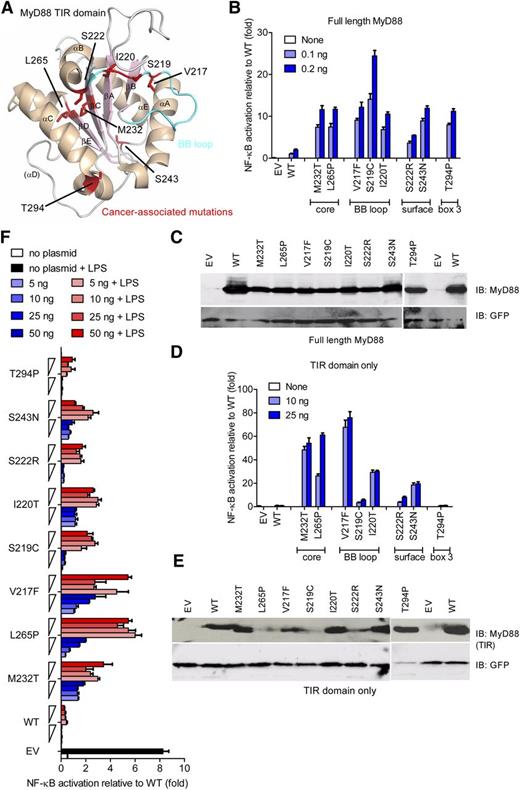
Most of the recurrent oncogenic MYD88 mutations3,28 map to the core of the TIR domain, adjacent to the functionally important BB loop, to the conserved box 3, or to the surface of the TIR domain. (See Figure 1A and supplemental Table 2 for a list of all mutants.) First, we assessed the activation potential of lymphoma-associated MyD88 mutants for the signaling pathways emanating from MyD88.1 Figure 1B shows activation of the NF-κB signaling pathway, compared with WT MyD88 constructs, which was increased upon the expression of point mutations corresponding to oncogenic mutations introduced into full-length MyD88 constructs, in line with overexpression studies in B-cell lines.3 Oncogenic mutations also led to increased IRF-dependent IFN-β promoter activation (data not shown). Interestingly, significantly higher NF-κB activation for MyD88 mutants was seen despite lower expression levels of mutants (Figure 1C).
Lymphoma-associated MyD88 mutants and their isolated TIR domains constitutively hyperactivate NF-κB signaling. (A) Structure of a human MyD88 TIR monomer (Protein Data Bank accession number 2JS7) with lymphoma-associated mutations highlighted in red. The α helices are shown in beige and β sheets in light pink. The BB loop is shown in cyan. (B-F) HEK293 cells were transfected with plasmids for full-length (B-C) or TIR-domain–only (D-F) constructs of lymphoma-associated MyD88 mutants, in parallel with NF-κB–inducible firefly luciferase and constitutive Renilla luciferase reporters, and TLR4 and MD-2 plasmids (F only). Cells were harvested 24 hours later (B-E) or stimulated with LPS (100 ng/mL) for another 24 hours (F). Luciferase activity was measured by DLA (B,D,F) or separately transfected cell lysates loaded on SDS polyacrylamide gel electrophoresis (PAGE) for immunoblot (C,E). For panels B, D, and F, data presented are means + standard deviation (SD) of triplicate samples, and for panels B to F, data shown are representative of at least 3 independent experiments. EV, empty vector; IB, immunoblot.
Lymphoma-associated MyD88 mutants and their isolated TIR domains constitutively hyperactivate NF-κB signaling. (A) Structure of a human MyD88 TIR monomer (Protein Data Bank accession number 2JS7) with lymphoma-associated mutations highlighted in red. The α helices are shown in beige and β sheets in light pink. The BB loop is shown in cyan. (B-F) HEK293 cells were transfected with plasmids for full-length (B-C) or TIR-domain–only (D-F) constructs of lymphoma-associated MyD88 mutants, in parallel with NF-κB–inducible firefly luciferase and constitutive Renilla luciferase reporters, and TLR4 and MD-2 plasmids (F only). Cells were harvested 24 hours later (B-E) or stimulated with LPS (100 ng/mL) for another 24 hours (F). Luciferase activity was measured by DLA (B,D,F) or separately transfected cell lysates loaded on SDS polyacrylamide gel electrophoresis (PAGE) for immunoblot (C,E). For panels B, D, and F, data presented are means + standard deviation (SD) of triplicate samples, and for panels B to F, data shown are representative of at least 3 independent experiments. EV, empty vector; IB, immunoblot.
We suspected that increased activity of MyD88 lymphoma-associated mutants is a consequence of conformational changes within the MyD88 TIR domain. Although the overexpressed MyD88 recruits IRAK1/2 and IRAK4 through heterotypic DD interaction and activates the NF-κB signaling pathway, the WT TIR domain alone does not participate in interactions with IRAK kinases.29 If the mutations disrupted the fold of the TIR domain, then these TIR domains should not be able to interact with other TIR-domain–containing proteins. An isolated WT TIR domain of MyD88 even inhibits activation of NF-κB.6 However, all MyD88 TIR-domain–only constructs except T294P were constitutively active, whereas the WT TIR domain was inactive (Figure 1D). This effect was not a result of elevated expression (Figure 1E) because the protein amount of most mutants was actually rather low compared with WT (eg, L265P), which confirms correct folding and a TIR-intrinsic hyperactive potential for lymphoma-associated mutants. Mutants located in the hydrophobic core and BB loop of the TIR domain exhibited the highest activation. Tethered MyD88 WT TIR domains exhibit inhibition of TLR signaling at low expression levels but constitutive activity at higher expression levels.22 We observed the similar dose-dependent response for lymphoma-associated MyD88 TIR-only constructs, namely inhibition of TLR4 signaling upon LPS stimulation at lower expression and increased activation at higher expression levels for the M232T, L265P, and V217F lymphoma-associated mutant (Figure 1F). Thus, lymphoma-associated TIR-domain mutants display a phenotype similar to that of a forced dimer of WT TIR domains, suggesting that a di/oligomeric MyD88 TIR-domain platform is a prerequisite for the constitutive activation.
The constitutive activity of TIR domains of lymphoma-associated mutants depends on TIR oligomerization and may involve WT MyD88
Because MyD88-IRAK DD interactions are required to mediate the recruitment of IRAKs and downstream signaling, we wondered how isolated TIR domains could exert the constitutive activation on their own. We hypothesized that mutant TIR domains might employ endogenously expressed WT MyD88 normally present in cells (as assessed by immunoblot and quantitative PCR [data not shown]) to hyperactivate downstream signaling. To test this hypothesis, we expressed full-length MyD88 or TIR-domain–only mutant constructs in HEK293-I3A cells, a MyD88-deficient cell line.30 The hyperactive phenotype of a full-length L265P mutant was reduced from 10-fold in HEK293 cells (Figure 1A) to 2- to 4-fold in I3A cells (Figure 2A-B; note differences in scale). Even more strikingly, NF-κB activation was virtually absent when the TIR-domain–only mutants were expressed in I3A cells (Figure 2C-D). This observation suggests that in this model system, mutated TIR domains form a scaffold for the recruitment of full-length WT MyD88 molecules that initiate formation of the signaling Myddosome complex recruiting IRAKs. This finding was confirmed by experiments using LUMIER, a technique recently used to quantify Myddosome interactions.24 In brief, the avidity of a protein-protein interaction is measured by coexpressing the 2 interacting proteins with Protein A (bait) and Renilla luciferase (prey) fusion tags (see figure legends and “Methods”) and by subsequent determination of the level of binding of the Renilla-fused partner upon Protein A purification. We discovered that oligomerization in both full-length (Figure 2E) and TIR-only constructs (Figure 2F) were most prominently increased for the heteromeric WT-L265P bait-prey pairs. L265P-L265P interactions were slightly weaker than WT-L265P interactions but nevertheless significantly stronger than WT-WT interactions. Interactions were generally weaker in I3A cells, particularly for TIR-domain–only constructs (supplemental Figure 1A-B), suggesting that strong interaction is promoted by endogenous WT MyD88. In agreement with the results on signaling, neither WT nor L265P TIR domains interacted with a DD-only construct (supplemental Figure 1C-D), confirming the key role of TIR-TIR interactions in MyD88 oligomerization.31 Additionally, siRNA knockdown of endogenous MyD88 in HEK293T cells strongly reduced the level of activation induced by the L265P TIR domain to background, suggesting that the hyperactive phenotype of L265P TIR-domain–only constructs in HEK293T (ie, MyD88 competent) cells is dependent on endogenous MyD88 (supplemental Figure 1E-G). These findings suggest that in vivo, lymphoma-associated MyD88 mutants may cooperate with WT MyD88 for NF-κB hyperactivation via constitutive TIR-TIR oligomer formation, in agreement with the finding that most MyD88-mutated DLBCL and WM cell lines and tumor samples are heterozygous for the mutation.3,28,32 Based on these data, we suggest that 1 allelic copy of mutated MyD88 would be sufficient to initiate potent NF-κB signaling in lymphoma cells through TIR-TIR oligomerization.
Augmented heterodimerization of lymphoma-associated mutants and WT MyD88 TIR domain. (A-D) MyD88-deficient HEK293-I3A cells were transfected with plasmids for full-length (A-B) or TIR-domain–only (C-D) constructs of lymphoma-associated MyD88 mutants, in parallel with NF-κB–inducible firefly luciferase and a constitutive Renilla luciferase reporters. Cells were harvested 48 hours later; 1 lysate aliquot was used for luciferase activity measurement by DLA (A,C) and another for immunoblot upon 15% SDS-PAGE separation (B,D). Data presented are means + SD of triplicate samples, and data shown are representative of at least 3 independent experiments. (E-F) The L265P mutation leads to increased TIR-TIR oligomerization. LUMIER luciferase analysis from HEK293T cells transfected with Protein A–tagged and Renilla luciferase–tagged WT or L265P mutant full-length (E) or TIR-domain–only (amino acid sequence 155-294) (F) MyD88 constructs. Forty-eight hours posttransfection, cells were lysed and raw luciferase measured in 10% of the lysate sample. The remainder was used for Protein A purification on immunoglobulin G magnetic beads and subsequent measurement of bound luciferase. Data represent ratios of bound vs raw luciferase for each transfection upon subtraction of background (Protein A–only control bait) combined from 7 identical experiments. Means ± SDs are shown and differences tested using a Mann-Whitney U t test. EV, empty vector; IB, immunoblot.
Augmented heterodimerization of lymphoma-associated mutants and WT MyD88 TIR domain. (A-D) MyD88-deficient HEK293-I3A cells were transfected with plasmids for full-length (A-B) or TIR-domain–only (C-D) constructs of lymphoma-associated MyD88 mutants, in parallel with NF-κB–inducible firefly luciferase and a constitutive Renilla luciferase reporters. Cells were harvested 48 hours later; 1 lysate aliquot was used for luciferase activity measurement by DLA (A,C) and another for immunoblot upon 15% SDS-PAGE separation (B,D). Data presented are means + SD of triplicate samples, and data shown are representative of at least 3 independent experiments. (E-F) The L265P mutation leads to increased TIR-TIR oligomerization. LUMIER luciferase analysis from HEK293T cells transfected with Protein A–tagged and Renilla luciferase–tagged WT or L265P mutant full-length (E) or TIR-domain–only (amino acid sequence 155-294) (F) MyD88 constructs. Forty-eight hours posttransfection, cells were lysed and raw luciferase measured in 10% of the lysate sample. The remainder was used for Protein A purification on immunoglobulin G magnetic beads and subsequent measurement of bound luciferase. Data represent ratios of bound vs raw luciferase for each transfection upon subtraction of background (Protein A–only control bait) combined from 7 identical experiments. Means ± SDs are shown and differences tested using a Mann-Whitney U t test. EV, empty vector; IB, immunoblot.
Lymphoma-associated mutations augment oligomerization and molecular aggregation
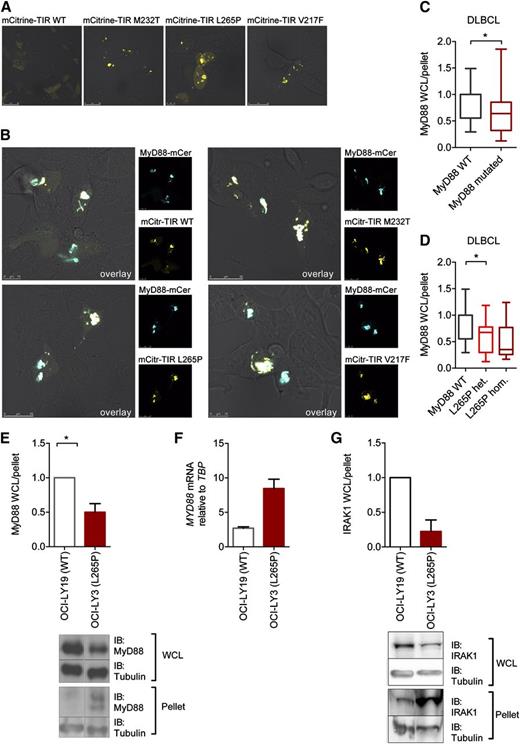
To investigate oligomerization of MyD88 TIR lymphoma-associated mutants in situ in cells, MyD88 mutants were linked to the fluorescent protein mCitrine. Whereas WT mCitrine TIR remained diffuse, mCitrine-TIR mutants (which were equally active as their counterparts without mCitrine; cf. supplemental Figure SA-B) strongly aggregated (Figure 3A), a phenomenon also observed in tethered WT MyD88 TIR domains.22 Dimerization-driven aggregation of mutant TIR domains as the nucleating platform for Myddosome assembly thus constitutes a plausible molecular mechanism underlying hyperactivation. All mCitrine-TIR-domain–only mutants colocalized with MyD88-mCerulean (Figure 3B), confirming that aggregated TIR domains create a scaffold for recruiting full-length MyD88 via their TIR domains.
Aggregation of constitutively active lymphoma-associated TIR mutants in lymphoma cell lines. (A-B) HEK293 cells were transfected with plasmids for mCitrine-TIR (yellow pseudocolor) or mutants and full-length MyD88-mCerulean (blue pseudocolor, B). Twenty-four hours later, cells were fixed and visualized under the confocal microscope at ×1000 magnification. Shown are representative confocal images of at least 3 independent experiments. (C-G) Augmented amount of MyD88 in aggregates in different DLBCL cell lines. Cell lysates were fractionated by centrifugation and lysate and pellet fraction analyzed by SDS-PAGE followed by anti-MyD88 (C-E) or IRAK1 (G) and anti-tubulin immunoblot (see blots in panels E and G). Relative MyD88 or IRAK1 and tubulin levels were quantified by charge-coupled device detection and plotted as the tubulin-normalized ratios of lysate (WCL) vs pellet MyD88 or IRAK1. (C-D) Data were combined from 9 experiments and are shown as min-to-max box-and-whisker plots. In panel C, WT MyD88 DLBCL cell lines (white, OCI-LY19, BJAB, U2392) were compared with MyD88-mutated cell lines (burgundy, OCI-LY10, TMD8, HBL1, OCI-LY3 with L265P; HLY1 with S119C, SUDHL2 with S122R). In panel D, cells were plotted according to L265P mutational status. White, WT MyD88 (OCI-LY19, BJAB, U2392); red, L265P heterozygous (OCI-LY10, TMD8, HBL1); burgundy, L265P homozygous (OCI-LY3). (E) Comparison of unmutated OCI-LY19 (white) vs L265P homozygous OCI-LY3 (red) lysate vs pellet ratio from 9 combined experiments and showing a representative immunoblot. (F) Differences in MyD88 lysate expression in L265P-mutated OCI-LY3 cells are not due to lower mRNA expression as assessed by quantitative PCR in OCI-LY19 (white) and OCI-LY3 (burgundy) cells. One representative of 3 independent experiments was done in triplicate (mean + upper limit). (G) MyD88-containing aggregates also contain IRAK1. Comparison of unmutated OCI-LY19 (white) vs L265P homozygous OCI-LY3 (red) lysate vs pellet ratio from 3 combined experiments and showing a representative immunoblot. IB, immunoblot; WCL, whole-cell lysate.
Aggregation of constitutively active lymphoma-associated TIR mutants in lymphoma cell lines. (A-B) HEK293 cells were transfected with plasmids for mCitrine-TIR (yellow pseudocolor) or mutants and full-length MyD88-mCerulean (blue pseudocolor, B). Twenty-four hours later, cells were fixed and visualized under the confocal microscope at ×1000 magnification. Shown are representative confocal images of at least 3 independent experiments. (C-G) Augmented amount of MyD88 in aggregates in different DLBCL cell lines. Cell lysates were fractionated by centrifugation and lysate and pellet fraction analyzed by SDS-PAGE followed by anti-MyD88 (C-E) or IRAK1 (G) and anti-tubulin immunoblot (see blots in panels E and G). Relative MyD88 or IRAK1 and tubulin levels were quantified by charge-coupled device detection and plotted as the tubulin-normalized ratios of lysate (WCL) vs pellet MyD88 or IRAK1. (C-D) Data were combined from 9 experiments and are shown as min-to-max box-and-whisker plots. In panel C, WT MyD88 DLBCL cell lines (white, OCI-LY19, BJAB, U2392) were compared with MyD88-mutated cell lines (burgundy, OCI-LY10, TMD8, HBL1, OCI-LY3 with L265P; HLY1 with S119C, SUDHL2 with S122R). In panel D, cells were plotted according to L265P mutational status. White, WT MyD88 (OCI-LY19, BJAB, U2392); red, L265P heterozygous (OCI-LY10, TMD8, HBL1); burgundy, L265P homozygous (OCI-LY3). (E) Comparison of unmutated OCI-LY19 (white) vs L265P homozygous OCI-LY3 (red) lysate vs pellet ratio from 9 combined experiments and showing a representative immunoblot. (F) Differences in MyD88 lysate expression in L265P-mutated OCI-LY3 cells are not due to lower mRNA expression as assessed by quantitative PCR in OCI-LY19 (white) and OCI-LY3 (burgundy) cells. One representative of 3 independent experiments was done in triplicate (mean + upper limit). (G) MyD88-containing aggregates also contain IRAK1. Comparison of unmutated OCI-LY19 (white) vs L265P homozygous OCI-LY3 (red) lysate vs pellet ratio from 3 combined experiments and showing a representative immunoblot. IB, immunoblot; WCL, whole-cell lysate.
We next sought to determine whether aggregation of MyD88 occurred in DLBCL cell lines with (n = 4) or without (n = 3) L265P mutation (see “Methods”) expressing MyD88 at natural levels. Cellular lysates were prepared and the HMW fraction was separated by centrifugation. Endogenous MyD88 levels were compared between lysates and pelleted HMW fractions by immunoblot and charge-coupled device quantification with tubulin levels as a normalization reference. Figure 3C shows significantly lower lysate:pellet ratios in MyD88-mutated DLBCL cell lines compared with nonmutated cell lines, thus indicating higher amounts of MyD88-containing aggregates. The same trend was also observed for groups of heterozygous (n = 3) and homozygous L265P-mutated (n = 1) vs WT MyD88 cell lines (n = 3; Figure 3D). An example for a WT MyD88 (OCI-LY19) and L265P homozygous cell line (OCI-LY3) is shown in Figure 3E. Of note, these differences were not attributable to differences in MYD88 mRNA levels (Figure 3F). MyD88-containing aggregates predominantly found in the L265P-mutated OCI-LY3 cells also contained elevated amounts of IRAK1 (Figure 3G), which is part of the Myddosome complex. Thus, the aggregates observed by confocal microscopy and through centrifugation/immunoblot analysis in cells containing mutated MyD88 are likely to correspond to Myddosome complexes. This was also confirmed by the fact that in transfected HEK293T cells, ECFP-MyD88/L265P and IRAK1-EGFP or IRAK4-EGFP colocalized, as assessed by confocal microscopy (supplemental Figure 2C-D) and confirmed by ImageStream bright-field microscopy analysis (data not shown). This conclusion is in agreement with previous findings that MyD88 L265P binds strongly to and utilizes IRAK1 for constitutive signal transduction.3,33 Our results illustrate an intrinsic tendency of lymphoma-associated mutations for oligomer/Myddosome formation both in the HEK293 cell model system as well as in DLBCL cells.
TIR-domain residues mutated in B-cell lymphoma are likely to have arisen by natural selection
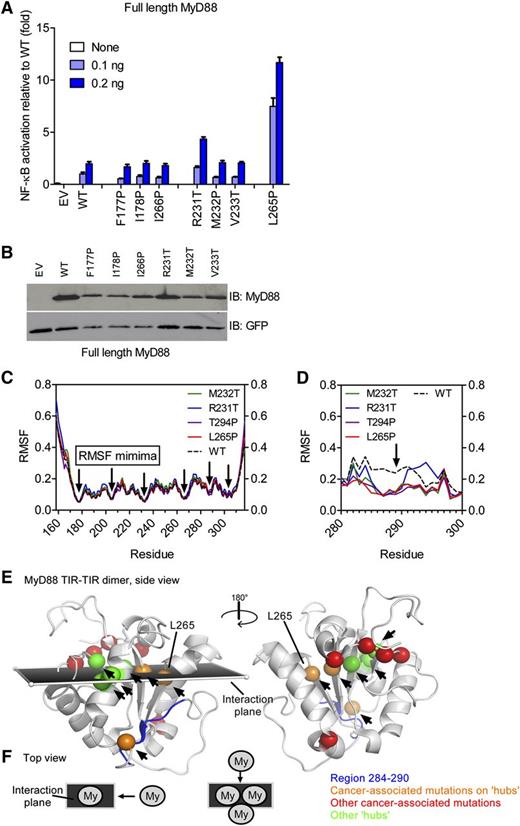
The underlying molecular mechanism for the aggregation of TIR domains is intrinsically linked to the structure of the MyD88 TIR domain. We therefore investigated how the tertiary structure of the TIR domain could be affected, especially because the mutated residues of most active mutants are buried and could only affect interacting residues through an allosteric effect. We sought to assess those differences by the functional analysis of designed mutations within the TIR domain and by molecular simulation. We investigated the mutations within the TIR domain at positions close to the positions of lymphoma-associated mutants and with similar amino acid changes that might cause similar constitutive activity. The selected designed mutants were located in β sheets (F177, I178, and I266) harboring substitutions into proline to disrupt the β sheet or substitutions at a lymphoma-specific site (M232) (see supplemental Table 1). Out of 6 tested mutations in the core of the TIR domain in the direct vicinity of lymphoma-associated mutations, only R231T led to an elevated activation, but mutations at positions 177, 178, 266, 232, and 233 did not affect NF-κB activation levels in a similar way as L265P, despite a comparable expression level (Figure 4A-B). Cellular localization of designed TIR-domain mutants fused to mCitrine demonstrated that, with the exception of R231T, designed mutants did not form aggregates and were diffusely distributed as the WT TIR domain, additionally supporting the correlation between aggregation and the hyperactivation phenotype (supplemental Figure 2E). Although lymphoma-associated M232T and L265P mutants are strongly hyperactive (Figure 1A,C), their designed “neighbors,” M232P and I266P (Figure 4A), were not. The lymphoma-associated substitutions are therefore likely to have emerged as a result of the selection in B cells, which is in line with a study showing that MyD88 L265P is an early driver for chronic lymphocytic leukemia oncogenesis and is found in nearly all subclones.34
Lymphoma-associated TIR-domain mutants are located at stable communication hubs in the TIR domain. (A-B) HEK293 cells were transfected with plasmids for designed MyD88 mutants and NF-κB–inducible firefly luciferase and constitutive Renilla luciferase reporter plasmids. Twenty-four hours later, cells were lysed and luciferase activity measured (A) or the lysates were run on SDS-PAGE and analyzed by anti-MyD88 immunoblot (B). (C-D) The MyD88 TIR domain contains highly stable RMSF minima. The 40-nanosecond (C) or 100-nanosecond (D) molecular dynamics simulations were done on WT (black dotted line) and mutant (colored as shown) MyD88 NMR structure 2js7 ensemble (composed of 20 structures each, 20 simulations each); shown are averaged data (C) or the single most representative conformer (D). RMSF over the TIR-domain residues were plotted. RMSF profiles are highly similar and show RMSF minima (black arrows pointing to residues 177, 204, 231/232, 265, and 303/304). (E-F) RMSF minima are shown as green or orange (in case they coincide with lymphoma-associated mutations) spheres and map to a plane describing MyD88 dimer formation (see schematic representation, panel F) as proposed by Bovijn et al.20 Communication hubs are indicated by arrows and coincide with RMSF minima or locate in the interaction plane within the MyD88 dimer. Some lymphoma-associated mutations directly map to hub positions (orange spheres) or locate within this plane (red spheres), with the exception of S143 and T294 (red). Hub positions for which so far no lymphoma-associated mutations have been reported are shown as green spheres. The region of higher flexibility in WT (black dotted line) vs mutated (colored as shown) TIR domains in molecular dynamics simulation (F) is shown in dark blue in panel E. EV, empty vector; IB, immunoblot.
Lymphoma-associated TIR-domain mutants are located at stable communication hubs in the TIR domain. (A-B) HEK293 cells were transfected with plasmids for designed MyD88 mutants and NF-κB–inducible firefly luciferase and constitutive Renilla luciferase reporter plasmids. Twenty-four hours later, cells were lysed and luciferase activity measured (A) or the lysates were run on SDS-PAGE and analyzed by anti-MyD88 immunoblot (B). (C-D) The MyD88 TIR domain contains highly stable RMSF minima. The 40-nanosecond (C) or 100-nanosecond (D) molecular dynamics simulations were done on WT (black dotted line) and mutant (colored as shown) MyD88 NMR structure 2js7 ensemble (composed of 20 structures each, 20 simulations each); shown are averaged data (C) or the single most representative conformer (D). RMSF over the TIR-domain residues were plotted. RMSF profiles are highly similar and show RMSF minima (black arrows pointing to residues 177, 204, 231/232, 265, and 303/304). (E-F) RMSF minima are shown as green or orange (in case they coincide with lymphoma-associated mutations) spheres and map to a plane describing MyD88 dimer formation (see schematic representation, panel F) as proposed by Bovijn et al.20 Communication hubs are indicated by arrows and coincide with RMSF minima or locate in the interaction plane within the MyD88 dimer. Some lymphoma-associated mutations directly map to hub positions (orange spheres) or locate within this plane (red spheres), with the exception of S143 and T294 (red). Hub positions for which so far no lymphoma-associated mutations have been reported are shown as green spheres. The region of higher flexibility in WT (black dotted line) vs mutated (colored as shown) TIR domains in molecular dynamics simulation (F) is shown in dark blue in panel E. EV, empty vector; IB, immunoblot.
Molecular simulation of TIR-domain mutations support the allosteric mechanism of activation
Circular dichroism spectroscopy of recombinant-mutated TIR domains revealed a preserved global secondary structure of mutants (data not shown). Therefore, hyperactivity results from subtler structural perturbations, but their aggregation propensity prevented high-resolution structural analysis, so we employed computational methods instead. The ensemble of 20 conformers or individual conformers of the WT and generated mutant MyD88 TIR were subjected to extensive molecular dynamics simulation analysis ranging from 40 to 100 nanoseconds (see “Methods”). In agreement with the circular dichroism data, backbone RMSF of <1 Å outside the extreme N and C termini (Figure 4C) indicated that the MyD88 TIR-domain mutant structures are as stable as the WT TIR domain. Thus, cancer-associated mutations are unlikely to act via a gross disturbance of the TIR fold as a whole. We noted 5 conserved local RMSF minima with <0.2 Å (ie, highly stable regions), corresponding to positions 177, 204, 231/232, 265, and 303/304 (supplemental Figure 3A). In the tertiary structure of the TIR domain, the positions of 4 RMSF minima, with the exception of A303/K304, are located next to each other on adjacent β strands along the proposed MyD88 dimerization axis19 (see Figure 4E-F). Recent analyses of the receptor TIR domains proposed residues of the central β strands form highly stable “hubs” within an evolutionarily conserved intradomain communication network, acting to relay protein-protein interactions from one end of the structure to the other.36 Thus, RMSF minima map to 5 communication hubs, H1 to H5.35 Strikingly, several lymphoma-associated mutations map exactly to the H3 (M232) or H4 (L265) positions, and, with the exception of S243N and T294P, all the remaining oncogenic mutations map to the interaction plane spanned by residues corresponding to RMSF minima (V217F, S219C, I220T, and S222R; Figure 4E), suggesting that lymphoma-associated mutations in fact modulate force transduction during oligomerization. Analysis of longer (100-nanosecond) molecular dynamics simulations confirmed the overall stability of the TIR fold in all mutants but revealed a highly localized difference in the RMSF between the WT MyD88 and all tested constitutively active lymphoma-associated mutants (L265P, M232T, and T294P), including the only active designed mutant (R231T) (Figure 4D): mutant TIR domains exhibited increased rigidity in the region 284 to 290 (corresponding to the βE strand, sequence TVCDYT), in comparison with the WT structure (Figure 4D). The biological significance of this difference is difficult to assess, but a recent structural analysis of the MyD88 TIR domain12 suggested that this segment participates in dimerization. Thus, by affecting regions involved in dimer formation and communication hubs in the TIR domain, lymphoma-associated mutations may facilitate the observed increased propensity for TIR oligomerization.
Discussion
Genomic studies of DLBCL cancer samples revealed several mutations in the core of the TIR domain that induced constitutive MyD88 signaling ability. Whereas others have noted the hyperactive effect of oncogenic-mutations and their effect on IRAK and TAK1 kinases,3,36 the underlying cause of these observations has remained elusive. Our data now provide a molecular explanation of this phenomenon. In agreement with other data,17 we show that lymphoma-associated mutants of MyD88 acquire increased oligomerization propensity in both HEK293T and DLBCL cells (Figure 2) due to the site-specific allosteric effect of selected mutations within the TIR domain. The increased oligomerization of WT and mutated MyD88 nucleates the formation of HMW, IRAK1-containing aggregates in DLBCL cells (Figure 3), which are most likely bona fide Myddosomes. Constitutive TIR oligomerization of mutated TIR domains thus provides the platform for the initiation of downstream signaling, similarly as tethered MyD88 TIR dimers.22 We propose that lymphoma-associated mutations emerge during B-cell development by providing an advantage in clonal selection,34 because they trigger the antiapoptotic NF-κB signaling that enables B-cell survival.37 Our data showing that mutated MyD88 may employ the WT MyD88 for NF-κB activation also provide an explanation for the observation that heterozygous mutation appears to be sufficient for oncogenesis as suggested by the L265P allelic status of the majority of DLBCL and WM. Mutation of a single allele would thus be sufficient to convey a survival signal and provide a selective advantage during clonal expansion. Our data are important for understanding the MyD88-driven oncogenesis and bear significant therapeutic implications. Although inhibition of hyperactivity at the level of IRAKs has been proposed,3 MyD88 dimerization may represent a target even closer to the origin of hyperactivity and amenable to blocking by decoy peptides or peptidomimetics,8,23,38,39 especially considering that not all MyD88-dependent signaling involves IRAKs.40,41 Indeed, a MyD88-blocking peptide23 significantly reduced TIR-TIR oligomerization (supplemental Figure 3B) and killed DLBCL cell lines in a mutation-dependent fashion (supplemental Figure 3C). Given the dim prognosis for many subtypes of B-cell malignancies harboring MyD88 mutations, targeting oncogenic MyD88 mutations at the level of MyD88 oligomerization seems an attractive option.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Access to the Paderborn Center for Parallel Computing (PC2) resources is gratefully acknowledged. The authors thank S. Dickhöfer and R. Bremšak for technical assistance and H. Wang and S. El Maadidi for helpful discussions. They also thank T. Espevik, C. Kirschning, J. Hiscott, and O. Griesbeck for plasmids for hTLR4, MyD88-mCerulean hMD-2, IFN-β luciferase, and mCitrine, respectively.
This work was supported by the Slovenian Research Agency and the Centre of Excellence EN-FIST (R.J., M.A., O.F., M.B., G.P., M.R., and J.M.), SFB 685 Immunotherapy and the University of Tübingen (O.-O.W., M.D.G., O.K., and A.W.), and FP7 program grant 283481 (SCI-BUS) (J.K. and O.K.).
Authorship
Contribution: M.A., O.-O.W., O.F., M.D.G., G.P., and M.B. performed experiments; J.M., M.R., M.A., C.S., O.K., J.K., O.-O.W., and A.N.R.W. analyzed data; R.J. and A.N.R.W. conceived and supervised the study in the respective laboratories; M.A., R.J., and A.N.R.W. wrote the manuscript; and all authors provided comments to the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alexander N. R. Weber, Interfaculty Institute for Cell Biology, Department of Immunology, University of Tübingen, Auf der Morgenstelle 15, 72076 Tübingen, Germany; e-mail: alexander.weber@uni-tuebingen.de; and Roman Jerala, Department of Biotechnology, National Institute of Chemistry, Hajdrihova 19, 1000 Ljubljana, Slovenia; e-mail: roman.jerala@ki.si.
References
Author notes
M.A. and O.-O.W. contributed equally to this study.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal