Key Points
Recurrent somatic mutations in MAP2K1 were identified in 33% of LCH lesions with wild-type BRAF. The mutant MAPK kinase 1 proteins activate ERK.
The ability of MAPK pathway inhibitors to suppress MAPK kinase and ERK phosphorylation in vitro was dependent on the specific LCH mutation.
Abstract
Langerhans cell histiocytosis (LCH) is a myeloproliferative disorder characterized by lesions composed of pathological CD207+ dendritic cells with an inflammatory infiltrate. BRAFV600E remains the only recurrent mutation reported in LCH. In order to evaluate the spectrum of somatic mutations in LCH, whole exome sequencing was performed on matched LCH and normal tissue samples obtained from 41 patients. Lesions from other histiocytic disorders, juvenile xanthogranuloma, Erdheim-Chester disease, and Rosai-Dorfman disease were also evaluated. All of the lesions from histiocytic disorders were characterized by an extremely low overall rate of somatic mutations. Notably, 33% (7/21) of LCH cases with wild-type BRAF and none (0/20) with BRAFV600E harbored somatic mutations in MAP2K1 (6 in-frame deletions and 1 missense mutation) that induced extracellular signal-regulated kinase (ERK) phosphorylation in vitro. Single cases of somatic mutations of the mitogen-activated protein kinase (MAPK) pathway genes ARAF and ERBB3 were also detected. The ability of MAPK pathway inhibitors to suppress MAPK kinase and ERK phosphorylation in cell culture and primary tumor models was dependent on the specific LCH mutation. The findings of this study support a model in which ERK activation is a universal end point in LCH arising from pathological activation of upstream signaling proteins.
Introduction
Langerhans cell histiocytosis (LCH) is a myeloproliferative disorder characterized by inflammatory lesions including pathological CD207+ dendritic cells (DCs).1 Despite indistinguishable histology between LCH lesions, clinical outcomes are highly variable, ranging from single lesions that resolve with curettage to highly aggressive disease that requires intensive chemotherapy.2 The current standard of care is empiric chemotherapy with risk stratification clinically determined by hematopoietic organ involvement and early response to therapy.3 Defining the etiology of LCH pathogenesis is hence essential to develop rational diagnostic and therapeutic strategies for this disease.
BRAF remains the only gene with somatic mutations reported in more than 1 patient with LCH, with the BRAFV600E point mutation identified in 50% to 65% of LCH lesions.1,4-7 BRAF is a member of the rapidly accelerated fibrosarcoma family of protein kinases and functions downstream of rat sarcoma in the mitogen-activated protein kinase (MAPK) signaling pathway, which also includes the MAPK kinases 1 and 2 (MEK1, 2) that activate extracellular signal-regulated kinases 1 and 2 (ERK1, 2) (reviewed in Poulikakos and Solit8 ). The high frequency of BRAF mutations in LCH, the presence of the mutation in myelomonocytic precursors in patients with high-risk LCH, and the ability of enforced expression of BRAFV600E in mice to recapitulate an LCH-like phenotype support a model in which LCH is a myeloid neoplasm that arises as the result of MAPK pathway activation at critical stages in myeloid differentiation.4,9 However, the mechanisms by which BRAF hyperactivation drives LCH pathogenesis remain to be established.
The MAPK pathway is frequently deregulated by mutations in a wide variety of human malignancies, including juvenile myelomonocytic leukemia, which is also defined as a myeloid neoplasia.10,11 It has been proposed that MAPK signaling might be activated in all LCH patients, even those with wild-type BRAF,4 although little is known about the spectrum of genetic alterations occurring in LCH. Advances in sequencing technologies have facilitated the genome-scale examination of human malignancies and identified the key genetic alterations in numerous cancer types.12 In order to define the extent and range of somatic mutations that underlie LCH pathogenesis, we performed whole exome sequencing (WES) on a series of patient-matched genomic DNA samples isolated from LCH lesions and peripheral white blood cells. A pilot series of more rare histiocytic disorders (juvenile xanthogranuloma [JXG], Erdheim-Chester disease [ECD], and Rosai-Dorfman disease [RDD]) was also included in order to characterize the genetic alterations present in these related diseases.
Methods
Study patients and samples
Biopsy tissue and blood samples were collected, processed, and frozen from 50 patients under a protocol approved by the Baylor College of Medicine Institutional Review Board. This series included 41 patients with LCH (including 2 LCH/ECD and 3 LCH/JXG hybrids), 4 patients with JXG, 4 patients with RDD, and 1 patient with ECD (supplemental Table 1; see the Blood Web site). The cohort of LCH patients was chosen to include approximately equal representation of lesions with known BRAFV600E (20 of 41, 49%) and wild-type BRAF (21 of 41, 51%) as previously determined by quantitative polymerase chain reaction. Disease assignments were based on review of diagnostic biopsies. Tissue specimens were selected that included >10% tumor infiltrate to accommodate technical limitations of WES for mutation detection.13,14 A second biopsy, obtained either from a synchronous lesion or at relapse, was available from 8 of the 50 patients and also submitted for WES. Peripheral white blood cells were used as the “normal” sample for analysis, with determination of somatic mutation status allowing for the low level of circulating mutated cells in patients with high-risk disease1 (supplemental Table 4). DNA was extracted from patient peripheral blood mononuclear cells and tissue biopsy samples using the QIAamp DNA micro kit (Qiagen Inc., Valencia, CA). Clinical data were recorded for each patient, including histologic diagnosis, patient age at diagnosis, clinical risk group, patient status at last contact, patient age at last contact, and treatment received before the study sample was obtained (supplemental Table 1).
WES
Whole exome capture was performed using the Baylor College of Medicine Human Genome Sequencing Center (BCM-HGSC) VCRome 2.1 design array (42 Mb, NimbleGen).15 Sequencing was conducted on the Illumina HiSequation 2000 platform utilizing the HGSC Mercury analysis pipeline (https://www.hgsc.bcm.edu/software/mercury).16 Sequencing runs generated ∼300 million to 400 million successful reads on each flow cell lane, yielding 8 to 11 Gb per sample. With these sequencing yields, samples achieved an average coverage of 106X, with 93.4% of the targeted exome bases covered to a depth >20X (supplemental Figure 1).
Ion AmpliSeq sequencing
Putative somatic mutations identified by WES were validated by sequencing on the Ion AmpliSeq platform. A custom Ion AmpliSeq panel was designed to cover the putative somatic variant sites identified by WES as well as all coding exons of 31 genes of interest (supplemental Table 2), consisting of 1078 amplicons (271 bp mean size; 244 bp median size) binned into 2 pools to avoid amplification of undesired targets. The 31 genes were selected based on the following criteria: (1) genes with more than 1 putative somatic mutation identified by WES (n = 6); (2) known cancer genes with 1 putative somatic mutation identified by WES (n = 10); and (3) MAPK-pathway related genes or genes identified in other MAPK-associated malignancies or myeloid neoplasias that were not found to be mutated in the initial WES (n = 15). Sequencing was performed on the Ion Proton platform. With sequencing yields averaging ∼886 Mb/sample run, 94.8% of the targeted bases were covered to a depth of >100X (supplemental Figure 2).
WES of purified CD207+ cells
For 19 LCH lesions, CD207+ cells were purified from viably preserved LCH lesions by flow cytometry as described previously.17 Genomic DNA was extracted as described previously, then amplified using NuGEN Ovation WGA kit (San Carlos, CA). WES was performed as described previously.
Identification of CNAs from WES data
Copy number alteration (CNA) analysis was performed using LOHcate, a method that detects CNA events in whole exome tumor sequence data via detecting enrichment in variant or reference allele quantities per site across polymorphic exonic sites. Illumina OmniExpress SNP arrays were also analyzed on a subset of 23 cases: 9 cases with BRAFV600E, 5 cases with MAP2K1 mutation, and 9 cases with neither mutation. Details are outlined in the supplemental Methods.
Expression constructs
Specific mutations were generated by QuickChange XL Site-Directed Mutagenesis Kit (Agilent Technologies, La Jolla, CA) in the full-length pEGFP-N1-MAP2K1 (#14746) vector (Addgene, Cambridge, MA) using primers in supplemental Table 3 and sequence verified. Expression plasmids encoding wild-type (#40775) and mutant (V600E) BRAF (#17544) were obtained from Addgene.
Cell culture, transfection, and treatment
Details of HEK293 transfection and treatment are outlined in the supplemental Methods.
Immunoblot analysis of MEK and ERK
Immunoblots were performed on lysate extracted from HEK293 cells transfected with expression vectors as well as from LCH biopsy samples with standard protocols. Details are outlined in the supplemental Methods.
Multispectral imaging flow cytometry
Viable LCH lesion cell suspensions were prepared as described previously.17 Multispectral imaging was performed according to standard methods, as outlined in the supplemental Methods.
Clinical correlation
Details of statistical analysis of clinical variables are outlined in the supplemental Methods.
Results
WES
WES of tumor and matched control samples from 41 LCH patients revealed a total of 91 putative somatic mutations targeting 63 genes. Resequencing of the corresponding tumor and control samples using the custom Ion AmpliSeq panel confirmed 52 of these mutations in 27 genes to be somatic (supplemental Table 4), resulting in a median of 1 mutation per LCH sample exome (range 0 to 5) and a mutation rate of 0.03 somatic mutations per Mb (supplemental Table 1). The somatic mutations identified consisted of 44 missense mutations, 6 in-frame deletions, and 2 nonsense mutations. In 9 of the LCH lesions analyzed, no somatic mutations were identified.
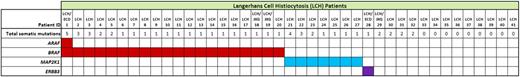
BRAFV600E mutations were confirmed in all 20 LCH lesions previously known to be BRAF mutant by quantitative polymerase chain reaction,1 at a median variant allele frequency of 21% by WES (range 4% to 41%) and 26% by AmpliSeq (range 4% to 40%) (Figure 1; supplemental Table 4). No other mutations in BRAF were identified. Notably, recurrent somatic mutations were also detected in MAP2K1 (which encodes the MEK1 protein) in 7 lesions (Figure 1; supplemental Table 4): 7/21 (33%) cases with wild-type BRAF and 0/20 (0%) cases with BRAFV600E mutations (P < .01, Fisher’s exact test). All MAP2K1 mutations identified were localized to exons 2 and 3 of the gene (Figure 2A; supplemental Table 4), which encode the autoregulatory domain and catalytic core of MEK1. Six of 7 MAP2K1 mutations discovered were small in-frame deletions (Table 1). The c.302_307del mutation was reported earlier in malignant melanoma18 and lung adenocarcinoma,19 and the p.Q56P mutation was previously reported in non–small cell lung cancer and gastric cancer (COSMIC database20 ). Although the c.159_173del, c.170_184del, and c.172_186del mutations have not yet been reported in COSMIC, they are in regions with frequent mutations in other neoplastic disorders, including melanoma21 and hairy cell leukemia variant (HCLv).22 BRAFV600E and MAP2K1 mutations were observed to be highly enriched in CD207-purified cells isolated from LCH lesions, with an average variant allele frequency of 49% (supplemental Table 5).
Key genetic alterations identified in MAPK pathway genes in LCH patients.
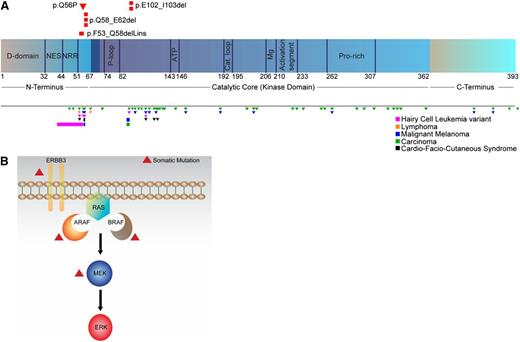
MAPK pathway mutations identified in LCH patients. (A) Location of somatic MAP2K1 mutations identified in LCH patients in this study (top) and reported in other malignancies in the COSMIC database and recent literature (bottom). Also depicted in black are germ-line mutations observed in cardio-facio-cutaneous (CFC) syndrome. Triangles and bars indicate missense and in-frame deletion mutations, respectively. Figure not to scale. (B) Somatic mutations identified in multiple MAPK pathway members in LCH patients. C, carboxyl; D, docking; Mg, magnesium positioning loop; N, peptidyl; NES, nuclear export signal; NRR, negative regulatory region; P, phosphate binding; Pro, proline.
MAPK pathway mutations identified in LCH patients. (A) Location of somatic MAP2K1 mutations identified in LCH patients in this study (top) and reported in other malignancies in the COSMIC database and recent literature (bottom). Also depicted in black are germ-line mutations observed in cardio-facio-cutaneous (CFC) syndrome. Triangles and bars indicate missense and in-frame deletion mutations, respectively. Figure not to scale. (B) Somatic mutations identified in multiple MAPK pathway members in LCH patients. C, carboxyl; D, docking; Mg, magnesium positioning loop; N, peptidyl; NES, nuclear export signal; NRR, negative regulatory region; P, phosphate binding; Pro, proline.
MAP2K1 mutations identified in LCH patients
| Patient ID . | Nucleotide (cDNA) . | Amino acid (protein) . |
|---|---|---|
| LCH-21 | c.302_307del | p.E102_I103del |
| LCH-22 | c.172_186del | p.Q58_E62del |
| LCH-23 | c.172_186del | p.Q58_E62del |
| LCH-24 | c.302_307del | p.E102_I103del |
| LCH-25 | c.170_184del | p.Q58_E62del |
| LCH-26 | c.159_173del | p.F53_Q58delinsL |
| LCH-27 | c.167A>C | p.Q56P |
| Patient ID . | Nucleotide (cDNA) . | Amino acid (protein) . |
|---|---|---|
| LCH-21 | c.302_307del | p.E102_I103del |
| LCH-22 | c.172_186del | p.Q58_E62del |
| LCH-23 | c.172_186del | p.Q58_E62del |
| LCH-24 | c.302_307del | p.E102_I103del |
| LCH-25 | c.170_184del | p.Q58_E62del |
| LCH-26 | c.159_173del | p.F53_Q58delinsL |
| LCH-27 | c.167A>C | p.Q56P |
cDNA, complementary DNA.
Somatic mutations were also identified in ARAF (p.T70M) in a BRAF-mutated tumor and the protein tyrosine kinase receptor ERBB3 (p.P921Q) in a BRAF wild-type tumor (Figures 1 and 2B; supplemental Table 4), resulting in a total of 29 LCH mutations targeting the MAPK pathway. The remaining 23 somatic mutations identified by WES in the 41 LCH samples were singletons in non-MAPK pathway members but did include genes in pathways that potentially impact MAPK signaling such as PICK1 and PIK3R2.23,24 No additional somatic mutations were found in the 41 LCH samples using the 31-gene Ion AmpliSeq panel of MAPK pathway members and related candidate genes (supplemental Table 2) despite an average target coverage of >3400X.
In addition to sequence alterations, we also analyzed the WES data for regions of chromosomal CNA by comparing the base pair level variant allele fractions in the normal and tumor samples using the CNA discovery tool LOHcate. No evidence for CNAs was found in the LCH samples, consistent with results of microarray analyses performed in a subset of 23 cases as well as previous observations of normal diploid LCH genomes25 (supplemental Figure 3).
A second biopsy sample was available from 8 patients (5 LCH, 1 LCH/JXG, and 2 JXG) and also subjected to WES. The second sample sequenced was obtained from a synchronous lesion in 3 cases and from a later recurrence in 5 cases (median 15.3 months; range 2.4 to 17.2 months). The validated WES results for the 2 samples were concordant for all 8 patients (supplemental Table 4).
No somatic mutations were identified by WES in the lesions obtained from RDD (n = 4) or ECD (n = 1) patients. In contrast, a total of 17 somatic mutations were found in the 4 JXG lesions analyzed (median of 4 mutations per case, range 0 to 9 mutations) (supplemental Table 4). Although no somatic mutations of BRAF were identified in JXG lesions, a germ-line NF1 consensus splice site alteration was detected in a patient known to have a clinical diagnosis of neurofibromatosis type 1.
Functional analysis of MAPK pathway mutations
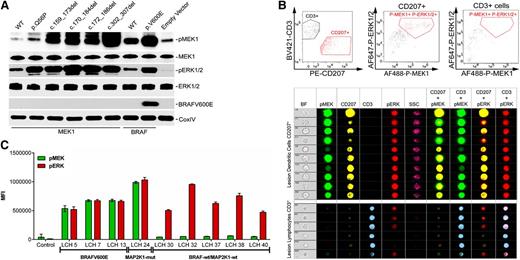
In silico analysis of the predicted 3-dimensional structures of wild-type and mutant MEK1 proteins revealed distinctive changes in the catalytic pocket and P loop of the mutant proteins (supplemental Figure 4). To assess the functional implications of the MAP2K1 mutations on the RAS-MAPK pathway, we analyzed the phosphorylation status of ERK1 and ERK2, the downstream targets of MAP2K1 and MAP2K2, in HEK293 cells transiently transfected with wild-type and mutant BRAF and MAP2K1 constructs (Figure 3). The ectopic expression of either of the deletion or point MAP2K1 mutants or BRAFV600E, but not the corresponding wild-type cDNAs, resulted in ERK1/2 phosphorylation, indicating that the mutants are constitutively active (Figure 3A). This was corroborated by imaging flow cytometry (IFC) analysis demonstrating increased MEK1 and ERK1/2 phosphorylation at steady-state in LCH lesion CD207+ cells (c.302_307del mutation) relative to control CD3+ cells from the same lesion (Figure 3B). IFC analyses on BRAFV600E mutated LCH patient samples also revealed high levels of steady-state phosphorylation of MEK1 and ERK1/2 in CD207+ cells purified from several different LCH lesions (Figure 3C). By comparison, in patient samples where no MAPK pathway gene mutations were detected in this series, MEK1 phosphorylation was variable, but ERK1/2 phosphorylation levels remained universally high (Figure 3C).
Functional characterization of the MAP2K1 mutants. (A) HEK293 cells were transiently transfected with expression plasmids encoding the indicated MAP2K1 and BRAF wild-type and mutant cDNAs, and corresponding lysates from cells maintained in serum were subjected to immunoblotting with the indicated antibodies. (B) Identification of functional implication of MAP2K1 mutations by IFC. Single cell suspension of LCH lesions were stained with Brilliant Violet (BV) 421-conjugated CD3, phycoerythrin (PE)-CD207, Alexa Fluor (AF) 488-phospho-MEK1, and AF647-phospho-ERK1/2 antibodies and analyzed by the gating strategy described in “Methods.” The top panels depict the distinctive CD3+ and CD207+ population sequestration (left) and overall p-MEK1+p-ERK1/2+ CD207+ (middle) or CD3+ (right) cells obtained by gating on 10 000 events. The middle and bottom panels show the representative images of random single cell events of 10 (138 through 2061) or 6 (308 through 4600) in CD207+ and CD3+ cells, respectively, identified by gating events with intermediate bright field (BF) area and BF aspect ratios close to 1. The indicated populations were then analyzed for coexpression of p-ERK1/2 or p-MEK1 in the CD207+ or CD3+ cells in the indicated channels. (C) Median fluorescent intensity (MFI) of p-MEK1 or p-ERK1/2 in CD207+ cells determined through IFC in patients harboring BRAFV600E (patients LCH5, LCH7, and LCH13) and MAP2K1 c.302_307del (patient LCH24) mutations, and patients with no somatic mutations (LCH30, LCH32, LCH37, LCH38, and LCH40).
Functional characterization of the MAP2K1 mutants. (A) HEK293 cells were transiently transfected with expression plasmids encoding the indicated MAP2K1 and BRAF wild-type and mutant cDNAs, and corresponding lysates from cells maintained in serum were subjected to immunoblotting with the indicated antibodies. (B) Identification of functional implication of MAP2K1 mutations by IFC. Single cell suspension of LCH lesions were stained with Brilliant Violet (BV) 421-conjugated CD3, phycoerythrin (PE)-CD207, Alexa Fluor (AF) 488-phospho-MEK1, and AF647-phospho-ERK1/2 antibodies and analyzed by the gating strategy described in “Methods.” The top panels depict the distinctive CD3+ and CD207+ population sequestration (left) and overall p-MEK1+p-ERK1/2+ CD207+ (middle) or CD3+ (right) cells obtained by gating on 10 000 events. The middle and bottom panels show the representative images of random single cell events of 10 (138 through 2061) or 6 (308 through 4600) in CD207+ and CD3+ cells, respectively, identified by gating events with intermediate bright field (BF) area and BF aspect ratios close to 1. The indicated populations were then analyzed for coexpression of p-ERK1/2 or p-MEK1 in the CD207+ or CD3+ cells in the indicated channels. (C) Median fluorescent intensity (MFI) of p-MEK1 or p-ERK1/2 in CD207+ cells determined through IFC in patients harboring BRAFV600E (patients LCH5, LCH7, and LCH13) and MAP2K1 c.302_307del (patient LCH24) mutations, and patients with no somatic mutations (LCH30, LCH32, LCH37, LCH38, and LCH40).
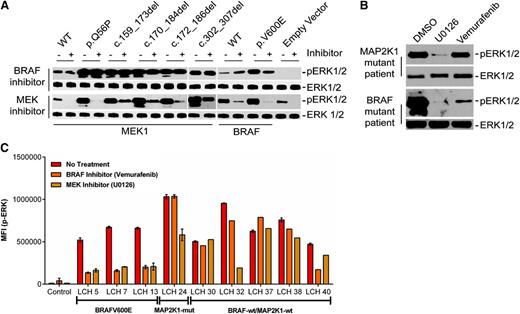
We next assessed the ability of MAPK inhibition to suppress pathway activation by LCH mutations with a small molecule BRAF inhibitor, vemurafenib, and the MEK inhibitor, U0126. (Figure 4). ERK1/2 phosphorylation was inhibited in both HEK293 cells transiently transfected with plasmids encoding BRAFV600E as well as BRAFV600E LCH lesion CD207+ cells by both U0126 and vemurafenib. By contrast, only U0126 decreased constitutive ERK1/2 phosphorylation by the ectopically expressed mutant MAP2K1 proteins. Similarly, U0126, but not vemurafenib, inhibited ERK phosphorylation in CD207+ cells from the MAP2K1 mutant patient sample (Figure 4A-B). IFC analyses further revealed that the samples in which no known mutation was identified exhibited variable responses to BRAF and MEK inhibition, in contrast to predicted responses from BRAF and MAP2K1 mutant samples (Figure 4C).26
Specific LCH mutations have differential sensitivity to BRAF and MEK inhibitors. (A,B) HE293 cells transiently transfected with expression plasmids encoding various cDNAs (A) or c.302_307del MAP2K1 lesion biopsy cell suspension (B) were treated with BRAF or MEK inhibitor for 1 hour, and corresponding lysates were subjected to immunoblotting with the indicated antibodies. (C) Median fluorescent intensity (MFI) of p-ERK1/2 in CD207+ cells determined through IFC analyses in patients harboring BRAFV600E (patients LCH5, LCH7, and LCH13) and MAP2K1 c.302_307del (patient LCH24) mutations, and patients with no somatic mutations (LCH30, LCH32, LCH37, LCH38, and LCH40) posttreatment with BRAF or MEK inhibitor.
Specific LCH mutations have differential sensitivity to BRAF and MEK inhibitors. (A,B) HE293 cells transiently transfected with expression plasmids encoding various cDNAs (A) or c.302_307del MAP2K1 lesion biopsy cell suspension (B) were treated with BRAF or MEK inhibitor for 1 hour, and corresponding lysates were subjected to immunoblotting with the indicated antibodies. (C) Median fluorescent intensity (MFI) of p-ERK1/2 in CD207+ cells determined through IFC analyses in patients harboring BRAFV600E (patients LCH5, LCH7, and LCH13) and MAP2K1 c.302_307del (patient LCH24) mutations, and patients with no somatic mutations (LCH30, LCH32, LCH37, LCH38, and LCH40) posttreatment with BRAF or MEK inhibitor.
Clinical correlations with somatic LCH mutations
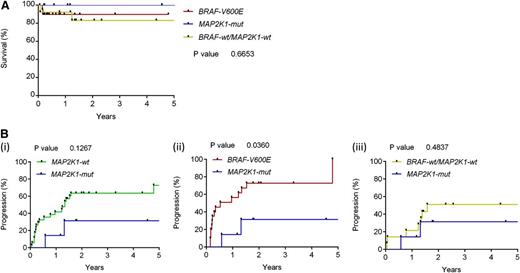
For this LCH cohort, the 3-year overall survival (OS) was 86.6% and 3-year progression-free survival was 41.7%. (Figure 5A). Genotype-specific progression-free survival for patients in this study is described in Figure 5B. In this series, there was no statistical increase or decrease in risk of recurrence in the MAP2K1 mutation group compared with the aggregate population. Patients with the BRAFV600E mutation were almost 3 times as likely to have a recurrence compared with those without the mutation, even after adjusting for age and gender (adjusted hazard ratio 2.9; P = .015; 95% CI, 1.2-6.8).1 No significant difference in OS because of BRAFV600E or MAP2K1 mutation was observed, and no significant differences in gender, age, presence of risk organ disease, or number of LCH lesions were noted when comparing genotypes (Table 2).
OS and disease progression of LCH patients in this series by tumor genotype. (A) Kaplan-Meier analysis of overall survival stratified by presence of BRAFV600E, MAP2K1 mutation, or neither mutation (P < .66). (B) Percentage with progression or recurrence: MAP2K1-wt vs MAP2K1-mut, P < .1267 (i); BRAFV600E vs MAP2K1-mut, P < .0360 (ii); and BRAF-wt/MAP2K1-wt vs MAP2K1-mut, P < .4837 (iii).
OS and disease progression of LCH patients in this series by tumor genotype. (A) Kaplan-Meier analysis of overall survival stratified by presence of BRAFV600E, MAP2K1 mutation, or neither mutation (P < .66). (B) Percentage with progression or recurrence: MAP2K1-wt vs MAP2K1-mut, P < .1267 (i); BRAFV600E vs MAP2K1-mut, P < .0360 (ii); and BRAF-wt/MAP2K1-wt vs MAP2K1-mut, P < .4837 (iii).
LCH patient characteristics
| . | Total (N = 41) N (%) . | BRAFV600E (N = 20) N (%) . | MAP2K1 mutation (N = 7) N (%) . | Neither mutation (N = 14) N (%) . | P . |
|---|---|---|---|---|---|
| Male | 22 (54) | 11 (55) | 3 (43) | 8 (57) | .814 |
| Female | 19 (46) | 9 (45) | 4 (57) | 6 (43) | |
| Age <2 y | 16 (39) | 9 (45) | 3 (43) | 4 (28) | .643 |
| Age 2-8 y | 16 (39) | 8 (40) | 3 (43) | 5 (36) | |
| Age >8 y | 9 (22) | 3 (15) | 1 (14) | 5 (36) | |
| Risk organ(+) | 8 (20) | 5 (25) | 1 (14) | 2 (14) | .687 |
| Risk organ(−) | 33 (80) | 15 (75) | 6 (86) | 12 (86) | |
| Multiple lesions | 27 (66) | 15 (75) | 3 (43) | 9 (64) | .300 |
| Single lesion | 14 (34) | 5 (25) | 4 (57) | 5 (36) |
| . | Total (N = 41) N (%) . | BRAFV600E (N = 20) N (%) . | MAP2K1 mutation (N = 7) N (%) . | Neither mutation (N = 14) N (%) . | P . |
|---|---|---|---|---|---|
| Male | 22 (54) | 11 (55) | 3 (43) | 8 (57) | .814 |
| Female | 19 (46) | 9 (45) | 4 (57) | 6 (43) | |
| Age <2 y | 16 (39) | 9 (45) | 3 (43) | 4 (28) | .643 |
| Age 2-8 y | 16 (39) | 8 (40) | 3 (43) | 5 (36) | |
| Age >8 y | 9 (22) | 3 (15) | 1 (14) | 5 (36) | |
| Risk organ(+) | 8 (20) | 5 (25) | 1 (14) | 2 (14) | .687 |
| Risk organ(−) | 33 (80) | 15 (75) | 6 (86) | 12 (86) | |
| Multiple lesions | 27 (66) | 15 (75) | 3 (43) | 9 (64) | .300 |
| Single lesion | 14 (34) | 5 (25) | 4 (57) | 5 (36) |
Discussion
The goal of this study was to use WES to define the landscape of somatic mutations in LCH and further characterize the key genes and pathways contributing to the pathogenesis of this disease. With rare exceptions, LCH lesions exhibit normal karyotypes and a paucity of gross chromosomal abnormalities.25 In addition, the cellular heterogeneity of LCH lesions previously limited the ability of Sanger sequencing methods to detect somatic mutations. More recently, the depth of sequencing coverage provided by next-generation sequencing technologies allowed the identification of somatic BRAFV600E mutations in the majority of LCH lesions,4 which has now been validated by several groups.1,5-7
Low mutation frequency in LCH
Our analysis revealed a remarkably low frequency of somatic mutations in LCH lesions with a median of 1 mutation per sample (0.03 mutations per Mb). This mutation rate is low even in comparison with those reported for other pediatric malignancies but is consistent with data for myeloproliferative neoplasms.27 In addition, analysis of the WES data as well as single nucleotide polymorphism arrays revealed a lack of CNAs. The absence of additional mutations observed in samples obtained from recurrent LCH lesions and the lack of correlation between mutation number and disease severity or death suggest that accumulation of novel mutations may not be a significant mechanism of disease progression in LCH, unlike other myeloproliferative neoplasms.28 Although it is possible that the heterogeneity of LCH lesions limited our ability to detect mutations by WES, no additional mutations were observed in the set of candidate genes sequenced at significantly greater depth using the targeted AmpliSeq panel or when sequencing CD207+ purified samples. The complete concordance of mutation calls between the paired LCH lesion samples sequenced provides further evidence for the validity of this WES analysis and suggests that biological reality rather than technical limitations underlies the low mutation rate identified in LCH lesions.
Identification of recurrent MAP2K1 mutations in LCH
The fact that LCH lesions harbor frequent somatic BRAFV600E mutations is now well-established.1,4-7 Although there have been case reports of other BRAF mutations (both somatic and germ line) in LCH patients as well as compound somatic ARAF mutations identified in a single patient, no recurrent mutations other than BRAFV600E were previously reported in LCH.7,29,30 A major finding of this study is the identification of recurrent activating somatic mutations in MAP2K1, the gene encoding MEK1 (a core MAPK pathway member downstream of BRAF), in patients with LCH. Somatic activating MAP2K1 mutations are rarer in human cancers than BRAF mutations but typically arise in the same spectrum of tumor types.31 MAP2K1 mutations are relatively uncommon in hematologic malignancies, although they have recently been described in ∼50% of HCLv cases.22 Like LCH, MAP2K1 and BRAF mutations appear to be mutually exclusive in HCL/ HCLv. By contrast, in malignant melanoma, MAP2K1 mutation may be present along with mutations in genes including BRAF at diagnosis or may develop as a resistance mechanism.32 Interestingly, germ-line mutations resulting in single amino acid substitutions in MAPK genes including MAP2K1 have been reported to occur in patients with CFC syndrome.33,34 CFC syndrome is characterized by craniofacial features, cardiac defects, ectodermal abnormalities, and developmental delay. Despite the hypothesized increase in ERK activation caused by CFC-associated mutations in BRAF and MAP2K1,33 there are no reports of CFC associated with LCH and few case reports of CFC associated with other malignancies.35-37
Characteristics of MAP2K1 mutations in LCH
Three observations regarding the MAP2K1 mutations identified by WES are particularly noteworthy. First, all mutations identified targeted exons 2 and 3 of the gene (Figure 2), which encode the negative regulatory domain, the P loop, and the catalytic core of MEK1 and are frequently targeted by somatic mutation in other malignancies, including melanoma and HCLv,22,31 and by germ-line mutation in CFC syndrome.33,34 Second, 6 of the 7 MAP2K1 mutations were in-frame deletions (Table 1), a rare mutation type that represents only 1.4% of total mutations in the COSMIC database and 3.5% of mutations reported for MAP2K1.20 In addition, 2 of these mutations were recurrent in our LCH cohort: p.Q58_E62del in 3 cases (resulting from 2 different mutations at the DNA level) and p.E102_I103del in 2 cases. Computer modeling supports the possibility that LCH-associated in-frame deletions around amino acids 53 to 62 could affect either the function of the negative regulatory region by disinhibiting enzymatic activity of MEK1. Similarly, mutations exposing the phosphate binding P loop could promote the constitutive activation (p.E102-I103 del) that was observed in this study in sample LCH24 (Figures 2-4 and supplemental Figure 4). Third, the MAP2K1 mutations identified were only seen in LCH lesions with wild-type BRAF, supporting the hypothesis that mutations in the 2 genes might serve as alternative mechanisms of MAPK pathway activation in LCH.
Functional effect of MAP2K1 mutations in LCH supports a model of universal ERK activation
Badalian-Very et al noted universal MEK and ERK activation in DCs within LCH lesions regardless of BRAF genotype based on immune fluorescence studies.4 The frequency of MAP2K1 mutations in LCH lesions (33% of BRAF wild-type lesions) and mutual exclusivity of these mutations with BRAFV600E support a generalized model of ERK activation in LCH pathogenesis. Further evidence for this concept was provided by ERK activation induced by LCH-associated mutations in vitro as well as universal ERK activation noted in primary lesion CD207+ cells by IFC (Figure 3C). Although not conclusive, the high BRAFV600E/MAP2K1 variant allele fractions observed in sorted CD207+ cells (supplemental Tables 4 and 5) support clonality of the pathological DCs bearing these somatic mutations. In fact, the vast majority of all somatic mutations identified in this series were highly enriched, supporting a model of genomically stable clones in LCH (supplemental Tables 4 and 5).
The prevalence of BRAFV600E mutations in LCH, as well as case reports of NRAS mutation in ECD, support general role for pathological MAPK signaling in histiocytic disorders.5,38,39 Similarly, JXG has been associated with neurofibromatosis and juvenile myelomonocytic leukemia, diseases defined by MAPK hyperactivity, as was observed in a case in this series (reviewed in Berres et al9 ). The increasingly frequent recognition of patients with lesions with distinct phenotypes (ECD in bone lesion and LCH in skin, for example) or with mixed LCH/JXG phenotype in a single lesion supports a common cell of origin with potential to differentiate into a range of terminal phenotypes. Of note, BRAFV600E mutations were identified in 2 mixed LCH/JXG phenotype cases. This series also included 1 case of LCH/ECD (BRAFV600E along with an ARAF mutation), 1 with an ERBB3 mutation, and a third with no MAPK mutations identified. Although there were only 4 cases in this series, it is notable that no mutations were identified in any case of RDD. Additional samples will be required to determine if somatic mutations may play a role in RDD.
MAPK pathway gene mutations other than BRAFV600E and MAP2K1 may also directly or indirectly impact the MAPK pathway in LCH (supplemental Table 4). The in vitro assays with purified CD207+ cells demonstrate that BRAFV600E and MAP2K1 LCH DCs respond to MEK and ERK inhibition in a predictable pattern. Unlike the Badalian-Very series,4 we found that in some cases MEK was relatively inactive. The samples in which no known mutation was identified demonstrated decreased baseline MEK activation relative to BRAFV600E and MAP2K1(c.302_307del) mutation cases as well as variable responses to BRAF and MEK inhibition (Figures 3C and 4C). The potential for alternative pathways to impact pathogenesis in some patients is also supported by variable clinical responses to protein kinase B inhibition.40
Etiology of LCH in nonmutated cases and future directions for research
The etiology of LCH in patients without observed BRAF or MAP2K1 mutations remains an unanswered question. Based on the observed in vitro activation of ERK in cases without identified BRAF or MAP2K1 mutations, we hypothesize that LCH is a disease characterized by universal activation of the MAPK pathway through a variety of genetic alterations, as in pilocytic astrocyoma,41 and that genetic alterations not detectable by WES remain to be identified in these lesions. Our WES analysis did reveal single LCH cases with potentially ERK-activating somatic mutations in ERBB3 and ARAF,42,43 but their functional significance remains to be proved. A larger series of LCH patients will be required to define the frequency of these rare mutations in MAPK pathway members. In addition, complementary methods of genomic analysis (eg, whole genome sequencing, RNA sequencing, and methylation studies) will be necessary to identify MAPK pathway alterations not captured by WES.
Clinical implications of LCH genotype
It is becoming increasingly clear that LCH represents a spectrum of conditions with a common histologic end point. A model is emerging in which clinical manifestations may be defined by both the specific mutation as well as the cell in which an ERK-activating genetic lesion arises.9 We previously observed that BRAFV600E confers a relative increase in risk of initial treatment failure using current therapies1 (note that most LCH patients were analyzed in both studies). Although the relatively small number of patients with MAP2K1 mutations, retrospective data collection, and heterogeneous patient population and treatment history limit the ability to generalize these findings, a novel observation in this cohort is the lack of increase in risk in initial treatment failure associated with MAP2K1 mutations compared with the aggregate, but a relative decrease in risk compared with BRAF-V600E (Figure 5B). These preliminary data will require validation through analysis of larger data sets of similarly treated LCH patients and emphasize the importance of collecting genotype data in future prospective trials to determine relative risks of treatment failure and responses to therapy. Our observation of differential responses to MEK and ERK inhibition in CD207+ cells of different genotypes (BRAFV600E, MAP2K1 mutant, or no MAPK pathway mutation identified) also has potentially significant clinical implications for the use of MAPK-directed therapies in patients with LCH. Additional functional studies in mouse models and a larger series of primary human LCH tumors will be required to determine the mutation-specific impact of ERK hyperactivation. As targeted agents make their way through early phase pediatric testing, the mutation status of patients with LCH will likely become central to therapeutic strategies; this study suggests that genotyping of LCH lesions may predict individual responses to specific therapies (Figure 4C).
In conclusion, this study defines LCH as a myeloid neoplasia with extremely low overall frequency of somatic mutations detectable by WES. The majority of cases have mutually exclusive activating ERK mutations, including BRAFV600E and MAP2K1 mutations. The low frequency of mutations and lack of novel mutations in serial samples is supportive of a model of pathogenesis in which the specific ERK-activating mutation and state of myeloid differentiation in which the cell arises define clinical manifestations of the disease.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Munu Bilgi, Susan Pilat, and Elizabeth Pacheco (Baylor College of Medicine, Texas Children’s Cancer Center) for data management support, and Aurora Alainis for technical support (Baylor College of Medicine, Texas Children’s Cancer Center).
This study was supported in part by funding from the Lester and Sue Smith Foundation, the Texas Children's Hospital Pediatric Center for Personal Cancer Genomics & Therapeutics, and the HistioCure Foundation (Texas Children's Hospital Histiocytosis Program); and by grants from the National Institutes of Health (R01 CA154489) (C.E.A., K.L.M.), National Institutes of Health National Cancer Institute Specialized Program of Research Excellence in Lymphoma (P50CA126752) (C.E.A.), National Institutes of Health National Human Genome Research Institute (U54 HG003273) (D.A.W.), National Institutes of Health National Cancer Institute and National Institute of Allergy and Infectious Diseases (R01 CA154947A, AI10008, AI089987) (M.M.), the German Research Association (Deutsche Forschungsgemeinschaft, BE 4818/1-1) (M.-L.B.), National Institutes of Health National Cancer Institute K12 (CA090433) (S.J.S.), and Howard Hughes Medical Institute to the Baylor College of Medicine Med into Grad Initiative (K.P.H.L.). We also appreciate the support of shared resources by a Dan L. Duncan Cancer Center support grant sponsored by the National Institutes of Health National Cancer Institute (P30CA125123).
Authorship
Contribution: R.C. and O.A.H. designed and performed research, collected data, analyzed and interpreted data, and wrote the manuscript; X.S. analyzed data; S.J.S. analyzed and interpreted data and wrote the manuscript; A.S., H.A., K.P.H.L., and K.R.C. performed research, collected data, and analyzed and interpreted data; L.T. designed and performed research, collected data, and analyzed and interpreted data; D.M.M., H.V.D., and J.H. performed research and collected data; N.D. designed and performed research and analyzed and interpreted data; L.W. designed and performed research, collected data, and analyzed and interpreted data; P.J.L. performed statistical analyses; M.J.H. analyzed and interpreted data; D.L.B. and K.C.D. performed research and analyzed and interpreted data; M.-L.B. and P.I.P. analyzed and interpreted data and wrote manuscript; and M.M., K.L.M., D.A.W., C.E.A., and D.W.P. designed research, analyzed and interpreted data, and wrote manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Carl Allen, Texas Children’s Hospital, 1102 Bates St; Suite 750.06, Houston, TX 77030; e-mail: ceallen@txch.org; and D. Williams Parsons, Texas Children’s Hospital, 1102 Bates St, Suite 1030.15, Houston, TX 77030; e-mail: dwparson@txch.org.
References
Author notes
R.C. and O.A.H. contributed equally to this study.