Abstract
Diamond-Blackfan anemia, Shwachman-Diamond syndrome, and dyskeratosis congenita are inherited syndromes characterized by marrow failure, congenital anomalies, and cancer predisposition. Genetic and molecular studies have uncovered distinct abnormalities in ribosome biogenesis underlying each of these 3 disorders. How defects in ribosomes, the essential organelles required for protein biosynthesis in all cells, cause tissue-specific abnormalities in human disease remains a question of fundamental scientific and medical importance. Here we review the overlapping and distinct clinical features of these 3 syndromes and discuss current knowledge regarding the ribosomal pathways disrupted in each of these disorders. We also explore the increasing complexity of ribosome biology and how this informs our understanding of developmental biology and human disease.
Introduction
The study of inherited bone marrow failure syndromes has yielded unique insights into global molecular pathways regulating hematopoiesis and clonal evolution. These clinically heterogeneous syndromes are all characterized by marrow failure, frequent physical anomalies, and cancer predisposition.1 Impairment of ribosome biogenesis is emerging as a common molecular pathogenic mechanism underlying many of these marrow failure syndromes. Ribosomes are ribonucleoprotein complexes that catalyze protein synthesis by translating the mRNA message into its cognate protein product. This basic cellular function is required by all cells and is essential for life. Previously, ribosomes had been largely relegated to the status of passive cellular drones in the ranks of the protein translational corps. The unexpected revelation that disruption of such an essential cellular function could preferentially affect specific tissues in human disease stimulated a reexamination of ribosome biology. The relevance of ribosome pathology for more common diseases was highlighted by the identification of acquired somatic mutations affecting ribosomal proteins in myelodysplastic syndrome (MDS) and leukemias arising in the general population. The question of how ribosomal abnormalities cause marrow failure and cancer predisposition is of fundamental biological and clinical interest.
In this review, we discuss the clinical, genetic, and molecular features of marrow failure ribosomopathies (Figure 1) and explore the clinical implications of recent advances in our understanding of ribosomal functions in cellular and developmental biology. There are prior excellent reviews for information regarding other nonhematologic ribosomal diseases.
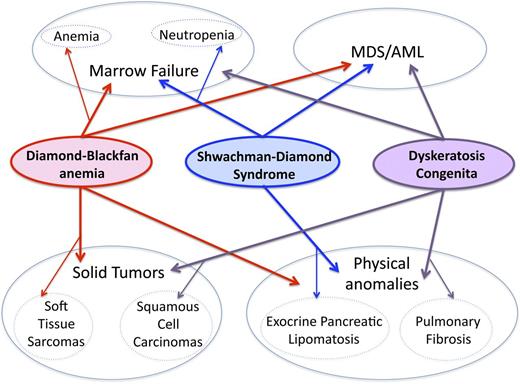
Overlapping and distinct clinical features of inherited marrow failure ribosomopathies. DBA, SDS, and DC are all characterized by marrow failure, predisposition to MDS/AML, and congenital anomalies. The primary feature of marrow failure in DBA is red-cell aplasia, although other hematologic lineages may also be variably affected. Although neutropenia is the most common feature of marrow failure in SDS, all 3 lineages may be depressed. Cellular and humoral immunologic abnormalities have been reported in DC and SDS. The spectrum of physical anomalies in these 3 syndromes shares both overlapping and distinct features.1 Exocrine pancreatic lipomatosis is characteristic of SDS, whereas pulmonary fibrosis is a common characteristic of DC. The risk of soft tissue sarcomas is increased in DBA, and the risk of squamous cell carcinomas of the oropharynx and gastrointestinal tract is elevated in DC. Data are insufficient to determine whether solid tumor risk is elevated in SDS. More detailed descriptions of clinical phenotypes have been reviewed.1
Overlapping and distinct clinical features of inherited marrow failure ribosomopathies. DBA, SDS, and DC are all characterized by marrow failure, predisposition to MDS/AML, and congenital anomalies. The primary feature of marrow failure in DBA is red-cell aplasia, although other hematologic lineages may also be variably affected. Although neutropenia is the most common feature of marrow failure in SDS, all 3 lineages may be depressed. Cellular and humoral immunologic abnormalities have been reported in DC and SDS. The spectrum of physical anomalies in these 3 syndromes shares both overlapping and distinct features.1 Exocrine pancreatic lipomatosis is characteristic of SDS, whereas pulmonary fibrosis is a common characteristic of DC. The risk of soft tissue sarcomas is increased in DBA, and the risk of squamous cell carcinomas of the oropharynx and gastrointestinal tract is elevated in DC. Data are insufficient to determine whether solid tumor risk is elevated in SDS. More detailed descriptions of clinical phenotypes have been reviewed.1
Overview of ribosome biogenesis and protein synthesis
Ribosome biogenesis is a highly-regulated, complex process.2 Eukaryotic ribosomes are comprised of 2 subunits: the small 40S subunit and the large 60S subunit. These subunits join together to form the active 80S ribosome. Ribosomes contain 4 structural ribosomal RNAs (rRNAs). The 40S subunit contains the 18S rRNA, and the large 60S subunit contains the 28S, 5.8S, and 5S rRNAs. These rRNAs are complexed with approximately 79 ribosomal proteins in eukaryotic ribosomes. Ribosome assembly is a complex and highly regulated process using a significant investment of cellular biosynthetic energy. More than 200 assembly factors and small nucleolar RNAs (snoRNAs) are required to synthesize ribosomes.
In the nucleolus, ribosomal RNA is initially transcribed into a long 47S pre-rRNA by RNA polymerase I. 5S rRNA is transcribed separately by RNA polymerase III. During this process, ribosomal proteins, nonribosomal assembly/processing factors, and snoRNAs associate with the nascent pre-rRNA to form the 90S preribosomal particle. The binding of ribosomal proteins to the maturing pre-ribosomal RNA is required for rRNA processing. The 47S pre-rRNA undergoes a series of carefully orchestrated cleavage and modification events, including methylation and pseudouridylation, to form the pre-40S and pre-60S precursor particles. These subunits are exported to the cytoplasm, where additional maturation steps take place before they are assembled in the active translating ribosomes. The most general mechanism of translation initiation, cap-dependent translation, requires the assembly of a complex of proteins, known as eukaryotic initiation factors (eIFs), to recruit the 40S ribosomal subunit on the 5′ end (5′cap structure or m7GTP) of the mRNA. The 40S bound to the ternary complex (eIF2-GTP-initiator methionyl tRNA [Met-tRNAiMet]) forms the 43S preinitiation complex and, with the help of the eIFs, scans the 5′ untranslated region (UTR) in search of a start codon (AUG). After scanning, the AUG codon base pairs with the anticodon of the initiator tRNA in a site of the ribosomal subunit known as P-site. This complex is joined by the 60S subunit to form an 80S ribosome competent for translation elongation. This is the mechanism engaged by the majority of mRNAs to initiate translation. Internal ribosome entry site (IRES)-dependent translation is an alternative mode of translation initiation and follows an RNA-based mechanism whereby the 40S is recruited directly by the IRES element independent of some, or even all, of the eIFs involved in the cap-dependent translation. The mechanism by which translation control is perturbed in ribosomopathies still remains an outstanding question.
Diamond-Blackfan anemia: clinical features
Diamond-Blackfan anemia (DBA) classically presents with red-cell aplasia in the first year of life, with a median age of presentation of 2 months, though, rarely, presentation may be delayed into adulthood.3 It remains unclear why DBA patients generally do not have development of severe anemia in utero during fetal development.4,5 The normochromic anemia is typically macrocytic with reticulocytopenia. The bone marrow is classically normocellular, with a paucity of erythroid precursors. The erythroid burst-forming units (BFU-E) and erythroid colony-forming units (CFU-E) in vitro are severely reduced, with relative sparing of the granulocyte-monocyte colony-forming units (CFU-GM).6 The erythrocyte adenosine deaminase levels are often elevated.7,8 Approximately 50% of DBA patients have physical anomalies such as craniofacial abnormalities, including clefting of the lip or palate, thumb abnormalities, cardiac malformations, and short stature.3 The risk of solid tumors, MDS, or leukemia is elevated in DBA. The cumulative incidence of malignancy was approximately 20% by age 46 years.9
Severe or symptomatic anemia is currently treated with corticosteroids or chronic red cell transfusions.10 Corticosteroids exert a cell-autonomous effect on hematopoietic stem cells to improve erythropoiesis.11 The mechanism whereby corticosteroids ameliorate anemia in DBA remains unclear. The subset of DBA patients who do not respond to corticosteroids or who require high doses with unacceptable toxicities receive chronic red cell transfusions. Transfusional iron overload remains a major cause of morbidity and mortality in DBA. Bone marrow transplant is the only curative treatment of marrow failure in DBA and is generally reserved for patients for whom corticosteroids or red cell transfusions are not viable options. A spontaneous remission rate of ∼20% by age 25 has been reported.3 The molecular pathophysiology of remission from anemia in DBA remains unknown, but such patients remain at risk for malignancy.
Diamond-Blackfan anemia: genetics
Heterozygous mutations resulting in haploinsufficiency have been identified for 11 genes encoding ribosomal proteins RPS19, RPL5, RPS10, RPL11, RPL35A, RPS26, RPS24, RPS17, RPS7, RPL26, and RPL15 (Figure 2).12 Mutations in ribosomal genes account for 60% to 70% of DBA cases. Of these ribosomal gene mutations, around 20% involve large deletions that require analysis of copy number variation for detection.13 Ribosomal gene mutations may be inherited in an autosomal dominant pattern or may arise spontaneously. X-linked mutations in the transcription factor GATA1, which is essential for erythropoiesis, have also been associated with DBA.14 Overall, it is estimated that approximately 35% of DBA patients remain as yet genetically undefined.
Mutations of key ribosome components underlying ribosomopathies. The DKC1 gene encoding dyskerin (rRNA pseudouridine synthase) is frequently mutated in X-DC. The SBDS gene encodes the SDBS protein and is found to be mutated in SDS. RP genes encoding ribosomal proteins belonging to both the large and small ribosomal subunits are found to be mutated in DBA.
Mutations of key ribosome components underlying ribosomopathies. The DKC1 gene encoding dyskerin (rRNA pseudouridine synthase) is frequently mutated in X-DC. The SBDS gene encodes the SDBS protein and is found to be mutated in SDS. RP genes encoding ribosomal proteins belonging to both the large and small ribosomal subunits are found to be mutated in DBA.
Diamond-Blackfan anemia: molecular pathophysiology
In general, the heterozygous DBA mutations result in loss of function in a single copy of a ribosomal protein gene. A dominant-negative mechanism has also been demonstrated in a murine model of DBA.15 The large number of ribosomal proteins mutated in DBA fail to cluster in any specific region of the ribosome. Haploinsufficiency or reduced expression of a ribosomal protein results in decreased levels of the cognate 40S or 60S subunit and a defect in processing of the ribosomal RNA precursor.16-19 The effect of decreased ribosomal protein activity in vivo and in a tissue-specific manner is still poorly understood. Interestingly, induced pluripotent stem cells derived from DBA patients recapitulate the aberrant rRNA processing, decreased ribosomal subunit levels, and impaired hematopoiesis that characterize DBA.20 Insights from and limitations of animal models of DBA have been recently reviewed.21
P53 activation
P53 activation has been observed in bone marrows from DBA patients, after depletion of ribosomal proteins in human erythroid progenitor cells, and in murine models.22,23 P53 is a stress response gene whose activation results in apoptosis or cell cycle arrest and appears to contribute to the pathogenesis of marrow failure in DBA. Because ribosomal protein haploinsufficiency results in reduced ribosomal protein gene expression in multiple different tissues in DBA, it remains to be demonstrated why erythroid precursors are particularly sensitive to p53 induction by ribosomal stress.24 What is the mechanism underlying the activation of P53 in DBA? The association of distinct ribosomal proteins with a critical regulator of P53 stability provides one working model. Specifically, several different ribosomal proteins, including RPS7, RPS14, RPL5, RPL11, RPL23, and RPL26, have been shown to interact with HDM2/MDM2.25,26 HDM2/MDM2 is a ubiquitin ligase that associates with p53 and targets p53 for degradation by proteosomes. Interaction of any of these ribosomal proteins with HDM2/MDM2 inhibits the ability of HDM2/MDM2 to target p53 for degradation with consequent stabilization and increased levels of p53, which in turn promotes apoptosis.25 Mice harboring an Mdm2C305F mutation that impairs Mdm2 binding to Rpl5/Rpl11 lost the p53 response to actinomycin D, an inducer of ribosome stress, while retaining a normal p53 response to DNA damage.27 The Mdm2C305F accelerated Myc-induced lymphomagenesis,27 suggesting that abrogation of the p53 response to ribosome stress provides a potential mechanism contributing to cancer predisposition. Advances in our understanding of ribosomal disorders in cancer have been recently reviewed.28
Clinical, genetic, and biochemical evidence points to a unique role for the ribosomal proteins RPL5 and RPL11. RPL5 and RPL11 appear to be central regulators of the MDM2/HDM2-p53 pathway. RPL5, RPL11, and the 5S ribosomal RNA (rRNA) form a nascent pre-ribosomal complex that coordinately inhibits HDM2 to upregulate p53.29 The formation of such a complex may explain the codependence of RPL5 and RPL11 on the activation of p53.30,31 DBA patients with RPL5 or RPL11 mutations are distinguished clinically by the higher incidence of congenital anomalies such as cleft palate and thumb abnormalities compared with other DBA patients.32 However, the observation that haploinsufficiency of RPL5 or RPL11 causes DBA indicates that additional pathways other than ribosomal stress activation of the HDM2-p53 pathway likely also contribute to DBA. Intriguingly, depletion of RPL5 or RPL11 impairs cell-cycle progression in a p53-independent manner.33,34
Mouse21 and zebrafish35 animal models also provide evidence for both p53-dependent and p53-independent pathways in DBA. Indeed, p53 activation appears to be a common downstream stress response pathway activated in response to a variety of different mechanisms underlying nonribosomal marrow failure syndromes such as anemia.36,37
Protein synthesis
The role of ribosomal proteins in gene expression at the translation level is still a key question in normal physiology and disease pathogenesis. In some DBA patients, global translation was reduced by 48% to 73% in lymphocytes.38 Furthermore, altered translation of 5′TOP mRNAs has been observed in response to deficiencies of 40S ribosomal subunit proteins, suggesting that translation of specific subsets of mRNAs might be preferentially altered by ribosomal abnormalities.39 Similarly, knockdown of Rps19 or Rpl11 by about 50% in fetal liver erythroblasts resulted in alterations in the polysome-associated mRNA pool, particularly for mRNAs containing an internal ribosome entry site (IRES) in their 5′UTR. Translational profiling of Epstein-Barr virus–immortalized lymphoblasts from DBA patients harboring mutations in RPS19 or RPL11 also showed differential polysome loading of a subset of transcripts.40 Differential translational control of specific mRNA subsets is an attractive hypothesis that provides a potential mechanism for tissue-specific phenotypes resulting from ribosomal protein gene haploinsufficiency.
mTOR signaling
The importance of translation control in DBA pathogenesis is also highlighted by recent evidence that the addition of the essential branched chain amino acid l-leucine to the lymphocytes of a subset of DBA patients increased global translation in vitro.38 A DBA patient with severe transfusion-dependent anemia received a trial of leucine with subsequent improvement in her hemoglobin levels and reticulocyte count free of transfusion support.41 Leucine also ameliorated the anemia and reduced p53 levels in a murine Rps19 model of DBA.42 Leucine treatment improved erythroid differentiation in zebrafish rps19 and rps14 morpholino knockdown models43 and in human CD34+ cells wherein RPS19 or RPS14 had been knocked down.43 These data suggest that the beneficial effect of l-leucine on DBA cells may be ascribed to its regulation of protein metabolism through activation of a key kinase known as mammalian target of rapacmycin (mTOR), a master regulator of cell growth and protein synthesis43-45 and the resulting stimulation of translation.46 The potential effect of leucine on protein translation of specific mRNAs in DBA has yet to be determined and clinical studies are ongoing.45
Acquired mutations in ribosomal protein genes: 5q– MDS
An acquired MDS characterized by a 5q– cytogenetic clonal abnormality shares clinical features with DBA.47 Clinically, 5q– syndrome presents with severe macrocytic anemia, thrombocytosis, and variable mild neutropenia. The bone marrow shows red cell aplasia and dysplastic hypolobulated small megakaryocytes. 5q– MDS typically follows a more indolent course, with a lower rate of progression to acute myelogenous leukemia (AML) compared with other MDS subtypes.
The common deleted region of 5q region contains the RPS14 gene, which encodes a protein subunit of the small 40S ribosome. A screen using small interfering RNAs to target each gene located within the common deleted region of 5q demonstrated that knockdown of RPS14 was sufficient to impair erythropoiesis, with relative sparing of the megakaryocytes, thus phenocopying 5q– syndrome.48 Introduction of the RPS14 gene rescued erythroid differentiation in primary patient bone marrow CD34+ cells.48 Additional genes and microRNAs contained within the 5q locus may additively contribute to the disease phenotype.47
The molecular pathophysiology of this acquired myelodysplastic syndrome involving haploinsufficiency of RPS14 shares overlap with that of DBA.49 Indeed, a small somatic deletion of 5q including RPS14 was identified in a 5-year-old girl who carried the clinical diagnosis of nonclassical DBA.50 Knockdown of RPS14 resulted in defective pre-18S rRNA processing and decreased levels of the 40S small ribosomal subunit.48 A mouse model haploinsufficient for the Cd74 – Nid67 regions syntenic to the commonly deleted region of 5q– resulted in a macrocytic anemia and dysplastic megakaryocytes.51 Elevated levels of p53 were noted in the bone marrows of the Cd74 – Nid67 haploinsufficient mice. Co-deletion of p53 rescued erythropoiesis in vitro. Altogether, these findings in 5q– MDS, wherein an acquired deletion of RPS14 causes erythroid failure, support the notion that a ribosomal protein gene mutation exerts an effect intrinsic to the hematopoietic stem cell.
Shwachman-Diamond syndrome: clinical features
Shwachman-Diamond syndrome (SDS) is classically characterized by marrow failure and exocrine pancreatic dysfunction.52,53 Patients typically present early in life with neutropenia and steatorrhea; however, a subset of patients have atypical or cryptic clinical presentations.54 Although neutropenia is the most common hematologic abnormality, anemia and thrombocytopenia are also common. Severe aplastic anemia may develop in a subset of patients. The bone marrow is typically mildly dysplastic and hypocellular. Abnormalities of neutrophil chemotaxis have been reported,55,56 though patients are able to form purulent abscesses and empyemas.57 Neutrophil respiratory burst activity is normal.58 Immunologic abnormalities of B- and T-cell numbers and function have been reported.59 SDS is a multisystem disorder with variable abnormalities in other organs including the skeletal, neurocognitive, endocrine, and cardiac systems.53 A variety of congenital anomalies have been reported.54 SDS patients are at increased risk for clonal cytogenetic abnormalities, MDS, and AML.60,61 The prognosis of leukemia in SDS is generally poor. Although early-onset solid tumors have been described in patients carrying the diagnosis of SDS,62-65 data are insufficient to determine whether SDS patients are at increased risk for solid tumors.
Hematologic complications such as severe or symptomatic marrow failure, MDS, or AML are treated with hematopoietic stem cell transplant. SDS patients are at increased risk for treatment-related complications, but outcomes with reduced-intensity conditioning regimens are promising.53 Supportive measures to manage cytopenias include granulocyte colony-stimulating factor for neutropenia and transfusion support of red cells or platelets. Exocrine pancreatic insufficiency is treated with pancreatic enzyme supplements.
Shwachman-Diamond syndrome: genetics
SDS is inherited in an autosomal recessive fashion. Approximately 90% of patients with the clinical features of SDS harbor biallelic mutations in the SBDS gene.66 Most SBDS mutations correspond to sequences found in an adjacent highly homologous pseudogene and likely result from gene conversion. The pseudogene contains mutations that disrupt protein production. The SBDS gene encodes an evolutionarily conserved protein and is expressed across a broad range of tissues. Although aplastic anemia has been described in a few patients harboring only a single SBDS mutation, it is currently unclear whether an increased risk of aplastic anemia is seen in obligate SBDS heterozygous carriers such as parents or grandparents.
Shwachman-Diamond syndrome: molecular pathophysiology
Knockdown of SBDS in hematopoietic cells impairs proliferation and hematopoietic colony formation.67-69 iPSCs derived from SDS patients manifest deficits in exocrine pancreatic and hematopoietic differentiation and enhanced apoptosis.70 Deletion of murine Sbds resulted in early embryonic lethality.71 Targeted knockdown of Sbds in mouse osteoprogenitor cells resulted in leukopenia and lymphopenia and dysplasias of the neutrophils and megakaryocytes.72 Observations of in vitro assays for abnormalities of marrow stroma from SDS patients have varied.73,74 A variety of cellular phenotypes has been observed in SDS, including mitotic spindle destabilization,75,76 Fas ligand–induced apoptosis,77 heightened cellular stress responses78 and Rac2-mediated monocyte migration,79 decreased mitochondrial membrane potential and oxygen consumption, and increased the production of reactive oxygen species.80,81
SBDS promotes the release of EIF6 from the pre-60S ribosome, which is required for the formation of a mature 80S functional ribosome82-84 (Figure 2). Human SBDS associates with the 60S large ribosomal subunit but not with mature polysomal ribosomes.85 Yeast deficient for Sdo1, the ortholog of SBDS, results in a slow-growth phenotype that is suppressed by Tif6 mutations that impair Tif6 binding to 60S ribosomes.82 Tif6, whose mammalian homolog is EIF6, is implicated in the maturation and nuclear export of the 60S subunit86,87 and sterically prevents premature joining of the 60S subunit to the 40S subunit.88-90 Complete abrogation of Sbds expression in animal models produces polysome profiles with half-mers, a pattern that arises when 40S subunits have not associated with the 60S subunit, and is consistent with a defect in ribosome joining.83 Half-mers have not been observed in cells from SDS patients, likely because of the retention of some scant SBDS expression in SDS patients.91 SDS patient cells exhibit impaired ribosome association in vitro.91 This observation was recapitulated with SBDS knockdown. The ribosome association defect was rescued by the expression of wild-type but not mutant SBDS cDNA. Knockdown of EIF6 improved ribosome association in SDS patient cells but did not improve hematopoietic colony formation of SBDS-deficient CD34+ cells.91
The release of eIF6 from pre-60S subunits was catalyzed in vitro by the addition of SBDS, EFL1, and GTP.83 Introduction of pathogenic SBDS mutations impaired EIF6 release. SBDS stimulated the GTPase activity of EFL1 in vitro. Kinetic studies of EFL1 in the presence or absence of SBDS suggest that SBDS may stabilize the binding of GTP to EFL1.92 Structural studies of the SBDS protein revealed a conserved internal flexible hinge region allowing rotation of the SBDS amino terminal domain.83 A direct interaction between recombinant SBDS and EFL1 alters the conformation of the interacting domain of EFL1.93 These experiments support a model wherein SBDS conformational changes couple ELF1 GTPase activity to EIF6 release from the pre-60S subunit.
An important role in malignancy for ribosomal maturation defects akin to those in SDS was highlighted by the demonstration in pediatric T-cell acute lymphoblastic leukemia of recurrent acquired mutations of arginine 98 or histidine 123 in RPL10.94 Expression of these RPL10 mutations resulted in reduced proliferation, impaired ribosome biogenesis, and abnormal nuclear accumulation of Nmd3 and Tif6 in yeast. These defects were ameliorated by introducing a mutation in Nmd3 that weakened its binding to the ribosome, suggesting that the RPL10 mutations affected the release of Nmd3 and Tif6.
Knockdown of SBDS expression using shRNAs in HEK293 cells resulted in alterations in mRNA transcript levels and mRNA polysome-loading, suggesting that disruption of ribosome biogenesis by SBDS may affect translation of specific mRNA subsets.95 Yeast models, wherein ribosomal joining is impaired by decreased levels of 60S subunits, manifest altered translation of specific mRNAs.96 Why distinct clinical phenotypes arise from mutations in ribosomal protein genes vs genes affecting ribosome assembly remains an intriguing question.
Dyskeratosis congenita: clinical features
Dyskeratosis congenita (DC) is an inherited marrow failure syndrome whose diagnosis was originally based on the clinical triad of dystrophic nails, mucosal leukoplakia, and a reticular or mottled rash.97 The phenotype of DC is now recognized to be broad, with many atypical or cryptic presentations. DC patients may also affect additional organ systems including the pulmonary, gastrointestinal, skeletal, neurologic, immunologic, and ophthalmologic systems.1,98 Congenital physical anomalies are also common. Cytopenias may involve any or all of the hematologic lineages. The bone marrow is typically hypocellular, with mild dysplasias at baseline. Patients with DC are at increased risk for MDS, AML, and solid tumors, namely adenocarcinomas of the head and neck and gastrointestinal tract.99
Marrow failure may respond to androgens such as oxymetholone. Androgens have been reported to stimulate telomerase.100-102 Premature telomere shortening is a key feature of DC.97 Hematopoietic stem cell transplant is the only curative therapy for the hematologic complications of DC, but it does not treat the life-threatening complications of other organ systems such as pulmonary fibrosis.103
Dyskeratosis congenita: genetics
To date, 9 genes have been reported to cause DC: DKC1, TINF2, TERC, TERT, WRAP53 (TCAB1), CTC1, RTEL1, NHP2, and NOP10.97 The mode of inheritance may be autosomal dominant (TINF2, TERC, TERT), autosomal recessive (TERT, WRAP53/TCAB1, CTC1, RTEL1, NHP2, NOP10), or X-linked (DKC1). Mutations in these 9 genes account for approximately 50% of patients who fulfill clinical criteria for DC.
Dyskeratosis congenita: molecular pathophysiology
Although disruption of telomere maintenance is central to the pathogenesis of DC, a subset of genes found mutated in DC plays an important role in both ribosome and telomere activity.104-106 Interestingly, approximately half of the genes mutated in DC (DKC1,NHP2,NOP10, and WRAP53/TCAB1) encode for proteins associated with abundant regulatory noncoding RNAs (ncRNAs) termed H/ACA small RNAs.107 H/ACA small RNAs lie at the nexus of several important RNA-based cellular processes implicated in telomere, ribosome, and splicing biology.107,108 One well-characterized example of an H/ACA small RNA found mutated and deregulated in several forms of DC is the telomerase RNA component (TERC) that plays an essential role in telomere maintenance.105,109 An outcome of deregulated TERC is the age-dependent shortening of telomeres, which is one common feature of DC.110 Importantly, the evolutionary conserved RNA-binding proteins encoded by DKC1,NHP2,NOP10, and WRAP53/TCAB1) associate with hundreds of additional H/ACA small RNAs to form H/ACA small ribonucleoprotein (RNP) complexes involved in modifying ribosomal RNA (rRNA) and small nuclear RNA (snRNA).107,111,112 Therefore, in addition to a role in telomere maintenance mediated by the stabilization of TERC, a large subset of genes mutated in DC exhibit additional RNA-based cellular functions.
Importantly, the DKC1 gene, encoding the pseudouridine synthase dyskerin, is mutated in the most common form of DC, X-linked dyskeratosis congenita (X-DC), as well as in the clinically severe variant of DC known as Hoyeraal-Hreidarsson (HH) syndrome.113,114 Notably, the most well-characterized role of dyskerin is the modification of pseudouridine residues on rRNA106,107,115-117 (Figure 2). The largest subgroup of dyskerin-associated H/ACA small RNAs, termed H/ACA small nucleolar RNAs (snoRNAs), is responsible for guiding dyskerin to convert uridine to pseudouridine at select nucleotides within the rRNA.118 Intriguingly, a recent study has identified remarkable heterogeneity in the expression of specific subsets of H/ACA snoRNAs in primary cells isolated from X-DC patients, including CD34+ hematopoietic progenitor cells.119 Specifically, these findings demonstrate that distinct DKC1 mutations lead to a significant decrease in the levels and activity of several H/ACA snoRNAs known to guide pseudouridine modifications on rRNA. Indeed, X-DC patient cells harbor site-specific defects in rRNA pseudouridine modifications at distinct residues, providing functional readouts of H/ACA snoRNA dysfunction in X-DC.119 Notably, it was also demonstrated that the pseudouridine synthase activity of dyskerin in modifying RNA is important for hematopoietic differentiation because expression of a catalytically inactive dyskerin mutant failed to rescue hematopoietic stem cell differentiation in X-DC patient cells when compared with catalytically active dyskerin.119 These exciting new findings highlight an important, yet perhaps unappreciated, role for impaired dyskerin pseudouridine synthase activity, H/ACA snoRNAs deregulation, and ribosome dysfunction in DC pathogenesis. For example, reduced levels of rRNA pseudouridylation appear to have no overall effect on global rates of protein synthesis; however, a role for pseudouridine modifications in maintaining translation fidelity has been established.120 In addition, it has also been demonstrated that rRNA pseudouridine modifications have an evolutionarily conserved role in the recruitment of structured RNA components, such as IRES elements, to the ribosome.120 Ultimately, these studies illustrate that pseudouridine modifications on rRNA guided by H/ACA snoRNAs may play an important role in modulating the expression of specific mRNAs such as those containing IRES elements. In agreement with this hypothesis, impairments in the translation of distinct mRNAs known to harbor IRES elements such as p53 and p27 were identified in primary X-DC patient cells and also in a mouse model of X-DC.119,121,122 Thus it seems likely that deregulated ribosome function may contribute to DC disease pathogenesis by altering the translation of specific mRNAs in DC patient cells. How ribosomal perturbations might act in concert with impaired telomere maintenance to contribute to the DC phenotype remains to be explored. These studies built the foundation for the hypothesis that alterations in the translation of distinct mRNAs are key determinants for the development of ribosomopathies.
Ribosomal complexity in cellular and developmental biology
Most of the ribosomal proteins appear to be dispensable for the ribosomal peptidyl transferase activity to catalyze peptide bone formation, which is largely dependent on the ribosomal RNA.123 Eukaryotic ribosomes contain additional ribosomal proteins and additional segments (expansion segments) within the rRNAs compared with prokaryotic ribosomes. Most of the additional ribosomal proteins and rRNA expansion segment found in eukaryotes are located over the surface of the ribosome apart from the conserved core catalytic elements of the ribosome; thus the roles of these additional proteins and RNA segments remain largely unclear. Variations in the composition and posttranslational modifications of ribosomal components raise the intriguing possibility of differential function of specialized ribosomes.124
Ribosomal proteins undergo a variety of posttranslational modifications, including phosphorylation, acetylation, methylation, ubiquitination, and O-linked β-D-N-acetylglucosamine (O-GlcNAc) modification.124 Ribosomal RNA is also subject to posttranscriptional modifications including pseudouridylation (see previous discussion) and methylation. Such modified residues are found within functionally important domains of the rRNA.125 Alterations in ribosome-associated proteins and mRNA structures may also contribute additional layers of translational regulation. Additional extraribosomal functions of ribosomal proteins have also been described.126 Nonredundant effects of disrupting specific ribosomal paralogues suggest that ribosomal proteins may serve specific cellular and developmental functions. For example, knockdown of rpl22 in zebrafish embryos results in T-cell developmental arrest, whereas knockdown of the paralogue rpl22l1 blocks the development of hematopoietic stem cells independent of p53 levels.127 Importantly, in mice, mutations in Rpl38 cause selective effects on axial skeletal patterning and the formation of the mammalian body plan.128 These tissue-specific phenotypes are mirrored by selective impairment in the translation of Hox genes, key vertebrate developmental regulators. These studies thereby link a key component of the ribosome to fundamental control of vertebrate embryonic development. They also suggest a deeper level of ribosome-mediated regulation in organogenesis that may be central to the understanding of congenital birth defects associated with ribosomopathies.
Summary
The inherited marrow failure syndromes provided the initial insights into the previously unexpected role of ribosomes in cellular and developmental processes contributing to human disease. The cancer predisposition of these inherited syndromes provides compelling evidence for an initiating or driving role for the observed ribosomal pathway mutations in cancers arising in the general population. The study of inherited marrow failure syndromes is revealing ribosomal functions in stress pathways and protein translational regulation regulating hematopoiesis and clonal evolution, and is opening up new avenues of investigation for targeted therapies.
Acknowledgments
The authors apologize to those whose work could not be included in this review because of space limitations. The reader is referred to additional primary literature within the cited reviews.
D.R. is a Leukemia & Lymphoma Society Scholar.
This work was supported by National Institutes of Health grants from the National Heart, Lung, and Blood Institute 5 R01 HL079582-11 (A.S.), the National Institute of Diabetes and Digestive and Kidney Diseases 5 P30 DK056465 (A.S.), and R01 DK098057 (D.R.), and the Seattle Children’s Hospital Butterfly Guild (A.S.).
Authorship
Contribution: D.R. and A.S. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Akiko Shimamura, Fred Hutchinson Cancer Research Center, 1100 Fairview Ave N, D2-100, Seattle, WA 98109; e-mail: ashimamu@fhcrc.org.