Key Points
Differential gene and miRNA expression analysis in PMF granulocytes identifies new biomarkers and putative therapeutic targets.
Activation of the miR-155/JARID2 axis in PMF CD34+ cells results in overproduction of MK precursors.
Abstract
Primary myelofibrosis (PMF) is a myeloproliferative neoplasm characterized by megakaryocyte (MK) hyperplasia, bone marrow fibrosis, and abnormal stem cell trafficking. PMF may be associated with somatic mutations in JAK2, MPL, or CALR. Previous studies have shown that abnormal MKs play a central role in the pathophysiology of PMF. In this work, we studied both gene and microRNA (miRNA) expression profiles in CD34+ cells from PMF patients. We identified several biomarkers and putative molecular targets such as FGR, LCN2, and OLFM4. By means of miRNA-gene expression integrative analysis, we found different regulatory networks involved in the dysregulation of transcriptional control and chromatin remodeling. In particular, we identified a network gathering several miRNAs with oncogenic potential (eg, miR-155-5p) and targeted genes whose abnormal function has been previously associated with myeloid neoplasms, including JARID2, NR4A3, CDC42, and HMGB3. Because the validation of miRNA-target interactions unveiled JARID2/miR-155-5p as the strongest relationship in the network, we studied the function of this axis in normal and PMF CD34+ cells. We showed that JARID2 downregulation mediated by miR-155-5p overexpression leads to increased in vitro formation of CD41+ MK precursors. These findings suggest that overexpression of miR-155-5p and the resulting downregulation of JARID2 may contribute to MK hyperplasia in PMF.
Introduction
Philadelphia-negative chronic myeloproliferative neoplasms (MPNs) are a heterogeneous group of clonal hematopoietic stem cell (HSC) disorders associated with overproduction of mature myeloid cells and include primary myelofibrosis (PMF).1,2 The molecular mechanisms underlying MPN pathogenesis were partially disclosed in 2005-2006 with the identification of somatic gain-of-function mutations of JAK2 and MPL,3,4 after which many other mutated genes were found,5,6 such as CALR.7,8 Despite the fact that the mutational landscape of MPNs has been extensively investigated, a comprehensive framework of the pathogenetic molecular mechanisms has not been fully elucidated.
MicroRNAs (miRNAs) are small noncoding RNAs involved in several biological processes such as differentiation,9 proliferation,10 and apoptosis11 through the posttranscriptional repression of their targets.12 Increasing evidence shows that deregulation of miRNAs plays an important role in both solid and hematologic malignancies.13,14 In particular, recent reports pointed to aberrant miRNA expression in MPNs, and specific miRNA signatures that distinguish MPN granulocytes from those of healthy donors have been described.15,16 However, because MPNs are stem cell–derived disorders, studies on disease mechanisms should be focused on a more primitive cell compartment. High-throughput analysis of miRNA expression levels in MPN CD34+ cells has been reported in only 2 studies, including a limited number of patients17,18 ; therefore, our understanding of the involvement of miRNAs in MPN pathogenesis is still limited.
Here, we focused our attention on the molecular mechanisms underlying PMF, the less common and likely the most pathogenetically complex of the MPNs. In order to define the role of miRNAs in the pathogenesis of PMF, we obtained both miRNA and gene expression profiles (miEP and GEP, respectively) in the same sample of CD34+ cells from 42 PMF patients and 31 healthy donors. Integration of these profiles allowed us to unveil how differentially expressed miRNAs (DEMs) potentially influence the dysregulation of several biological processes in PMF by interacting with their target genes.
In particular, we described a regulatory network involving several upregulated miRNAs with oncogenic potential (oncomiRs) and their target messenger RNAs (mRNAs), including CDC42, HMGB3, and NR4A3, which were previously characterized in knockout (K/O) murine models of myeloproliferative disorders, and the chromatin remodeler JARID2.
We also demonstrated that miR-155-5p affects the in vitro expansion of the megakaryocyte (MK) lineage through the modulation of JARID2 expression, suggesting that the miR-155-5p/JARID2 axis might contribute to the abnormal megakaryopoiesis typical of PMF.
Materials and methods
Patients and samples
Forty-two patients with a diagnosis of PMF in a typical fibrotic stage of the disease according to the World Health Organization19 were included in the microarray analysis. Their characteristics are reported in supplemental Table 1, available on the Blood Web site. PMF CD34+ cells were purified from peripheral blood (PB). In addition, 16 PB samples and 15 bone marrow (BM) samples were collected from normal donors. Moreover, an independent cohort of 36 PMF patients, 12 healthy donors, and 26 cord blood (CB) samples was selected for validation studies.
All subjects provided informed written consent, and the study was performed under the local Institutional Review Board’s approved protocol. The study was conducted in accordance with the Declaration of Helsinki.
The presence of the JAK2V617F mutation and the allele burden were determined via quantitative reverse transcription polymerase chain reaction (qRT-PCR), as previously described.20
GEP and miEP integrative analysis
GEP and miEP were performed on the same RNA preparation using the Affymetrix technology (HG-U219 Array Strip and miRNA 2.0 arrays) as detailed in the supplemental Methods.
Differentially expressed genes (DEGs) and DEMs were then selected following a supervised approach with the analysis of variance (ANOVA) module included in the Partek GS package. In particular, we selected all the probe sets with a fold change contrast ≥2 for DEGs or ≥1.5 for DEMs in the pairwise comparison of PMF vs controls and a false discovery rate (q value) <0.05.
To construct the regulatory networks of the functional miRNA-target interactions, in silico integrative analysis (IA) was performed by using Ingenuity Pathway Analysis (IPA) software (version 8.6; Ingenuity Systems, Redwood City, CA; http://www.ingenuity.com), which combines computationally predicted targets with gene expression data. Briefly, the lists of DEMs and DEGs were separately uploaded on the software; then, the putative targets of DEMs were identified within the DEG list by microRNA Target Filter, according to at least 1 out of 4 different databases (TargetScan, miRecords, Tarbase, or Ingenuity expert findings); finally, pairs with anticorrelated expression trends were filtered and selected for further analysis to build the regulatory networks.
Electroporation of CD34+ cells
The electroporation program of CD34+ cells was based on a previously published protocol,21 which was optimized to be performed on the 4D-Nucleofector System (Lonza) (see supplemental Results). Briefly, each sample was electroporated 3 times once every 24 hours with a mix of 3 Silencer Select small interfering RNAs (siRNAs) targeting human JARID2 (supplemental Table 2) (Life Technologies), starting from the day after CD34+ cell purification.22 For each electroporation, 4 × 105 CD34+ cells were resuspended in 100 µL of P3 Primary Cell Solution (Lonza), containing 3 µg of siRNA mix, and pulsed with the program DS112. To exclude nonspecific effects caused by interfering RNA (RNAi) nucleofection, a sample transfected with a nontargeting siRNA negative control (NegCTR; Silencer Select Negative Control #2 siRNA; Life Technologies) was included.
As described for siRNA transfections, the number of nucleofections and the quantities of miRNA mimics/inhibitors were modified from a previously described protocol23 to best fit the properties of the second-generation of miRNA mimics/inhibitors (Life Technologies). Briefly, CD34+ cells were nucleofected twice, once every 24 hours, with 3 µg of mirVana miR-155-5p mimic or mirVana miRNA mimic Negative Control #1 (Neg-mimic), by using the previously mentioned electroporation protocol DS112. PMF and CB CD34+ cells were nucleofected 4 times, once every 24 hours, with 3 µg of mirVana miR-155-5p inhibitor or mirVana miRNA inhibitor Negative Control #1 (NegINH), by using the electroporation protocol DS112.
Cells were analyzed 24 and 48 hours after the last nucleofection for both cell viability and JARID2 or miR-155-5p expression.
Statistical analysis
The SPSS software (StatSoft; Tulsa, OK) was used for statistical analysis of clinical correlation. Comparison between groups was performed by using the Student t test, and the chosen level of significance was P < .05 with a 2-sided test.
The statistics used for data analysis in silencing/overexpression experiments and 3′ untranslated region (UTR) luciferase reporter assays were based on 2-tailed Student t tests for averages comparison in paired samples. Data were analyzed by using Microsoft Excel (Microsoft Office, 2008 release) and are reported as mean ± standard error of the mean (SEM). P < .05 was considered significant.
Results
Gene expression profile of CD34+ cells from PMF patients
We performed mRNA expression profiling in CD34+ cells from 42 PMF patients (n = 23 JAK2V617F-positive and n = 19 wild-type JAK2) and 31 healthy donors (n = 15 from the BM and n = 16 from the PB). All microarray data (GEP and miEP) were submitted to the Gene Expression Omnibus repository (http://www.ncbi.nlm.nih.gov/geo) and can be downloaded as series GSE41812 and GSE53482.
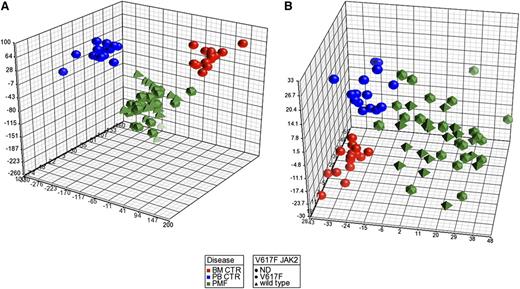
After data preprocessing, to explore the relationships between samples, we performed a principal component analysis (PCA). Figure 1A shows that the PMF samples clustered together and were clearly separated from both the BM and PB control samples. Of note, unsupervised analysis was unable to ungroup JAK2V617F and wild-type JAK2 patients.
PCA of gene and miRNA expression microarray data. The PCA graph of global gene expression data (A) and PCA graph of global miRNA expression data (B) were computed using Partek GS, version 6.6; BM control samples are shown as red spheres; PB control samples are shown as blue spheres; PMF samples are shown in green. PMF JAK2 wild-type samples are shown as pyramids, and PMF JAK2V617F samples are shown as prisms.
PCA of gene and miRNA expression microarray data. The PCA graph of global gene expression data (A) and PCA graph of global miRNA expression data (B) were computed using Partek GS, version 6.6; BM control samples are shown as red spheres; PB control samples are shown as blue spheres; PMF samples are shown in green. PMF JAK2 wild-type samples are shown as pyramids, and PMF JAK2V617F samples are shown as prisms.
Next, using an ANOVA-based supervised approach for comparing PMF samples with both BM and PB controls, we identified 718 DEGs. Supplemental Table 3 shows the 100 most up- and downregulated genes.
Array data confirmed the abnormal expression of several genes (ie, WT1, NFE2, CXCR4, and CD9) previously identified as deregulated in PMF CD34+ cells by our group in a different cohort of PMF patients.24 Moreover, PMF samples exhibited increased levels of several putative cancer markers, such as ANGPT1, CEACAM8, and CP, previously reported to be associated with poor prognosis in hematologic and solid neoplasms, as well as genes associated with BM fibrosis (LEPR, MMP9, and TIMP3) and aberrant migration (TM4SF1, RHOB, ARHGAP18, and MMP8). In addition, PMF samples showed a deregulated expression pattern of a number of transcription factors and chromatin remodelers involved in myeloid and MK commitment, either downregulated (ie, JARID2, RUNX2, KLF3, and AFF3) or upregulated (ie, FHL2, MAF, and IKZF2; for a list of pertinent references, see supplemental Table 4). A selected list of DEGs chosen for their biological significance is presented in Table 1.
DEGs selected by biological significance
| Gene symbol . | Gene ontology function . | Notes . | Fold change . |
|---|---|---|---|
| ANGPT1 | Secreted protein | Overexpressed in AML, CML, and MDSs | 3.1 |
| ANXA3 | Cytoplasmatic protein | Negative prognostic factor for prostate cancer | 10.4 |
| ARHGAP18 | Cytoplasmatic protein | Involved in cell spreading and motility | 3.0 |
| CD9 | Membrane protein | Involved in platelet activation and aggregation; involved in BM remodeling in PMF | 2.2 |
| CEACAM8 | Membrane protein | Overexpressed in imatinib-resistant CML cells | 5.5 |
| CP | Enzyme | Overexpressed in AML; negative prognostic factor for renal carcinoma | 2.6 |
| DEFA1 | Secreted protein | Overexpressed in imatinib-resistant CML cells; biomarker for diagnosis of CRCA | 60.2 |
| FGR | Kinase | Involved in cell migration | 1.9 |
| FHL2 | Transcription factor | Promotes myeloid proliferation; overexpressed in AML | 2.7 |
| IDH1 | Cytoplasmatic protein | Mutated in MPNs | 2.4 |
| IFI27 | Membrane protein | Involved in defense and immunity | 2.4 |
| IFIH1 | Cytoplasmatic protein | Involved in defense and immunity | 2.3 |
| IKZF2 | Transcription factor | Overexpressed in in Hodgkin lymphoma and ALL | 3.6 |
| ITGB3 | Membrane protein | Involved in platelet activation and aggregation | 2.5 |
| LCN2 | Secreted protein | Expression induced by BCR-ABL protein; negative prognostic factor for breast cancer | 7.0 |
| LEPR | Membrane protein | Overexpressed in AML and PMF; involved in fibrosis | 9.7 |
| MAF | Transcription factor | Negative prognostic factor for MM | 8.0 |
| MEF2C | Transcription factor | Involved in MK differentiation | 2.3 |
| MMP9 | Extracellular matrix protein | Involved in the development of fibrosis | 3.8 |
| MYC | Transcription factor | Involved in MK differentiation; cancer marker | 2.8 |
| NFE2 | Transcription factor | Involved in MK differentiation; overexpressed in PMF | 2.0 |
| OLFM4 | Secreted protein | Negative prognostic factor for colorectal, breast, and lung cancer | 3.5 |
| PF4 | Secreted protein | Involved in platelet activation and aggregation | 3.4 |
| PIM1 | Transcription factor | Overexpressed in PMF | 2.6 |
| RHOB | Cytoplasmatic protein | Involved in cell spreading and motility | 3.7 |
| TIMP3 | Extracellular matrix protein | Involved in the development of fibrosis | 4.0 |
| TM4SF1 | Membrane protein | Involved in cell spreading and motility | 4.4 |
| VWF | Secreted protein | Highly expressed by early MKs; involved in platelet adhesion | 4.4 |
| WT1 | Transcription factor | Negative prognostic factor in AML; associated with high severity score in PMF | 2.0 |
| AFF3 | Transcription factor | Fusion with MLL gene in ALL and with RUNX1 gene, partner of MLL | −2.2 |
| ARHGEF7 | Cytoplasmatic protein | Involved in cell migration, attachment, and cell spreading | −2.2 |
| ARID4A | Nuclear protein | Involved in chromatin remodeling; K/O mice develop myelofibrosis | −2.3 |
| BRWD1 | Nuclear protein | Involved in chromatin remodeling | −1.8 |
| CDC42 | Cytoplasmatic protein | Cdc42-deficient mice developed a fatal myeloproliferative disorder | −4.9 |
| CEBPD | Transcription factor | Myeloid commitment regulator | −2.2 |
| CEBPG | Transcription factor | Myeloid commitment regulator | −2.5 |
| CXCR4 | Membrane protein | Involved in BM homing | −2.5 |
| EIF2AK3 | Kinase | The ablation in tumor cells results in accumulation of ROS | −3.0 |
| FOXO1 | Transcription factor | Involved in OXS response; negative prognostic factor for AML | −2.3 |
| HMGB3 | Nuclear protein | Involved in chromatin remodeling; required for the proper balance between HSC self-renewal and differentiation | −2.5 |
| IRF4 | Transcription factor | Downregulated in CML | −2.7 |
| IRF8 | Transcription factor | Downregulated in CML | −6.4 |
| JARID2 | Nuclear protein | Involved in chromatin remodeling and in AML progression | −2.5 |
| KLF3 | Transcription factor | Downregulated in AML; K/O mice display abnormalities in hematopoiesis | −3.6 |
| MAFF | Transcription factor | Myeloid commitment regulator | −2.0 |
| MEF2D | Transcription factor | Involved in myogenic differentiation; fusion with DAZAP1 gene in ALL | −2.7 |
| MLL5 | Transcription factor | Frequently deleted in human myeloid malignancies | −2.7 |
| MXD1 | Transcription factor | Involved in regulation of cell proliferation; antagonizes MYC gene | −3.4 |
| NR4A3 | Nuclear protein | Involved in chromatin remodeling; hypoallelic mice display MDS/MPNs features | −2.4 |
| NUP98 | Transcription factor | Involved in fusions with different partner genes in patients with hematopoietic malignancies | −2.5 |
| PHC3 | Nuclear protein | Component of polycomb repressive complex | −2.1 |
| PURB | Nuclear protein | Deleted in MDS and AML | −2.5 |
| RUNX2 | Transcription factor | Involved in hematopoietic and osteogenic lineages differentiation | −2.1 |
| SF3B1 | Nuclear protein | Mutated in MDS and in MDS/MPNs | −2.0 |
| SMAD7 | Transcription factor | Involved in fibrosis; downregulated in MDSs | −7.2 |
| TCF4 | Transcription factor | Myeloid commitment regulator | −2.4 |
| TLE4 | Transcription factor | Deleted in AML | −2.2 |
| TP53INP1 | Nuclear protein | Loss of expression in several cancers; inactivation correlates with increased cell migration | −2.5 |
| Gene symbol . | Gene ontology function . | Notes . | Fold change . |
|---|---|---|---|
| ANGPT1 | Secreted protein | Overexpressed in AML, CML, and MDSs | 3.1 |
| ANXA3 | Cytoplasmatic protein | Negative prognostic factor for prostate cancer | 10.4 |
| ARHGAP18 | Cytoplasmatic protein | Involved in cell spreading and motility | 3.0 |
| CD9 | Membrane protein | Involved in platelet activation and aggregation; involved in BM remodeling in PMF | 2.2 |
| CEACAM8 | Membrane protein | Overexpressed in imatinib-resistant CML cells | 5.5 |
| CP | Enzyme | Overexpressed in AML; negative prognostic factor for renal carcinoma | 2.6 |
| DEFA1 | Secreted protein | Overexpressed in imatinib-resistant CML cells; biomarker for diagnosis of CRCA | 60.2 |
| FGR | Kinase | Involved in cell migration | 1.9 |
| FHL2 | Transcription factor | Promotes myeloid proliferation; overexpressed in AML | 2.7 |
| IDH1 | Cytoplasmatic protein | Mutated in MPNs | 2.4 |
| IFI27 | Membrane protein | Involved in defense and immunity | 2.4 |
| IFIH1 | Cytoplasmatic protein | Involved in defense and immunity | 2.3 |
| IKZF2 | Transcription factor | Overexpressed in in Hodgkin lymphoma and ALL | 3.6 |
| ITGB3 | Membrane protein | Involved in platelet activation and aggregation | 2.5 |
| LCN2 | Secreted protein | Expression induced by BCR-ABL protein; negative prognostic factor for breast cancer | 7.0 |
| LEPR | Membrane protein | Overexpressed in AML and PMF; involved in fibrosis | 9.7 |
| MAF | Transcription factor | Negative prognostic factor for MM | 8.0 |
| MEF2C | Transcription factor | Involved in MK differentiation | 2.3 |
| MMP9 | Extracellular matrix protein | Involved in the development of fibrosis | 3.8 |
| MYC | Transcription factor | Involved in MK differentiation; cancer marker | 2.8 |
| NFE2 | Transcription factor | Involved in MK differentiation; overexpressed in PMF | 2.0 |
| OLFM4 | Secreted protein | Negative prognostic factor for colorectal, breast, and lung cancer | 3.5 |
| PF4 | Secreted protein | Involved in platelet activation and aggregation | 3.4 |
| PIM1 | Transcription factor | Overexpressed in PMF | 2.6 |
| RHOB | Cytoplasmatic protein | Involved in cell spreading and motility | 3.7 |
| TIMP3 | Extracellular matrix protein | Involved in the development of fibrosis | 4.0 |
| TM4SF1 | Membrane protein | Involved in cell spreading and motility | 4.4 |
| VWF | Secreted protein | Highly expressed by early MKs; involved in platelet adhesion | 4.4 |
| WT1 | Transcription factor | Negative prognostic factor in AML; associated with high severity score in PMF | 2.0 |
| AFF3 | Transcription factor | Fusion with MLL gene in ALL and with RUNX1 gene, partner of MLL | −2.2 |
| ARHGEF7 | Cytoplasmatic protein | Involved in cell migration, attachment, and cell spreading | −2.2 |
| ARID4A | Nuclear protein | Involved in chromatin remodeling; K/O mice develop myelofibrosis | −2.3 |
| BRWD1 | Nuclear protein | Involved in chromatin remodeling | −1.8 |
| CDC42 | Cytoplasmatic protein | Cdc42-deficient mice developed a fatal myeloproliferative disorder | −4.9 |
| CEBPD | Transcription factor | Myeloid commitment regulator | −2.2 |
| CEBPG | Transcription factor | Myeloid commitment regulator | −2.5 |
| CXCR4 | Membrane protein | Involved in BM homing | −2.5 |
| EIF2AK3 | Kinase | The ablation in tumor cells results in accumulation of ROS | −3.0 |
| FOXO1 | Transcription factor | Involved in OXS response; negative prognostic factor for AML | −2.3 |
| HMGB3 | Nuclear protein | Involved in chromatin remodeling; required for the proper balance between HSC self-renewal and differentiation | −2.5 |
| IRF4 | Transcription factor | Downregulated in CML | −2.7 |
| IRF8 | Transcription factor | Downregulated in CML | −6.4 |
| JARID2 | Nuclear protein | Involved in chromatin remodeling and in AML progression | −2.5 |
| KLF3 | Transcription factor | Downregulated in AML; K/O mice display abnormalities in hematopoiesis | −3.6 |
| MAFF | Transcription factor | Myeloid commitment regulator | −2.0 |
| MEF2D | Transcription factor | Involved in myogenic differentiation; fusion with DAZAP1 gene in ALL | −2.7 |
| MLL5 | Transcription factor | Frequently deleted in human myeloid malignancies | −2.7 |
| MXD1 | Transcription factor | Involved in regulation of cell proliferation; antagonizes MYC gene | −3.4 |
| NR4A3 | Nuclear protein | Involved in chromatin remodeling; hypoallelic mice display MDS/MPNs features | −2.4 |
| NUP98 | Transcription factor | Involved in fusions with different partner genes in patients with hematopoietic malignancies | −2.5 |
| PHC3 | Nuclear protein | Component of polycomb repressive complex | −2.1 |
| PURB | Nuclear protein | Deleted in MDS and AML | −2.5 |
| RUNX2 | Transcription factor | Involved in hematopoietic and osteogenic lineages differentiation | −2.1 |
| SF3B1 | Nuclear protein | Mutated in MDS and in MDS/MPNs | −2.0 |
| SMAD7 | Transcription factor | Involved in fibrosis; downregulated in MDSs | −7.2 |
| TCF4 | Transcription factor | Myeloid commitment regulator | −2.4 |
| TLE4 | Transcription factor | Deleted in AML | −2.2 |
| TP53INP1 | Nuclear protein | Loss of expression in several cancers; inactivation correlates with increased cell migration | −2.5 |
ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; BCR-ABL, breakpoint cluster region gene-Abelson murine leukemia viral oncogene homolog 1; CML, chronic myeloid leukemia; CRCA, colorectal cancer; MDS, myelodysplastic syndromes; MM, multiple myeloma; OXS, oxidative stress; ROS, reactive oxygen species.
To validate the array data, we designed a TaqMan low-density array containing 63+GAPDH TaqMan gene expression assays (supplemental Table 5). The selection of genes was based on either the highest absolute fold-change contrast and/or their putative role in PMF pathogenesis. TaqMan assays were carried out in an independent cohort of CD34+ cells from 10 PMF patients and 8 healthy subjects (n = 4 BM, n = 4 PB) and enabled validation of the expression of 50 out of 63 genes (79.4%) (supplemental Table 6).
miRNA expression profile of CD34+ cells from PMF patients
To draw a comprehensive picture of miRNA deregulation and its relationship with differential gene expression in PMF cells, we performed miEP in the same sample set, using the Affymetrix miRNA 2.0 arrays. As already described for GEP, after an initial preprocessing of data aimed at removing the noise, we performed a PCA to explore the relationships between samples. Figure 1B depicts the PCA graph, which shows the presence of the cluster of PMF samples that is clearly distinct from the PB and BM samples; moreover, PCA did not show any difference between mutated JAK2V617F and wild-type JAK2 patients, similar to the GEP PCA.
Next, by using an ANOVA-based supervised approach as described in “Materials and methods,” we selected 76 DEMs (supplemental Table 7). In particular, we found several upregulated miRNAs associated with hematologic malignancies, or known as oncomiRs (ie, miR-155-5p, miR-21-5p, miR-29a-3p, and miRNAs belonging to the miR-17-92 cluster).25 By contrast, among the downregulated miRNAs, we found miR-378c, which is described as a tumor suppressor gene in gastric cancer.26 Furthermore, overexpressed miRNAs previously identified as being involved in MK commitment were also found (ie, miR-146b-5p and miR-34a-5p).27
To validate the array data, we used TaqMan single miRNA assays to assess the expression of 46 DEMs, which were selected based on either their relatively high differential expression or their biological role in cancer-related processes or myeloid differentiation (supplemental Table 8). TaqMan assays were carried out in an independent cohort of CD34+ cells from 10 PMF patients and 8 healthy subjects (n = 4 BM, n = 4 PB) and confirmed the deregulated expression of 34 out of 46 miRNAs (73.9%) (supplemental Table 9).
Validation of a gene set on granulocytes and serum from PMF patients
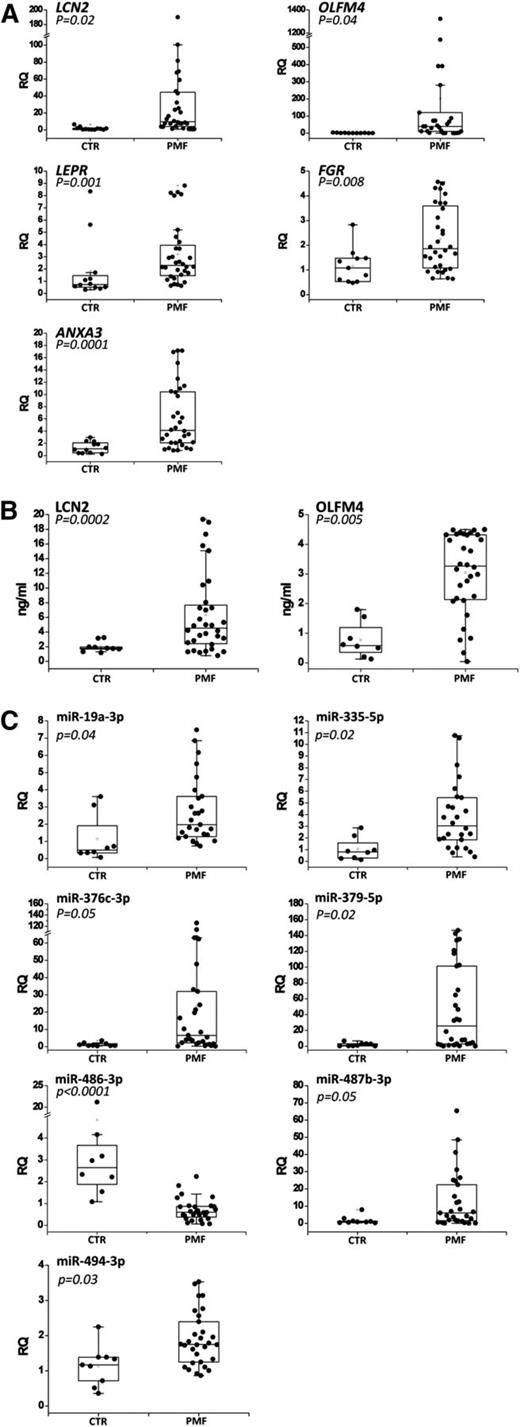
Among the 50 validated genes described previously, we selected a set of 7 genes (OLFM4, LCN2, LEPR, FGR, ANXA3, CEACAM8, and DEF1A) out of those most upregulated to validate their expression in granulocytes, which represent a more appropriate source for diagnostic/prognostic purposes. Using qRT-PCR, we observed that OLFM4, LCN2, LEPR, FGR, and ANXA3 mRNA levels were significantly increased in PMF granulocytes (n = 32) compared with healthy controls (n = 12) (Figure 2A), whereas CEACAM8 and DEF1A expression was not statistically modulated between the 2 groups (data not shown).
Validation of the selected genes and miRNAs in granulocytes and plasma from PMF patients. (A) Expression of the 7 selected genes in granulocytes from PMF patients and healthy donors. Gene expression levels were measured by qRT-PCR starting from granulocyte total RNA and were expressed as relative quantity (RQ). Boxes represent the interquartile range that contains 50% of the subjects, the horizontal line in the box marks the median, and the bars show the range of values. Data are representative of 32 PMF and 12 control (CTR) samples. (B) Serum levels of 2 secreted proteins (LCN2 and OLFM4) in PMF patients and healthy donors. Protein levels were measured by enzyme-linked immunosorbent assay (ELISA) and were expressed as ng/mL. Boxes represent the interquartile range that contains 50% of the subjects, the horizontal line in the box marks the median, and the bars show the range of values. Data are representative of 30 PMF and 8 CTR samples. (C) Expression levels of the 8 selected miRNAs in granulocytes from PMF patients and healthy donors. The miRNA expression levels were measured by qRT-PCR starting from granulocyte total RNA and were expressed as RQ. Boxes represent the interquartile range that contains 50% of the subjects, the horizontal line in the box marks the median, and the bars show the range of values. Data are representative of 32 PMF and 12 CTR samples. *P < .05 vs CTR.
Validation of the selected genes and miRNAs in granulocytes and plasma from PMF patients. (A) Expression of the 7 selected genes in granulocytes from PMF patients and healthy donors. Gene expression levels were measured by qRT-PCR starting from granulocyte total RNA and were expressed as relative quantity (RQ). Boxes represent the interquartile range that contains 50% of the subjects, the horizontal line in the box marks the median, and the bars show the range of values. Data are representative of 32 PMF and 12 control (CTR) samples. (B) Serum levels of 2 secreted proteins (LCN2 and OLFM4) in PMF patients and healthy donors. Protein levels were measured by enzyme-linked immunosorbent assay (ELISA) and were expressed as ng/mL. Boxes represent the interquartile range that contains 50% of the subjects, the horizontal line in the box marks the median, and the bars show the range of values. Data are representative of 30 PMF and 8 CTR samples. (C) Expression levels of the 8 selected miRNAs in granulocytes from PMF patients and healthy donors. The miRNA expression levels were measured by qRT-PCR starting from granulocyte total RNA and were expressed as RQ. Boxes represent the interquartile range that contains 50% of the subjects, the horizontal line in the box marks the median, and the bars show the range of values. Data are representative of 32 PMF and 12 CTR samples. *P < .05 vs CTR.
Because OLFM4 and LCN2 genes encode for 2 secreted proteins, we assessed OLFM4 and LCN2 protein levels in the serum of the same PMF patients (n = 32) and healthy donors (n = 8) by means of an ELISA. As shown in Figure 2B, the levels of OLFM4 and LCN2 secreted proteins were significantly higher in PMF patients than in healthy donors. Of note, the median concentration of OLFM4 serum protein was fivefold higher in PMF patients than in controls.
Validation of the selected miRNAs in the granulocytes from PMF patients
Among the validated miRNAs in PMF CD34+ cells, we selected the 16 most upregulated ones (supplemental Table 9) to test their expression in the granulocytes from the same PMF patients (n = 30) and healthy donors (n = 8) previously used for mRNA expression validation. We demonstrated that the levels of miR-19a-3p, miR-335-5p, miR-379-5p, miR-376c-3p, miR-487b-3p, and miR-494-3p were significantly increased in PMF granulocytes compared with controls; whereas miR-486-3p expression was significantly decreased in PMF granulocytes, in contrast to the results obtained in CD34+ cells (Figure 2C). The levels of the remaining miRNAs were not statistically modulated between PMF and controls (data not shown).
GEP and miEP integrative analysis
Because each miRNA can target many mRNAs while a single mRNA can be targeted by multiple miRNAs, we performed IA by means of IPA to untangle this combinatorial complexity. Based on IPA, we selected DEM-DEG pairs with an anticorrelated expression pattern. In particular, 56 out of the 76 DEMs have at least 1 anticorrelated target among DEGs; whereas 445 out of the 718 DEGs have at least 1 anticorrelated targeting DEM. There were 1167 anticorrelated miRNA-target pairs finally generated.
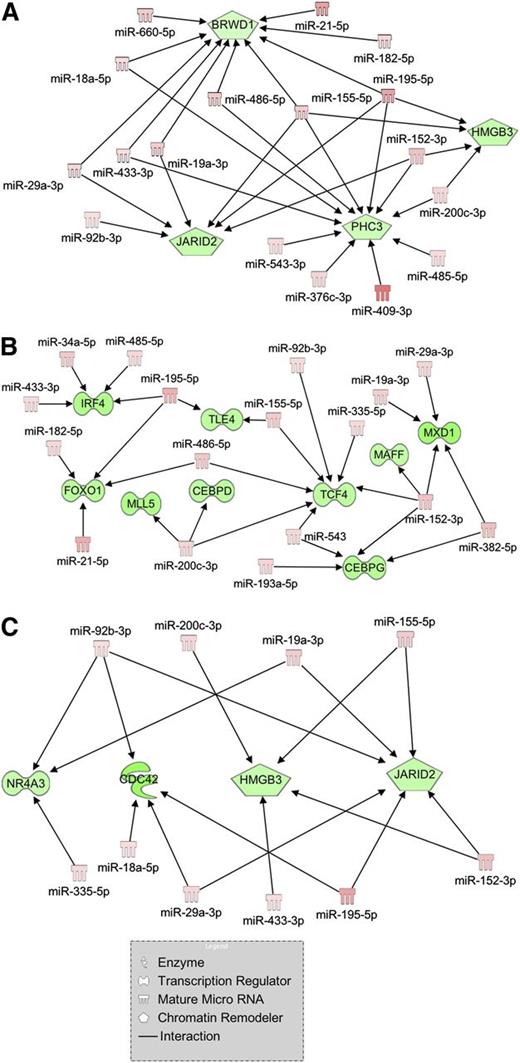
Among the interaction networks uncovered by IPA, we focused our attention on 3 of them because of their enrichment in genes and miRNAs involved in myeloproliferative disorders and/or in hematopoietic differentiation. Figure 3A shows several miRNAs that are highly expressed in PMF, such as miR-18a-5p, miR-29a-3p, miR-433-3p, miR-19a-3p, miR-155-5p, miR-195-5p, miR-200c-3p, and miR-152-3p, and their downregulated targets (BRWD1, JARID2, PHC3, and HMGB3) implicated in chromatin remodeling, a process severely impaired in MPNs. The second network (Figure 3B) displays several upregulated miRNAs involved in myeloid differentiation (miR-155-5p, miR-21-5p, and miR-29a-3p)28 and their interactions with downregulated transcription factors, known as leukemic tumor suppressors (TLE4, MLL5, and FOXO1) or myeloid commitment regulators (CEBPD, CEBPG, MAFF, TCF4, and MXD1).
Regulatory networks of mRNA-miRNA interactions built through IPA. Visualization of the regulatory networks enriched for chromatin remodeling genes (A), myeloid transcription factors (B), and myeloproliferative disorder-related genes (C). Red filling means upregulation, and green filling indicates downregulation.
Regulatory networks of mRNA-miRNA interactions built through IPA. Visualization of the regulatory networks enriched for chromatin remodeling genes (A), myeloid transcription factors (B), and myeloproliferative disorder-related genes (C). Red filling means upregulation, and green filling indicates downregulation.
Finally, through IA we could identify a regulatory network gathering a high number of upregulated oncomiRs (ie, miR-155-5p, miR-29a-3p, miR-92b-3p, miR-19a-3p, and miR-18a-5p) targeting anticorrelated mRNAs whose downregulation or deletion has been related to hematopoietic disorders. In particular, hypoallelic NR4A3 or CDC42 K/O in mice leads to a myeloproliferative disorder,29,30 and JARID2 is a frequently deleted gene in leukemic transformation of chronic myeloid malignancies31 ; the other target, HMGB3, is instead described as a regulator of self-renewal/differentiation balance in murine HSCs32 (Figure 3C).
miRNA-mRNA interaction validation by luciferase reporter assays
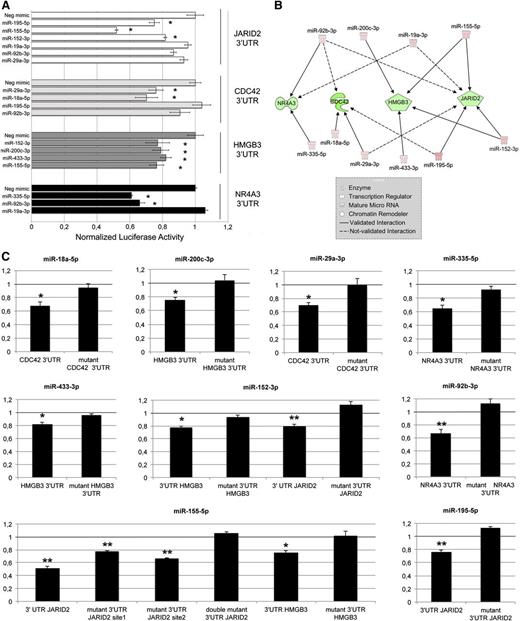
Because the network shown in Figure 3C contained the highest number of oncomiRs and targets involved in malignant hematopoiesis, it was selected to validate every putative miRNA/target pair by assessing 3′UTR luciferase activity upon miRNA overexpression; K562 cells were selected as an in vitro system providing a hematopoietic background. As shown in Figure 4A, our data demonstrated that the following 3′UTR-miRNA interactions were statistically significant: JARID2 3′UTR by miR-152-3p, miR-195-5p, and miR-155-5p (further details on JARID2 3′UTR/miR-155-5p interaction are provided in the supplemental Results); CDC42 3′UTR by miR-29a-3p and miR-18a-5p; HMGB3 3′UTR by miR-152-3p, miR-200c-3p, miR-433-3p, and miR-155-5p; NR4A3 3′UTR by miR-335-5p and miR-92b-3p. Conversely, the interactions between the remaining miRNA-3′UTR target pairs were not confirmed. Figure 4B graphically recapitulates all the confirmed and nonconfirmed interactions of the IA network previously shown. Figure 4C clearly shows that mutations in the miRNA binding site of 3′UTR targets prevented the decrease in luciferase reporter activity by miRNAs, which conversely occurs in their wild-type counterparts. Collectively, the luciferase assay data support the good predictive power by IA, as demonstrated by the 11/17 (64.7%) successful predictions for the selected network.
Validation of 3′UTR-miRNA interactions. (A) Normalized luciferase activity of K562 cells nucleofected with the indicated miRNA mimics and 3′UTR luciferase reporter vectors. Firefly luciferase activity was measured 48 hours after nucleofection and normalized to Renilla luciferase activity. Values are reported as mean ± SEM; *P < .05 vs miRNA mimic negative control (Neg-mimic). Results come from 3 independent experiments performed in duplicate. (B) Graphical representation of mRNA-miRNA interactions validated by means of 3′UTR luciferase reporter assays. Solid lines (validated interactions); dashed lines (not validated interactions). (C) Results of the luciferase reporter assays performed with wild-type and mutant 3′UTR. Assays were carried out only for the 3′UTR-miRNA interactions previously validated (A). Each bar represents the luciferase activity upon miRNA overexpression normalized on the value of the same 3′UTR luciferase vector upon Neg-mimic transfection. Values are reported as mean ± SEM; *P < .05 vs mutant 3′UTR. Results come from 3 independent experiments performed in duplicate.
Validation of 3′UTR-miRNA interactions. (A) Normalized luciferase activity of K562 cells nucleofected with the indicated miRNA mimics and 3′UTR luciferase reporter vectors. Firefly luciferase activity was measured 48 hours after nucleofection and normalized to Renilla luciferase activity. Values are reported as mean ± SEM; *P < .05 vs miRNA mimic negative control (Neg-mimic). Results come from 3 independent experiments performed in duplicate. (B) Graphical representation of mRNA-miRNA interactions validated by means of 3′UTR luciferase reporter assays. Solid lines (validated interactions); dashed lines (not validated interactions). (C) Results of the luciferase reporter assays performed with wild-type and mutant 3′UTR. Assays were carried out only for the 3′UTR-miRNA interactions previously validated (A). Each bar represents the luciferase activity upon miRNA overexpression normalized on the value of the same 3′UTR luciferase vector upon Neg-mimic transfection. Values are reported as mean ± SEM; *P < .05 vs mutant 3′UTR. Results come from 3 independent experiments performed in duplicate.
Silencing of JARID2 in normal CD34+ cells
Because the contribution of JARID2 to PMF pathogenesis has never been investigated, we performed RNAi-mediated gene silencing experiments on normal CD34+ cells. First, we optimized the CD34+ cell nucleofection protocol for the Amaxa 4D-Nucleofector System technology (see supplemental Results).
CD34+ cells were transfected with a mixture of 3 Silencer Select siRNAs targeting JARID2 mRNA (supplemental Table 2) and with a nontargeting siRNA as NegCTR. The expression level of JARID2 in control samples and JARID2-siRNA cells was assessed by qRT-PCR at 24 and 48 hours after the last nucleofection (Figure 5A).
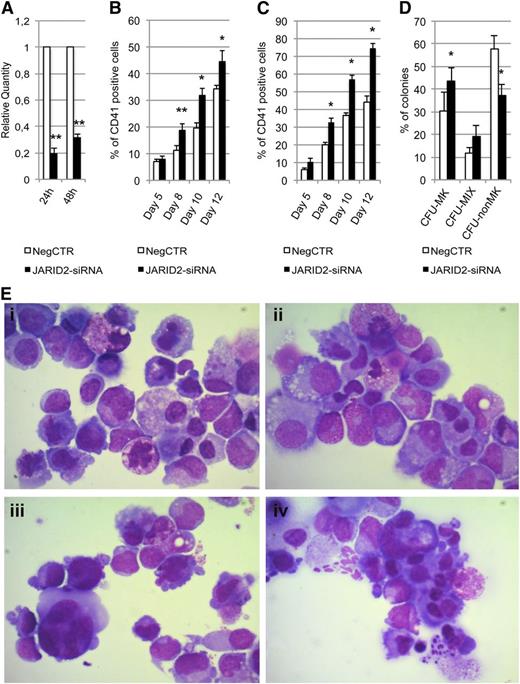
Effect of JARID2 silencing on normal CD34+ cell differentiation. (A) Expression levels of JARID2 at 24 and 48 hours after the last nucleofection were measured by qRT-PCR, and data are reported as RQ. (B-C) Results of the statistical analysis on the percentage of CD41+ cells performed by flow cytometry at days 5, 8, 10, and 12 after the last nucleofection on serum-free multilineage and MK unilineage cultures. (D) Results of the statistical analysis of collagen-based clonogenic assay. The cells were plated 24 hours after the last nucleofection and scored after 12 days. (E) Morphologic analysis of NegCTR (i-ii) and JARID2-siRNA (iii-iv) samples after May-Grünwald-Giemsa (MGG) staining at days 8 and 10 of MK unilineage culture after the last nucleofection in a representative experiment. Magnification, ×1000. Values are reported as mean ± SEM. **P < .01 vs NegCTR; *P < .05 vs NegCTR. The results come from 5 independent experiments.
Effect of JARID2 silencing on normal CD34+ cell differentiation. (A) Expression levels of JARID2 at 24 and 48 hours after the last nucleofection were measured by qRT-PCR, and data are reported as RQ. (B-C) Results of the statistical analysis on the percentage of CD41+ cells performed by flow cytometry at days 5, 8, 10, and 12 after the last nucleofection on serum-free multilineage and MK unilineage cultures. (D) Results of the statistical analysis of collagen-based clonogenic assay. The cells were plated 24 hours after the last nucleofection and scored after 12 days. (E) Morphologic analysis of NegCTR (i-ii) and JARID2-siRNA (iii-iv) samples after May-Grünwald-Giemsa (MGG) staining at days 8 and 10 of MK unilineage culture after the last nucleofection in a representative experiment. Magnification, ×1000. Values are reported as mean ± SEM. **P < .01 vs NegCTR; *P < .05 vs NegCTR. The results come from 5 independent experiments.
Flow cytometric analysis of the CD41 MK marker performed on serum-free multilineage culture at days 8, 10, and 12 showed that JARID2 inhibition induces a significant increase in the MK fraction compared with the NegCTR sample (Figure 5B). Unilineage MK differentiation culture experiments further confirmed these results (Figure 5C). Flow cytometric analysis of granulocytic, monomacrophagic, and erythrocyte differentiation markers did not highlight any significant modulation between JARID2-siRNA CD34+ cells and the NegCTR sample (data not shown). The methylcellulose assay indicated a 1.5-fold increase in the clonogenic efficiency of JARID2-siRNA CD34+ cells vs the NegCTR sample, whereas there was no significant difference in the percentage of erythroid and myeloid colonies (data not shown).
Next, we examined the effect of JARID2 silencing on MK commitment by plating NegCTR and JARID2-siRNA CD34+ cells in a collagen-based serum-free semisolid culture medium that supports the growth of MK progenitors in vitro. The results, reported in Figure 5D, demonstrated that JARID2 silencing induces a remarkable increase in colony forming unit (CFU)-MKs and a strong decrease of non-MK colonies (CFU non-MK) compared with the NegCTR sample. Moreover, morphologic evaluation of MGG-stained cytospins of thrombopoietin-treated cells at days 8 and 10 after the last nucleofection clearly displayed a considerable enrichment in MK precursors at different stages of maturation in JARID2-siRNA cells compared with NegCTR (Figure 5E). Finally, to better characterize the changes in gene expression induced by JARID2 gene silencing, we performed mRNA profiling in NegCTR and JARID2-siRNA CD34+ cells (see supplemental Results and supplemental Figure 1).
miR-155-5p overexpression and silencing in normal and PMF CD34+ cells
Next, we asked whether any JARID2-targeting miRNA could reproduce its silencing effects on MK differentiation. Thus, we decided to overexpress miR-155-5p in normal CD34+ cells because it was highlighted by the luciferase reporter assay as the strongest regulator of the JARID2 3′UTR (Figure 4A). Furthermore, a significant negative correlation was observed between miR-155-5p and JARID2 expression levels across the whole microarray data set (r = −0.55, P = 5.29 × 10−7) (supplemental Figure 2).
CD34+ cells were transfected either with miR-155-5p mimic or with Neg-mimic. By means of qRT-PCR, we observed that the JARID2 mRNA level was downregulated upon miR-155-5p overexpression (RQ ± SEM, 34.7 ± 11.1, P < .05) at 24 and 48 hours after the last nucleofection (Figure 6Ai).
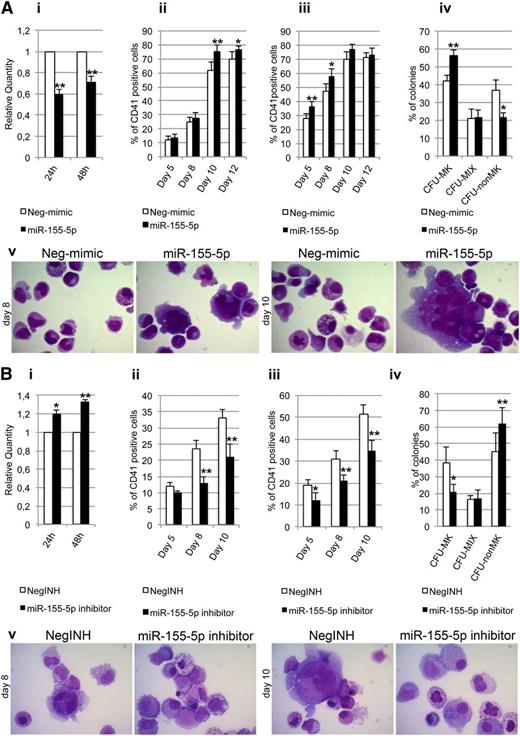
Transfection of miR155-5p mimic and inhibitor in normal and PMF CD34+ cells. (A) Effect of miR155-5p mimic transfection on normal CD34+ cell differentiation. (i) Expression levels of JARID2 at 24 and 48 hours after the last nucleofection, measured by qRT-PCR and reported as RQ. (ii-iii) Results of statistical analysis on the percentage of CD41+ cells performed by flow cytometry at days 5, 8, 10, and 12 after the last nucleofection on serum-free multilineage and MK unilineage cultures. (iv) Results of the statistical analysis of the collagen-based clonogenic assay. The cells were plated 24 hours after the last nucleofection and scored after 12 days. (v) Morphologic analysis of Neg-mimic and miR-155-5p nucleofected cells after MGG staining at days 8 and 10 of MK unilineage serum-free liquid culture after the last nucleofection in a representative experiment. Magnification ×1000. Values are reported as mean ± SEM. **P < .01 vs Neg-mimic; *P < .05 vs Neg-mimic. The results come from 5 independent experiments. (B) Effect of miR155-5p downregulation in PMF CD34+ cells. (i) Expression levels of JARID2 in PMF CD34+ cells at 24 and 48 hours after the last nucleofection of miR-155-5p inhibitor. The JARID2 expression was measured by qRT-PCR, and data are reported as RQ. (ii-iii) Percentage of viable CD41+ cells assessed by flow cytometry at days 5, 8, and 10 after the last nucleofection on serum-free multilineage and MK unilineage cultures. (iv) Results of the statistical analysis of the collagen-based clonogenic assay. The cells were plated 24 hours after the last nucleofection and scored after 12 days. (v) Morphologic analysis of negative control inhibitor (NegINH) and miR-155-5p inhibitor treated cells after MGG staining at days 8 and 10 of MK unilineage serum-free liquid culture after the last nucleofection in a representative experiment. Magnification ×1000. Values are reported as mean ± SEM. **P < .01 vs NegINH; *P < .05 vs NegINH. The results come from 4 independent experiments.
Transfection of miR155-5p mimic and inhibitor in normal and PMF CD34+ cells. (A) Effect of miR155-5p mimic transfection on normal CD34+ cell differentiation. (i) Expression levels of JARID2 at 24 and 48 hours after the last nucleofection, measured by qRT-PCR and reported as RQ. (ii-iii) Results of statistical analysis on the percentage of CD41+ cells performed by flow cytometry at days 5, 8, 10, and 12 after the last nucleofection on serum-free multilineage and MK unilineage cultures. (iv) Results of the statistical analysis of the collagen-based clonogenic assay. The cells were plated 24 hours after the last nucleofection and scored after 12 days. (v) Morphologic analysis of Neg-mimic and miR-155-5p nucleofected cells after MGG staining at days 8 and 10 of MK unilineage serum-free liquid culture after the last nucleofection in a representative experiment. Magnification ×1000. Values are reported as mean ± SEM. **P < .01 vs Neg-mimic; *P < .05 vs Neg-mimic. The results come from 5 independent experiments. (B) Effect of miR155-5p downregulation in PMF CD34+ cells. (i) Expression levels of JARID2 in PMF CD34+ cells at 24 and 48 hours after the last nucleofection of miR-155-5p inhibitor. The JARID2 expression was measured by qRT-PCR, and data are reported as RQ. (ii-iii) Percentage of viable CD41+ cells assessed by flow cytometry at days 5, 8, and 10 after the last nucleofection on serum-free multilineage and MK unilineage cultures. (iv) Results of the statistical analysis of the collagen-based clonogenic assay. The cells were plated 24 hours after the last nucleofection and scored after 12 days. (v) Morphologic analysis of negative control inhibitor (NegINH) and miR-155-5p inhibitor treated cells after MGG staining at days 8 and 10 of MK unilineage serum-free liquid culture after the last nucleofection in a representative experiment. Magnification ×1000. Values are reported as mean ± SEM. **P < .01 vs NegINH; *P < .05 vs NegINH. The results come from 4 independent experiments.
As observed for the JARID2 knockdown, miR-155-5p overexpression led to a significant increase of the percentage of CD41+ cells at days 10 and 12 after the last nucleofection in serum-free multilineage culture (Figure 6Aii). Similar results were obtained under unilineage MK differentiation culture conditions (Figure 6Aiii). Furthermore, the collagen-based assay showed that miR-155-5p overexpression causes a significant increase in the CFU-MK percentage coupled with a strong decrease of non-MK colonies (Figure 6Aiv). Finally, the morphologic analysis of MGG-stained cytospins showed a remarkable enrichment in MK precursors at different stages of maturation in miR-155-5p–overexpressing cells compared with controls at days 10 and 12 after the last nucleofection (Figure 6Av).
Furthermore, we aimed at assessing whether miR-155-5p downregulation in PMF CD34+ cells could reduce the expansion MK lineage. First, we evaluated JARID2 levels upon miR-155-5p silencing, observing a statistically significant increase at 24 and 48 hours after the last nucleofection (Figure 6Bi). Strikingly, knockdown of miR-155-5p impaired the ability of PMF CD34+ cells to give rise to CD41+ cells, in both multilineage and MK unilineage cultures (Figure 6Bii-iii). Those observations were further confirmed by decrease of CFU-MK colonies in collagen-based assays (Figure 6Biv) and by decrease of MK progenitors in MGG-stained cytospins (Figure 6Bv).
Functional validation of the miR-155-5p/JARID2 axis in MK lineage expansion
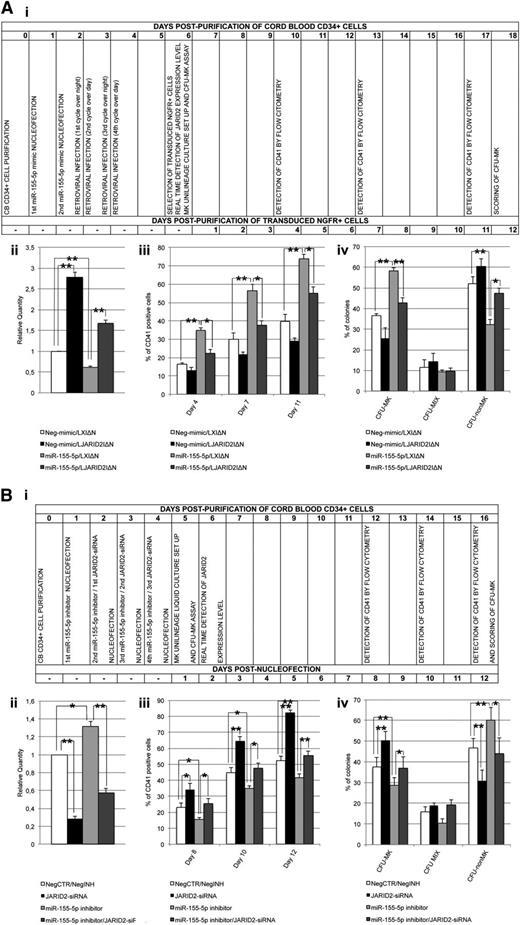
In order to demonstrate that miR-155-5p affects megakaryocytopoiesis by means of JARID2 modulation, we concurrently overexpressed JARID2 and miR-155-5p in normal CD34+ cells. To this aim, CB CD34+ cells were nucleofected twice with miR-155-5p or Neg-mimic and consecutively transduced with the retroviral vector expressing JARID2 gene (LJARID2I∆N) or empty vector (LXI∆N) (see timing flowchart in Figure 7Ai and supplemental Methods).33 To assess JARID2 expression, qRT-PCR was performed after NGFR+ cell purification (Figure 7Aii). As shown in Figure 7Aiii, the fraction of CD41+ cells in the MK unilineage culture decreased in miR-155-5p/LJARID2I∆N compared with miR-155-5p/LXI∆N cells at days 4, 7, and 11 postpurification. Moreover, the CFU-MK assay results showed that JARID2 overexpression in miR-155-5p cells causes a significant decrease in the CFU-MK percentage compared with miR-155-5p/LXI∆N cells (Figure 7Aiv).
Simultaneous overexpression or inhibition of JARID2 and miR-155-5p in normal CD34+ cells. (A) Rescue of JARID2 expression in miR-155-5p–overexpressing cells. (i) Flowchart listing the experiment timing (expressed in days) after NGFR+ cell purification. (ii) Expression levels of JARID2 in CB CD34+ cells after NGFR+ cells purification. The JARID2 expression was measured by qRT-PCR, and data are reported as RQ. (iii) Percentage of viable CD41+ cells in the MK unilineage culture assessed by flow cytometry at days 4, 7, and 11 after NGFR+ cell purification. (iv) Results of the statistical analysis of the collagen-based clonogenic assay. The cells were plated 24 hours after NGFR+ cell purification and scored after 12 days. Values are reported as mean ± SEM. **P < .01; *P < .05. The results come from 3 independent experiments. (B) Simultaneous downregulation of JARID2 and miR155-5p in CB CD34+ cells. (i) Flowchart reporting the experiment timing (expressed in days) after the last nucleofection. (ii) Expression levels of JARID2 in CB CD34+ cells at 48 hours after the last nucleofection. The JARID2 expression was measured by qRT-PCR and data are reported as RQ. (iii) Percentage of viable CD41+ cells in the MK unilineage culture assessed by flow cytometry at days 8, 10, and 12 after the last nucleofection. (iv) Results of the statistical analysis of the collagen-based clonogenic assay. The cells were plated 24 hours after the last nucleofection and scored after 12 days. Values are reported as mean ± SEM. **P < .01; *P < .05. The results come from 3 independent experiments.
Simultaneous overexpression or inhibition of JARID2 and miR-155-5p in normal CD34+ cells. (A) Rescue of JARID2 expression in miR-155-5p–overexpressing cells. (i) Flowchart listing the experiment timing (expressed in days) after NGFR+ cell purification. (ii) Expression levels of JARID2 in CB CD34+ cells after NGFR+ cells purification. The JARID2 expression was measured by qRT-PCR, and data are reported as RQ. (iii) Percentage of viable CD41+ cells in the MK unilineage culture assessed by flow cytometry at days 4, 7, and 11 after NGFR+ cell purification. (iv) Results of the statistical analysis of the collagen-based clonogenic assay. The cells were plated 24 hours after NGFR+ cell purification and scored after 12 days. Values are reported as mean ± SEM. **P < .01; *P < .05. The results come from 3 independent experiments. (B) Simultaneous downregulation of JARID2 and miR155-5p in CB CD34+ cells. (i) Flowchart reporting the experiment timing (expressed in days) after the last nucleofection. (ii) Expression levels of JARID2 in CB CD34+ cells at 48 hours after the last nucleofection. The JARID2 expression was measured by qRT-PCR and data are reported as RQ. (iii) Percentage of viable CD41+ cells in the MK unilineage culture assessed by flow cytometry at days 8, 10, and 12 after the last nucleofection. (iv) Results of the statistical analysis of the collagen-based clonogenic assay. The cells were plated 24 hours after the last nucleofection and scored after 12 days. Values are reported as mean ± SEM. **P < .01; *P < .05. The results come from 3 independent experiments.
Furthermore, to confirm that miR-155-5p–driven megakaryopoiesis depends on JARID2 mRNA levels, we simultaneusly silenced miR-155-5p and JARID2 in normal CD34+ cells (see timing flowchart in Figure 7Bi and supplemental Methods). The expression level of JARID2 was assessed by qRT-PCR at 48 hours after the last nucleofection (Figure 7Bii). Flow cytometric analysis of CD41 expression performed on the MK unilineage culture at days 8, 10, and 12 after the last nucleofection showed a decrease in the MK fraction in miR-155-5p-silenced cells compared with NegCTR/negative control inhibitor (NegINH) cells (Figure 7Biii). As expected, the simultaneous JARID2 knockdown could rescue the MK differentiation unbalance in miR-155-5p silenced cells. In agreement with the flow cytometry data, the results of MK assay highlighted that the concurrent downregulation of miR-155-5p and JARID2 could prevent the impairment of MK differentiation observed in both independently silenced miR-155-5p and JARID2 samples (Figure 7Biv).
Discussion
In this study, we aimed at characterizing the role of miRNAs in PMF pathogenesis. To this end, we integrated GEP and miEP of PMF CD34+ cells, demonstrating how differentially expressed miRNAs affect the gene expression pattern in PMF cells. In particular, miRNA-mRNA expression data integration led to the identification of several networks, one of which was further investigated in terms of interactions between miRNAs and the 3′UTRs of their putative targets. Finally, we demonstrated that the overexpression of miR-155-5p in normal CD34+ cells, as well as the silencing of its validated target, JARID2, determines the expansion of the megakaryocytic lineage. Conversely, we showed that miR-155-5p inhibition in PMF CD34+ cells impaired the MK commitment. Overall, this work sheds light on the influence of deregulated miRNAs on gene expression regulation in PMF CD34+ cells and on their contribution to features typical of PMF, such as a hyperplastic megakaryopoiesis. In parallel, the assessment of the expression levels of several DEGs in PMF granulocytes highlighted a number of possible disease markers, which might eventually become relevant for target therapy approaches, such as membrane protein- and kinase-coding genes.
Because miRNAs were recently demonstrated to be deregulated in MPN cells,15-17 interest of their involvement in pathogenetic mechanisms is progressively growing. However, most of the miRNA and mRNA expression studies performed to date have been conducted using terminally differentiated hematopoietic cells, namely granulocytes or MKs.15,16,34,35 Because MPNs are considered to arise from the HSC compartment,36 understanding of the pathogenetic molecular mechanisms should best be assessed by studying CD34+ cells. So far, only 2 studies have reported data on miRNA expression profiling in a very small number of MPN CD34+ samples,17,18 and no integrated miRNA-mRNA expression analysis was available until now. Here, we provide the results of an extensive study that profiled both gene and miRNA expression in the same CD34+ cell sample from 42 PMF patients.
GEP and miEP analysis showed that PMF CD34+ cells present a different expression pattern compared with BM and unmobilized PB CD34+ cells; of note, PCA was unable to separate PMF patients according to JAK2 mutational status (Figures 1A-B). Differential expression analysis enabled the identification of several deregulated miRNAs and mRNAs suitable as biomarkers or as putative molecular targets for diagnostic or prognostic purposes. Therefore, the most upregulated genes and miRNAs were monitored on PMF granulocytes because they could represent a more suitable cell source for clinical practice, whereas secreted protein levels were assessed in the patients’ sera. We identified a set of 5 genes (ie, LCN2, OLFM4, ANXA3, FGR, and LEPR) (Figure 2A) and 6 miRNAs (ie, miR-19a-3p, miR-335-5p, miR-376c-3p, miR-379-5p, miR-487b-3p, and miR-494-3p) (Figure 2B) whose expression levels are aberrant in PMF granulocytes as well as in CD34+ cells. Evidence in PMF of a higher mRNA expression of the leptin receptor (LEPR), previously reported in AML,37 as well as of Src kinase FGR,38 could also be useful to drive the future design of targeted drugs. Of note, LEPR and Src kinase inhibitors are already being used in preclinical or clinical trials.39,40 In addition, we demonstrated that OLFM4 and LCN2 secreted protein levels could be considered as PMF biomarkers because they were significantly high in patients’ sera compared with those of healthy controls. Strikingly, >80% of the PMF patients presented with higher OLFM4 protein levels compared with all the evaluated controls.
The present study has mainly provided information about the molecular mechanisms underlying PMF pathogenesis. Indeed, data analysis clearly showed that several genes involved in adhesion or migration processes (TM4SF1, RHOB, ARHGAP18, and MMP8) as well as fibrogenic potential (LEPR, MMP9, and TIMP3) are deregulated in PMF CD34+ cells. Interestingly, regulators of megakaryocytic commitment were also upregulated (ie, NFE-2, MEF2C, miR-146b-5p, and miR-34a-5p). Furthermore, we found an increased expression of genes or miRNAs with oncogenic potential, that is, CEACAM8, ANGPT1, miR-29a-3p, and miRNAs belonging to the miR-17-92 cluster25,41 (supplemental Table 4).
Because one of the main mechanisms through which miRNAs act is degradation of specific targets,42 miRNAs and their targets are expected to display anticorrelated expression. Hence, we performed IA to select more reliable interactions among those predicted in silico. As chromatin remodeling is one of the main processes involved in PMF pathogenesis, we determined whether a DEM could affect this pathway. Figure 3A shows that chromatin remodeler genes such as PHC3 and HMGB3 could be downregulated by a network of upregulated targeting miRNAs (ie, miR-18a-5p, miR-433-3p, miR-195-5p, miR-200c-3p, and miR-152-3p). We also observed that low expression of different hematopoietic transcription factors as well as leukemia suppressors (ie, CEBP, FOXO1, and MLL5) is, at least in part, caused by several highly expressed oncomiRs such as miR-21-5p and miR-29a-3p, as depicted in Figure 3B. Finally, a single regulatory network collecting the highest number of oncomiRs and target genes involved in malignant hematopoiesis is shown in Figure 3C. Of note, by means of 3′UTR luciferase reporter assays, we were able to confirm 11/17 (64.7%) putative interactions for the previously mentioned network, highlighting the good predictive power of IA in identifying true miRNA-target interactions (Figure 4). Specifically, the upregulation of several targeting miRNAs explains the negative regulation of genes like CDC42 and NR4A3, whose downregulation leads to myeloproliferative disorders in murine models,29,30 as well as HMGB3, which codes for a regulator of the self-renewal/differentiation balance in murine HSCs.32 Moreover, upregulated miR-155-5p, miR-195-5p, and miR-152-3p share the experimentally observed target JARID2, a chromatin remodeler that is a member of the Jumonji family of transcription factors belonging to the polycomb repressive complex 2.43 Of note, Puda and colleagues demonstrated that JARID2 is frequently deleted in leukemic transformation of chronic myeloid malignancies.31 However, although the function of JARID2 in hematopoiesis has been already partially unraveled in mouse embryo development studies, there are no data about its role in human primary hematopoietic CD34+ cells.44 Thus, we decided to investigate the effect of JARID2 downregulation in hematopoiesis by means of RNAi-mediated silencing in human normal CD34+ cells. Our findings support the contributing role of JARID2 deficiency in the expansion of the MK lineage both in liquid and in semisolid culture, as shown in Figure 5. Because the most effective miRNA in downregulating JARID2 3′UTR-luciferase activity was miR-155-5p, whose enforced expression causes a myeloproliferative disorder in mice,45 we accordingly overexpressed this miRNA in hematopoietic CD34+ cells. Here, we demonstrated that miR-155-5p overexpression downregulates JARID2 mRNA, thus supporting the expansion of the MK compartment (Figure 6A). In parallel, miRNA inhibition in PMF CD34+ cells clearly showed that miR-155-5p may play an important role in the increased megakaryopoiesis observed in PMF patients (Figure 6B). Finally, the restoration of JARID2 expression level in miR-155-5p–overexpressing CD34+ cells impaired the expansion of MK lineage induced by miR-155-5p (Figure 7A); this unbalance could be prevented by simultaneusly silencing JARID2 and miR-155-5p as well, thus definitively demonstrating that miR155-5p affects megakaryopoiesis via JARID2 modulation (Figure 7B). Overall, this interaction could explain the MK hyperplasia observed in BM biopsies of PMF patients19 and the high proliferative potential of MKs derived from PMF CD34+ cells reported in studies in vitro.46 Moreover, the analysis of miR-155-5p and JARID2 mRNA levels in ET CD34+ cells revealed that this regulatory axis specifically works in PMF CD34+ cells (see supplemental Figure 3 and supplemental Results).
Taken together, integration of GEP and miEP uncovered regulatory networks in which aberrantly expressed miRNAs and genes interact, thereby elucidating some of the pathogenetic characteristics of PMF. Finally, IA has proved to be a good approach for identifying reliable mRNA/miRNA interactions that could contribute to PMF pathogenesis, such as the JARID2-miR-155-5p axis, which is involved in hyperplastic megakaryopoiesis.
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by AIRC project number 10005 “Special Program Molecular Clinical Oncology 5x1000” to Associazione Italiana per la Ricerca sul Cancro Gruppo Italiano Malattie Mieloproliferative (AGIMM) (http://www.progettoagimm.it), AIRC project number 12055 and 9034, and Italian Ministry of University & Research (FIRB Project 2011, project number RBAP11CZLK, and PRIN 2010-11, project number 2010NYKNS7); Project Tecnopolo (Regione Emilia Romagna http://www.aster.it/tiki-index.php?page=Tecnopoli).
Authorship
Contribution: A.M.V. and R.M. designed the study; P.G., V.R., D.P., C.B., S. Salmoiraghi, G.B., M.C., A.R., and A.M.V. enrolled patients; R.Z., R.N., V.P., and T.F. performed and analyzed microarray and real time-PCR data; P.G., C.M., and T.F. performed ELISA assays; R.Z., R.N., V.P., A.B., S.B., and E.T. performed data analysis; V.P., S. Salati, G.S., Z.P., and S. Ruberti performed gene silencing and miRNA overexpression experiments, R.N., E.B., S. Ruberti, and S. Rontauroli carried out luciferase reporter assays; and R.Z., R.N., V.P., E.T., S.F., A.M.V., and R.M. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
A complete list of the members of the AGIMM investigators are given at http://www.progettoagimm.it/Progetto/elenco_en.shtml.
Correspondence: Rossella Manfredini, Centre for Regenerative Medicine “Stefano Ferrari,” University of Modena and Reggio Emilia, via Gottardi 100, 41125 Modena, Italy; e-mail: rossella.manfredini@unimore.it.
References
Author notes
R.N., R.Z., and V.P. contributed equally to this study.
A.M.V. and R.M. are cosenior authors.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal