Key Points
The natural inhibitor of neutrophil elastase, SLPI, is severely reduced in severe congenital neutropenia patients.
SLPI controls myeloid differentiation by regulation of NFκB, ERK1/2:LEF-1, and c-myc activation.
Abstract
We identified diminished levels of the natural inhibitor of neutrophil elastase (NE), secretory leukocyte protease inhibitor (SLPI), in myeloid cells and plasma of patients with severe congenital neutropenia (CN). We further found that downregulation of SLPI in CD34+ bone marrow (BM) hematopoietic progenitors from healthy individuals resulted in markedly reduced in vitro myeloid differentiation accompanied by cell-cycle arrest and elevated apoptosis. Reciprocal regulation of SLPI by NE is well documented, and we previously demonstrated diminished NE levels in CN patients. Here, we found that transduction of myeloid cells with wild-type NE or treatment with exogenous NE increased SLPI messenger RNA and protein levels, whereas transduction of mutant forms of NE or inhibition of NE resulted in downregulation of SLPI. An analysis of the mechanisms underlying the diminished myeloid differentiation caused by reduced SLPI levels revealed that downregulation of SLPI with short hairpin RNA (shRNA) upregulated nuclear factor κB levels and reduced phospho–extracellular signal-regulated kinase (ERK1/2)–mediated phosphorylation and activation of the transcription factor lymphoid enhancer-binding factor-1 (LEF-1). Notably, microarray analyses revealed severe defects in signaling cascades regulating the cell cycle, including c-Myc–downstream signaling, in myeloid cells transduced with SLPI shRNA. Taken together, these results indicate that SLPI controls the proliferation, differentiation, and cell cycle of myeloid cells.
Introduction
Secretory leukocyte protease inhibitor (SLPI), an 11.7-kDa cationic serine protease, is a nonglycosylated protein composed of 2 highly homologous domains of 53 and 54 amino acids and a large number of positively charged residues.1-3 It is expressed by a variety of cells, including granulocytes, monocytes/macrophages, and epithelial cells. Jacobsen et al have reported that SLPI messenger RNA (mRNA) expression is highest in granulocyte progenitor cell myelocytes/metamyelocytes.4 SLPI is one of the natural regulators of neutrophil elastase (NE).5,6 Dose-dependent reciprocal feedback regulation of NE and SLPI is well documented.1,7,8 Autosomal-dominant mutations in the elastase, neutrophil expressed (ELANE) encoding the NE protein have been reported in the majority of patients with severe congenital neutropenia (CN).9 CN is a primary immunodeficiency syndrome characterized by the absence of mature granulocytes in the BM and peripheral blood owing to the inability of BM granulocytic progenitor cells to differentiate into mature granulocytes at the stage of promyelocytes/myelocytes.10 Elevated apoptosis and cell-cycle arrest in the G0/G1 phase have been described in promyelocytes of CN patients.11-14 It is unclear how mutations in ELANE contribute to the failures in granulocytic differentiation, cell-cycle arrest, and elevated apoptosis. We and others have suggested activation of the unfolded protein response (UPR) by misfolded mutated NE protein in myeloid progenitors as one possible mechanism.15-17 However, it is unknown whether activation of the UPR is the only mechanism responsible for the differentiation block in CN myeloid progenitors.
Recently, we found that expression levels of NE in myeloid cells and plasma of CN patients were severely downregulated independently of ELANE mutational status.18 We also found markedly reduced levels of the myeloid-specific transcription factors lymphoid enhancer-binding factor 1 (LEF-1) and CCAAT/enhancer binding protein α (C/EBPα) in myeloid progenitors of CN patients harboring either ELANE or HCLS1 associated protein X-1 (HAX1) mutations.19 ELANE mRNA expression is regulated by the binding of both LEF-1 and C/EBPα at the ELANE gene promoter; because the expression of these transcription factors was reduced, ELANE mRNA could not be activated.19,20 It remains unclear how diminished NE levels contribute to the defective granulopoiesis in CN patients. NE is a member of the chymotrypsin superfamily of serine proteinases expressed by neutrophils, monocytes, and mast cells.21 Intracellularly, NE degrades foreign particles in phagosomes. Upon cell activation, NE is secreted and degrades extracellular matrix proteins and some chemokine and cytokine receptors, including granulocyte colony-stimulating factor receptor (G-CSFR), granulocyte-macrophage colony-stimulating factor (GM-CSF) receptor, C-X-C chemokine receptor type 4, and stromal cell-derived factor 1.22-25 NE also upregulates expression of cytokines such as interleukin-6 (IL-6), inerleukin-8, transforming growth factor-β (TGF-β), and GM-CSF.26-28
We hypothesized that mutated NE or reduced expression of wild-type NE and its antiprotease, SLPI, might dysregulate intracellular signaling systems, ultimately resulting in defective myeloid differentiation, cell-cycle arrest, and apoptosis of myeloid BM progenitors observed in CN patients. SLPI expression is modulated by diverse stimuli, including lipopolysaccharide, NE, phorbol myristate acetate, and both pro- and anti-inflammatory cytokines.1,7,8,29 In addition to its antiprotease activity, SLPI has been shown to exert anti-inflammatory functions through downregulation of tumor necrosis factor-α (TNF-α).29 SLPI also exhibits antiapoptotic activity through binding to annexin II on the cell surface30 and has been shown to protect granulocytes from apoptosis.31 Intriguingly, SLPI is also known to modulate intracellular signal transduction pathways, such as nuclear factor κB (NF-κB) and extracellular signal-regulated kinase (ERK).32-34 In this latter context, it has been shown that SLPI is involved in the activation (phosphorylation) of ERK1/2 protein,33 which plays an important role in G-CSF–triggered myeloid differentiation.35,36 Conversely, SLPI inhibits NF-κB activation in monocytes. It does this through 2 mechanisms: (1) by inhibiting the degradation of the NF-κB–binding inhibitory protein IκBα, an action that depends on the antiprotease activity of SLPI, and (2) by directly binding to NF-κB consensus sites on DNA and preventing p65 binding.32 These actions reflect the ability of SLPI to rapidly enter cells and localize to the cytoplasm and nucleus. SLPI also has an impact on cell proliferation and the cell cycle through upregulation of cyclin D1 mRNA expression and downregulation of antiproliferative factors such as insulin-like growth factor binding protein-3.37 SLPI is also involved in the regulation of growth factors, augmenting hepatocyte growth factor production by lung fibroblasts.38
We detected severely reduced levels of SLPI in myeloid cells and plasma of CN patients owing to mutated or defective ELANE expression and demonstrated that a lack of SLPI led to diminished myeloid differentiation in vitro. Thus, we identified and characterized a new and unexpected mechanism of the regulation of myeloid differentiation by the protease inhibitor SLPI.
Materials and methods
Patients and controls
Twelve patients with CN harboring either ELANE (n = 8) or HAX1 (n = 4) mutations, 3 with other idiopathic neutropenia (IN), and 3 healthy volunteers participated in this study. All patients with neutropenia were treated long-term (>1 year) with G-CSF in a dose between 2 and 7.5 μg/kg per day at the time of study. None of the patients have developed acute myeloid leukemia or myelodysplastic syndrome so far. Three healthy volunteers received G-CSF in a dose of 5 μg/kg per day for 2 days before BM was taken. BM samples were collected in association with the annual follow-up recommended by the Severe Chronic Neutropenia International Registry. Approval for this study was obtained from the Hannover Medical School’s institutional review board, and informed consent was obtained in accordance with the Declaration of Helsinki.
Purification and separation of CD34+ and CD33+ progenitor cells
BM aspirates were obtained from the posterior iliac crest by standard techniques. BM mononuclear cells were isolated by Ficoll-Hypaque gradient centrifugation (Amersham Bioscience). For short hairpin RNA (shRNA) experiments, G-CSF–primed CD34+ cells were harvested by leukapheresis from healthy volunteers. CD33+ and CD34+ cells were positively selected with the use of sequential immunomagnetic labeling and sorting according to the manufacturer’s protocol (Miltenyi Biotec). Cells were counted, and viability was assessed by trypan blue dye exclusion. Purity of sorted cells was >96% as tested by fluorescence-activated cell sorter (FACS) analysis and by hematoxylin/eosin staining of cytospin preparations.
qRT-PCR
For quantitative real-time reverse-transcription polymerase chain reaction (qRT-PCR), we isolated RNA with the use of the QIAGEN RNeasy Mini Kit with the manufacturer’s protocol, amplified complementary DNA (cDNA) with random hexamer primer (Fermentas), and measured mRNA expression with the SYBR green quantitative PCR kit (QIAGEN). Target gene mRNA expression was normalized to β-actin and represented as arbitrary units. Primer sequences are available on request.
Western blot analysis
We used the following antibodies: goat polyclonal anti-NE (Santa Cruz Biotechnology, cat. sc-9520), rabbit polyclonal anti-SLPI (Abcam, cat. ab46763), rabbit monoclonal anti–LEF-1 (Cell Signaling, cat. 2286), rabbit polyclonal anti-phospho–LEF-1 (Eurogentec), rabbit polyclonal anti-ERK1 (Santa Cruz Biotechnology, cat. sc-93), mouse monoclonal anti–phospho-ERK1/2 (Cell Signaling, cat. 9106), rabbit monoclonal anti–β-actin (Cell Signaling, cat. 4970), rabbit monoclonal anti–NF-κB p65 (Cell Signaling, cat. 8242), rabbit monoclonal anti–IκBα (Cell Signaling, cat. 4812), and secondary anti-goat, anti-rabbit, or anti–mouse horseradish-peroxidase–conjugated antibody (Santa Cruz Biotechnology). We obtained whole-cell lysates either through lysis of a defined number of cells in lysis buffer or through direct disruption in Laemmli loading buffer followed by brief sonication. We separated proteins by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and probed the blots with primary antibody either for 1 hour at room temperature or overnight at 4°C followed by washing.
Analysis of the SLPI protein by ELISA
The concentrations of SLPI in plasma were assayed in triplicate in appropriately diluted samples with an SLPI-specific enzyme-linked immunoabsorbent assay (ELISA) kit (R&D Systems). The detection limit of the assay was 25 pg/mL (SLPI).
In vitro proliferation and granulocytic differentiation experiments
We cultured 1 × 105 transduced and sorted red fluorescent protein (RFP)+ or green fluorescent protein (GFP)+ CD34+ cells in Stemline II hematopoietic stem cell expansion medium (Sigma) supplemented with 20 ng/mL of interleukin-3 (IL-3), 20 ng/mL of IL-6, 20 ng/mL of thrombopoietin, 50 ng/mL of stem cell factor (SCF), and 50 ng/mL of Flt3 ligand (R&D Systems). We cultured NB4 cells (1 × 105 per well) in supplemented RPMI 1640/10% fetal calf serum medium. To assess proliferation and cell cycle, we counted viable cells by trypan blue dye exclusion in a hemocytometer and measured 5-bromo-2′-deoxyuridine (BrdU) uptake using the BrdU Flow Kit (BD Pharmingen). We determined the percentage of apoptotic cells using annexin V/APC conjugate (BD Pharmingen). For granulocytic differentiation, we cultured 1 × 105 GFP+- or RFP+-transduced CD34+ cells in supplemented RPMI 1640/10% fetal calf serum medium supplemented with G-CSF (5 ng/mL), SCF (5 ng/mL), IL-3 (5 ng/mL), and GM-CSF (5 ng/mL) for 7 days and in the presence of G-CSF only (10 ng/mL) for an additional 7 days. Granulocytic differentiation of transduced NB4 cells was induced with all-trans retinoic acid (ATRA) at 1 μM concentration. We characterized granulocytic differentiation by FACS analysis with anti-CD15 (BD Pharmingen, cat. 561716), anti-CD11b (BD Pharmingen, cat. 553312), and anti-CD16 antibody (BD Pharmingen, cat. 561304).
Colony-forming unit (CFU) assay
Human CD34+ cells were transduced with SLPI-specific or control GFP-tagged shRNA lentiviral constructs; GFP+ cells were sorted and plated in 1 mL methylcellulose medium (5 × 103/dish; Methocult H4230; Stemcell Technologies) supplemented with 10 ng/mL of G-CSF, 10 ng/mL of GM-CSF, 10 ng/mL of IL-3, 10 ng/mL of SCF, and 1 U of erythropoietin (R&D Systems). After 10 days of culture, the numbers of CFU-G, CFU-GM, and burst-forming unit-erythroid (BFU-E) colonies were calculated.
Statistical analysis
Statistical analysis was performed with the SPSS Version 9.0 statistical package (SPSS). To analyze differences in mean values between 2 groups, a 2-sided unpaired Student t test was used. For the analysis of >2 groups, analysis of variance followed by Student t test was applied.
Results
Reduced SLPI mRNA and protein levels in CN patients harboring ELANE mutations
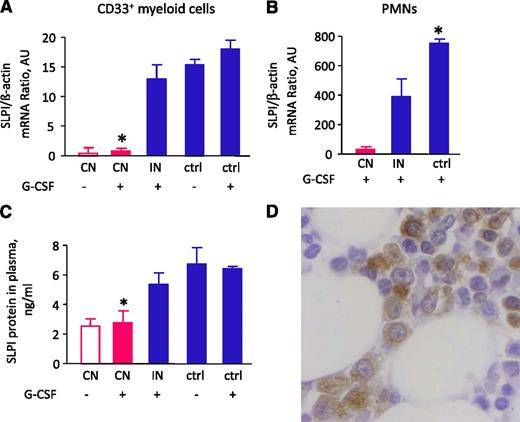
We investigated whether ELANE mutations or diminished ELANE expression affect the intracellular signaling program that operates during myeloid differentiation of hematopoietic cells. A search for proteins that are regulated by NE and show diminished expression in CD33+ myeloid cells of CN patients identified SLPI, which was severely decreased at mRNA and/or protein levels in BM CD33+ myeloid cells, polymorphonuclear neutrophils (PMNs), and plasma of CN patients carrying mutations in ELANE or HAX1 compared with G-CSF–treated healthy individuals (Figure 1A-C). Using immunohistochemistry of BM biopsies of healthy individuals, we confirmed expression of SLPI protein in myeloid cells (Figure 1D).
SLPI mRNA expression in myeloid cells and PMNs and SLPI protein levels in plasma are severely downregulated in patients with CN. (A) SLPI mRNA expression in BM CD33+ cells of studied groups treated and untreated with G-CSF. CN, severe congenital neutropenia; IN, idiopathic neutropenia; ctrl, healthy control. SLPI mRNA expression, normalized to β-actin and presented as arbitrary units (AUs), was measured by qRT-PCR; data represent mean ± standard deviation (SD) of triplicates. (B) SLPI mRNA expression in PMNs from studied groups treated with G-CSF. SLPI mRNA expression is normalized to that of β-actin and presented in AUs; data represent mean ± SD measured in triplicate (*P < .05; **P < .01). (C) SLPI plasma levels were measured in healthy volunteers and the groups of patients indicated above using SLPI-specific ELISA. Data represent mean ± SD and are derived from 2 independent experiments, each in triplicate (*P < .05). (D) Representative micrograph of a BM section from a healthy individual showing immunostaining for SLPI (brown) and counterstaining with hematoxylin (blue). The image shows that myeloid cells express SLPI protein.
SLPI mRNA expression in myeloid cells and PMNs and SLPI protein levels in plasma are severely downregulated in patients with CN. (A) SLPI mRNA expression in BM CD33+ cells of studied groups treated and untreated with G-CSF. CN, severe congenital neutropenia; IN, idiopathic neutropenia; ctrl, healthy control. SLPI mRNA expression, normalized to β-actin and presented as arbitrary units (AUs), was measured by qRT-PCR; data represent mean ± standard deviation (SD) of triplicates. (B) SLPI mRNA expression in PMNs from studied groups treated with G-CSF. SLPI mRNA expression is normalized to that of β-actin and presented in AUs; data represent mean ± SD measured in triplicate (*P < .05; **P < .01). (C) SLPI plasma levels were measured in healthy volunteers and the groups of patients indicated above using SLPI-specific ELISA. Data represent mean ± SD and are derived from 2 independent experiments, each in triplicate (*P < .05). (D) Representative micrograph of a BM section from a healthy individual showing immunostaining for SLPI (brown) and counterstaining with hematoxylin (blue). The image shows that myeloid cells express SLPI protein.
Feedback regulation of SLPI by NE
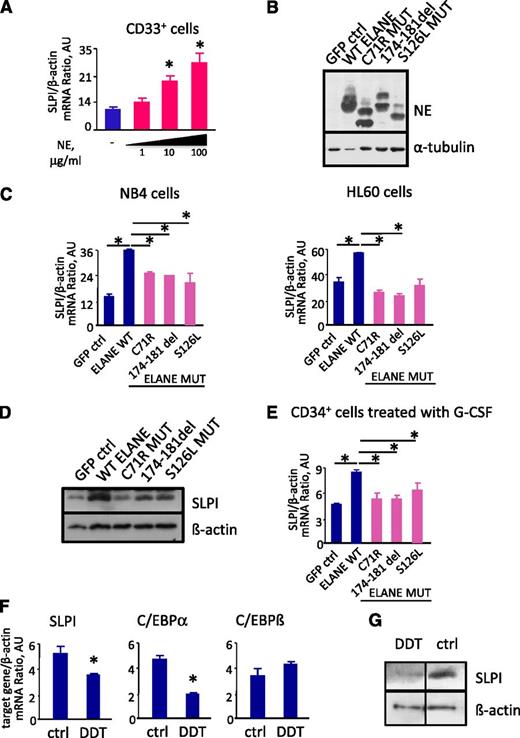
We next examined whether NE regulates SLPI in hematopoietic cells. Indeed, we found that treatment with purified human NE activated SLPI mRNA synthesis in BM CD33+ myeloid cells (Figure 2A). Moreover, transduction of CD34+ hematopoietic progenitor cells or the myeloid cell lines NB4 and HL60 with wild-type (WT) ELANE cDNA resulted in a substantial increase in SLPI mRNA and protein content (Figure 2B-E). We also studied whether ELANE mutants described in CN patients could induce SLPI expression. We used 3 ELANE mutants: C71R and 174-181del, which are frequent mutations found only in CN patients, and S126L, which is common to both CN and cyclic neutropenia patients. We found that all studied ELANE mutants failed to induce SLPI mRNA and protein expression (Figure 2B-E). These data clearly show that WT ELANE induces SLPI expression, whereas mutant ELANE variants do not.
NE regulates SLPI expression levels. (A) SLPI mRNA expression in BM CD33+ cells isolated from healthy individuals untreated (−) or treated with 1, 10, and 100 μg/mL of human purified neutrophils NE. SLPI mRNA expression, normalized to that of β-actin and expressed in arbitrary units (AUs), was measured by qRT-PCR; data are mean ± SD of triplicates (*P < .05). (B) Representative western blot images showing NE protein expression in total lysates from NB4 cells transduced with control (ctrl) cDNA, WT ELANE cDNA, or ELANE mutant cDNA. α-Tubulin staining was used as a loading control. (C) β-Actin–normalized SLPI mRNA expression, in arbitrary units (AU), in NB4 and HL-60 cell lines transduced with control, WT ELANE, or ELANE mutant cDNA, was measured by qRT-PCR; data represent mean ± SD of triplicates (*P < .05 vs control lv samples). (D) Western blot analysis of SLPI expression in NB4 cells transduced with control cDNA, WT ELANE cDNA, or ELANE mutant cDNA. β-Actin was used as a loading control. Representative images are depicted. (E) SLPI mRNA expression in CD34+ cells transduced with control, WT ELANE, or ELANE mutant cDNA and treated with G-CSF. mRNA expression, normalized to that of β-actin and presented in AUs, was measured by qRT-PCR; data represent means ± SDs of triplicates (*P < .05). (F) NB4 cells were untreated (ctrl) or treated with 2 mM of DDT for 4 hours. mRNA expression of indicated genes, normalized to that of β-actin and presented in AUs, was measured by qRT-PCR; data represent mean ± SD of triplicates (*P < .05). (G) Representative western blot images showing SLPI protein expression in total lysates from NB4 cells untreated (ctrl) or treated with DDT for 4 hours. β-Actin staining was used as a loading control.
NE regulates SLPI expression levels. (A) SLPI mRNA expression in BM CD33+ cells isolated from healthy individuals untreated (−) or treated with 1, 10, and 100 μg/mL of human purified neutrophils NE. SLPI mRNA expression, normalized to that of β-actin and expressed in arbitrary units (AUs), was measured by qRT-PCR; data are mean ± SD of triplicates (*P < .05). (B) Representative western blot images showing NE protein expression in total lysates from NB4 cells transduced with control (ctrl) cDNA, WT ELANE cDNA, or ELANE mutant cDNA. α-Tubulin staining was used as a loading control. (C) β-Actin–normalized SLPI mRNA expression, in arbitrary units (AU), in NB4 and HL-60 cell lines transduced with control, WT ELANE, or ELANE mutant cDNA, was measured by qRT-PCR; data represent mean ± SD of triplicates (*P < .05 vs control lv samples). (D) Western blot analysis of SLPI expression in NB4 cells transduced with control cDNA, WT ELANE cDNA, or ELANE mutant cDNA. β-Actin was used as a loading control. Representative images are depicted. (E) SLPI mRNA expression in CD34+ cells transduced with control, WT ELANE, or ELANE mutant cDNA and treated with G-CSF. mRNA expression, normalized to that of β-actin and presented in AUs, was measured by qRT-PCR; data represent means ± SDs of triplicates (*P < .05). (F) NB4 cells were untreated (ctrl) or treated with 2 mM of DDT for 4 hours. mRNA expression of indicated genes, normalized to that of β-actin and presented in AUs, was measured by qRT-PCR; data represent mean ± SD of triplicates (*P < .05). (G) Representative western blot images showing SLPI protein expression in total lysates from NB4 cells untreated (ctrl) or treated with DDT for 4 hours. β-Actin staining was used as a loading control.
It is known that mutated ELANE induces UPR and endoplasmic reticulum (ER) stress in hematopoietic cells. To evaluate whether SLPI is a downstream target of UPR effectors, we treated NB4 cells with dichlorodiphenyltrichloroethane (DDT) to induce UPR and measured SLPI mRNA and protein levels. Indeed, we found markedly reduced SLPI mRNA and protein levels in cells treated with DDT, in comparison with control samples (Figure 2F-G). We also found diminished mRNA expression of C/EBPα and slightly elevated levels of C/EBPβ transcription factors (Figure 2F). Moreover, mRNA expression levels of caspase-9 and of the antiapoptotic factor MCL1 were also dramatically activated by treatment of cells with DDT (supplemental Figure 1A). Interestingly, we previously demonstrated a similar expression pattern of C/EBPα, C/EBPβ, caspase-9, and MCL1 in myeloid cells of CN patients harboring ELANE mutations.13,39 Therefore, we concluded that one of the mechanisms of the defective signal transduction programs downstream of mutated ELANE, including downregulation of SLPI, could be activation of UPR.
To analyze whether the diminished ELANE levels seen in CN patients10 have any effect on SLPI expression, we created shRNA constructs that specifically target ELANE and transduced the myeloid cell lines NB4 and HL60 as well as CD34+ primary hematopoietic cells with these constructs. Indeed, we found that inhibition of ELANE expression led to inhibition of SLPI mRNA and protein synthesis compared with control cells transduced with a scrambled shRNA construct (Figure 3A-D).
Inhibition of NE leads to diminished levels of SLPI. (A-D) The leukemia cell lines NB4 and HL-60 and BM CD34+ cells were transduced with lentiviral constructs of 2 different ELANE-specific RFP-tagged shRNAs (ELANE shRNA E3 and ELANE shRNA E7) or with irrelevant shRNA (ctrl shRNA). On day 4 of culture, RFP+ cells were sorted and analyzed. (A) Representative western blot images showing NE protein expression in total lysates from NB4 cells transduced with shRNA constructs, as indicated above. α-Tubulin staining was used as a loading control. (B) β-Actin–normalized SLPI mRNA expression, in arbitrary units (AUs), in NB4 and HL-60 cell lines transduced with 2 different ELANE-specific shRNAs (ELANE shRNA E3 and ELANE shRNA E7) or with irrelevant shRNA (ctrl shRNA) was measured by qRT-PCR; data represent mean ± SD of triplicates (*P < .05 vs control lv samples). (C) Representative western blot images of SLPI in NB4 cells transduced with shRNA constructs, as indicated above. β-Actin was used as a loading control. (D) β-Actin–normalized SLPI mRNA expression, in AUs, in BM CD34+ cells transduced with 2 different ELANE-specific shRNAs (ELANE shRNA E3 and ELANE shRNA E7) or with irrelevant shRNA (ctrl shRNA) was measured by qRT-PCR; data represent mean ± SD of triplicates (*P < .05 vs control lv samples). (E) SLPI mRNA expression in NB4 cells, transduced with HAX- or HCLS1-specific shRNAs or with irrelevant shRNA (ctrl shRNA). mRNA expression, normalized to that of β-actin and presented in AUs, was measured by qRT-PCR; data represent mean ± SD of triplicates (*P < .05). (F) β-Actin–normalized SLPI mRNA expression, in AUs, in the NB4 cell line, transduced with LEF-1–specific shRNA or irrelevant shRNA (ctrl shRNA) was measured by qRT-PCR; data represent mean ± SD of triplicates (*P < .05 vs control lv samples). (G) Three LEF-1 binding sites on the promoter (blue ovals) were mapped to the region of the SLPI promoter using in silico analysis. Quantitative ChIP assay of nuclear extracts of NB4 cells and PCR products were amplified using qRT-PCR with primer pairs flanking the C/EBPα binding sites (b. s. 1; b. s. 2; b. s. 3) of the SLPI promoter to assess C/EBPα binding to the SLPI promoter. Ab, antibody; C/EBPα, ChIP with anti-C/EBPα antibody; isotype, isotype antibody control. Bar graphs represent % of ChIP to input chromatin of target regions. Data are mean ± SD derived from 2 independent experiments.
Inhibition of NE leads to diminished levels of SLPI. (A-D) The leukemia cell lines NB4 and HL-60 and BM CD34+ cells were transduced with lentiviral constructs of 2 different ELANE-specific RFP-tagged shRNAs (ELANE shRNA E3 and ELANE shRNA E7) or with irrelevant shRNA (ctrl shRNA). On day 4 of culture, RFP+ cells were sorted and analyzed. (A) Representative western blot images showing NE protein expression in total lysates from NB4 cells transduced with shRNA constructs, as indicated above. α-Tubulin staining was used as a loading control. (B) β-Actin–normalized SLPI mRNA expression, in arbitrary units (AUs), in NB4 and HL-60 cell lines transduced with 2 different ELANE-specific shRNAs (ELANE shRNA E3 and ELANE shRNA E7) or with irrelevant shRNA (ctrl shRNA) was measured by qRT-PCR; data represent mean ± SD of triplicates (*P < .05 vs control lv samples). (C) Representative western blot images of SLPI in NB4 cells transduced with shRNA constructs, as indicated above. β-Actin was used as a loading control. (D) β-Actin–normalized SLPI mRNA expression, in AUs, in BM CD34+ cells transduced with 2 different ELANE-specific shRNAs (ELANE shRNA E3 and ELANE shRNA E7) or with irrelevant shRNA (ctrl shRNA) was measured by qRT-PCR; data represent mean ± SD of triplicates (*P < .05 vs control lv samples). (E) SLPI mRNA expression in NB4 cells, transduced with HAX- or HCLS1-specific shRNAs or with irrelevant shRNA (ctrl shRNA). mRNA expression, normalized to that of β-actin and presented in AUs, was measured by qRT-PCR; data represent mean ± SD of triplicates (*P < .05). (F) β-Actin–normalized SLPI mRNA expression, in AUs, in the NB4 cell line, transduced with LEF-1–specific shRNA or irrelevant shRNA (ctrl shRNA) was measured by qRT-PCR; data represent mean ± SD of triplicates (*P < .05 vs control lv samples). (G) Three LEF-1 binding sites on the promoter (blue ovals) were mapped to the region of the SLPI promoter using in silico analysis. Quantitative ChIP assay of nuclear extracts of NB4 cells and PCR products were amplified using qRT-PCR with primer pairs flanking the C/EBPα binding sites (b. s. 1; b. s. 2; b. s. 3) of the SLPI promoter to assess C/EBPα binding to the SLPI promoter. Ab, antibody; C/EBPα, ChIP with anti-C/EBPα antibody; isotype, isotype antibody control. Bar graphs represent % of ChIP to input chromatin of target regions. Data are mean ± SD derived from 2 independent experiments.
Taken together, these results indicate that diminished ELANE/NE levels as well as mutations in ELANE result in reduced levels of SLPI mRNA and protein.
SLPI expression was downregulated in both groups of CN patients harboring either ELANE or HAX1 mutations. Previously, we found that hematopoietic-specific interaction partner of HAX1, HCLS1 protein, was downregulated in myeloid cells of CN patients.40 Therefore, we next analyzed whether diminished levels of HAX1 or HCLS1 led to inhibition of SLPI levels. Indeed, we found that SLPI mRNA expression was markedly reduced in NB4 cells transduced with HAX1 or HCLS1 shRNA constructs, in comparison with control-shRNA–transduced cells (Figure 3E).
We also demonstrated diminished levels of transcription factors LEF-1 and C/EBPα, which are direct target genes of LEF-1, in myeloid cells of CN patients.19 To assess if SPLI mRNA expression could be regulated by LEF-1 transcription factor in myeloid cells, we transduced NB4 cells with LEF-1 shRNA or control shRNA and measured SLPI mRNA levels. Indeed, we found reduced SLPI mRNA expression in LEF-1-shRNA–transduced cells, as compared with control-shRNA–transduced samples (Figure 3F). In silico analysis of the SLPI promoter revealed 3 binding sites of C/EBPα, which is a target gene of LEF-1. A quantitative chromatin immunoprecipitation (ChIP) assay in NB4 cells confirmed binding of C/EBPα to the SLPI promoter (Figure 3G). Taken together, these findings suggest that SLPI mRNA expression in myeloid cells is regulated by LEF-1–triggered activation of C/EBPα transcription factor and subsequent binding of C/EBPα to the SLPI promoter.
Induction of cell-cycle arrest and apoptosis and abrogation of myeloid differentiation by downregulation of SLPI
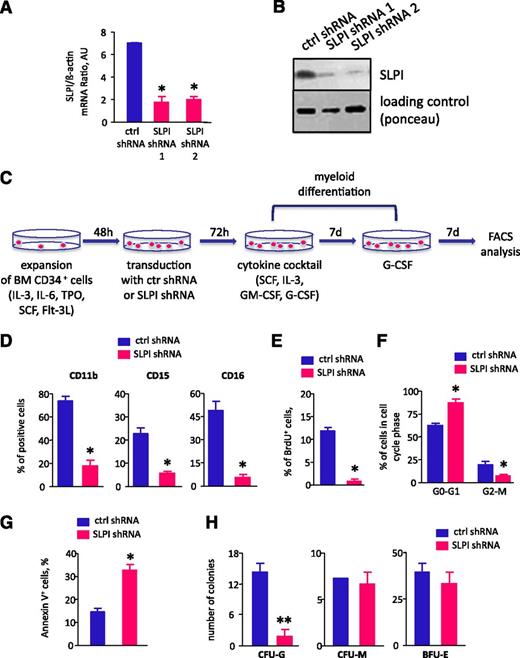
We further analyzed whether diminished levels of SLPI are associated with the “maturation arrest” of myeloid cells seen in CN patients. We inhibited SLPI in CD34+ BM hematopoietic progenitor cells and in the NB4 cell line using lentivirus-mediated transduction of SLPI-specific shRNA and analyzed G-CSF– or ATRA-induced myeloid differentiation, respectively. We found that G-CSF–induced myeloid differentiation was significantly diminished in CD34+ cells transduced with SLPI-specific shRNA compared with that in control-shRNA–transduced cells (Figure 4A-D). Similar results were observed for ATRA-dependent differentiation of the myeloid cell line NB4. These experiments showed a marked reduction in the numbers of CD11b+ and CD15+ cells after ATRA treatment in SLPI-shRNA–transduced NB4 cells compared with control-shRNA–transduced cells (supplemental Figure 2A). Using a CFU assay, we demonstrated dramatically diminished numbers of CFU-G colonies and unaffected formation of CFU-M and BFU-E colonies in CD34+ cells transduced with SLPI shRNA, in comparison with control-shRNA–transduced cells (Figure 4H).
SLPI is essential for granulocytic differentiation in vitro. CD34+ BM cells were transduced with lentiviral constructs of 2 different SLPI-specific RFP-tagged shRNAs (SLPI shRNA 1 and SLPI shRNA 2) or with irrelevant shRNA (ctrl shRNA). On day 4 of culture, RFP+ cells were sorted and in vitro granulocytic differentiation was tested. (A) β-Actin-normalized SLPI mRNA expression, in arbitrary units (AUs), in sorted RFP+ NB4 cells transduced with shRNA constructs as indicated above was measured by qRT-PCR; data represent mean ± SD of triplicates (*P < .05 vs control lv samples). (B) Representative western blot images showing SLPI protein expression in total lysates from RFP+ cells transduced with shRNA constructs as indicated above. Ponceau staining is depicted as a loading control. (C-D) Granulocytic differentiation of transduced CD34+ cells was tested using a liquid culture method. The percentage of CD11b+, CD15+, and CD16+ cells in the RFP+ cell population on day 14 of culture is presented. Data represent mean ± SD of triplicates of the median of 2 different SLPI shRNAs (SLPI shRNA1 and SLPI shRNA2) (*P < .05). (E-F) CD34+ BM cells, transduced with SLPI-specific or control-GFP–tagged shRNA constructs, were labeled with BrdU for 30 minutes, after which the percentage of BrdU+ cells (E) and cell-cycle profile (F) were assessed using a BrdU Flow Kit and FACS analysis, respectively. Data represent the median of 2 different samples of cells transduced with either SLPI-specific shRNA 1 or shRNA 2 (mean ± SD of triplicates of 3 experiments; *P < .05). (G) The apoptosis of CD34+ BM cells, transduced with SLPI-specific or control-GFP–tagged shRNA, was quantified by flow cytometry using an annexin V apoptosis detection kit. Data are expressed as the percentage of annexin V+ cells in the GFP+ cell population. Data represent the median of 2 different samples of cells transduced with either SLPI-specific shRNA 1 or shRNA 2 (mean ± SD of triplicates of 3 experiments; *P < .05). (H) CD34+ cells from healthy individuals (n = 3) were transduced with SLPI-specific or control-GFP–tagged shRNA lentiviral constructs, and GFP+ cells were sorted on day 4 of culture. The indicated CFU assays were performed. Data are mean ± SD and are derived from 2 independent experiments, each in duplicate (**P < .01).
SLPI is essential for granulocytic differentiation in vitro. CD34+ BM cells were transduced with lentiviral constructs of 2 different SLPI-specific RFP-tagged shRNAs (SLPI shRNA 1 and SLPI shRNA 2) or with irrelevant shRNA (ctrl shRNA). On day 4 of culture, RFP+ cells were sorted and in vitro granulocytic differentiation was tested. (A) β-Actin-normalized SLPI mRNA expression, in arbitrary units (AUs), in sorted RFP+ NB4 cells transduced with shRNA constructs as indicated above was measured by qRT-PCR; data represent mean ± SD of triplicates (*P < .05 vs control lv samples). (B) Representative western blot images showing SLPI protein expression in total lysates from RFP+ cells transduced with shRNA constructs as indicated above. Ponceau staining is depicted as a loading control. (C-D) Granulocytic differentiation of transduced CD34+ cells was tested using a liquid culture method. The percentage of CD11b+, CD15+, and CD16+ cells in the RFP+ cell population on day 14 of culture is presented. Data represent mean ± SD of triplicates of the median of 2 different SLPI shRNAs (SLPI shRNA1 and SLPI shRNA2) (*P < .05). (E-F) CD34+ BM cells, transduced with SLPI-specific or control-GFP–tagged shRNA constructs, were labeled with BrdU for 30 minutes, after which the percentage of BrdU+ cells (E) and cell-cycle profile (F) were assessed using a BrdU Flow Kit and FACS analysis, respectively. Data represent the median of 2 different samples of cells transduced with either SLPI-specific shRNA 1 or shRNA 2 (mean ± SD of triplicates of 3 experiments; *P < .05). (G) The apoptosis of CD34+ BM cells, transduced with SLPI-specific or control-GFP–tagged shRNA, was quantified by flow cytometry using an annexin V apoptosis detection kit. Data are expressed as the percentage of annexin V+ cells in the GFP+ cell population. Data represent the median of 2 different samples of cells transduced with either SLPI-specific shRNA 1 or shRNA 2 (mean ± SD of triplicates of 3 experiments; *P < .05). (H) CD34+ cells from healthy individuals (n = 3) were transduced with SLPI-specific or control-GFP–tagged shRNA lentiviral constructs, and GFP+ cells were sorted on day 4 of culture. The indicated CFU assays were performed. Data are mean ± SD and are derived from 2 independent experiments, each in duplicate (**P < .01).
Consistent with the diminished myeloid differentiation of SLPI-shRNA–transduced CD34+ cells, the number of BrdU+ proliferating cells was dramatically reduced in these cells compared with cells in the control-shRNA–transduced group (Figure 4E). The reduction in proliferation observed following transduction of CD34+ cells with SLPI shRNA was caused by cell-cycle arrest of cells in G0/G1 phase (Figure 4F). In addition, we detected a significant elevation in the number of annexin V–positive apoptotic cells in the SLPI-shRNA–transduced group compared with control-shRNA–transduced CD34+ cells (Figure 4G).
Overexpression of ELANE did not affected myeloid differentiation of CD34+ cells upon treatment with G-CSF (supplemental Figure 3A). In the contrast, inhibition of ELANE by shRNA had negative effect on G-CSF–triggered myeloid differentiation (supplemental Figure 3B). At the same time, transduction of CD34+ cells with ELANE mutants led to reduced proliferation and elevated apoptosis (supplemental Figure 4A).
Diminished SLPI expression leads to elevated NF-κB levels and to diminished phospho-ERK1/2–mediated LEF-1 phosphorylation
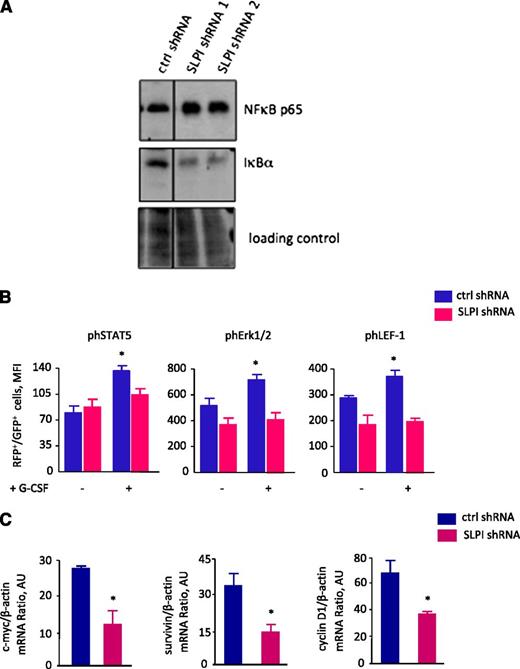
It has been demonstrated that SLPI regulates TNF-α and NF-κB signaling cascades.29,32 Therefore, we evaluated whether these pathways are disrupted by inhibition of SLPI in hematopoietic cells. We measured TNF-α levels in culture supernatants of CD34+ cells transduced with SLPI or control shRNA after induction of granulopoiesis by treatment of cells with G-CSF for 14 days. We did not found any differences in TNF-α levels between control- and SLPI-shRNA–transduced groups (data not shown). At the same time, we identified diminished IκBα and elevated NF-κB p65 protein levels in NB4 cells transduced with SLPI shRNA, in comparison with control-shRNA–transduced cells (Figure 5A).
SLPI is involved in signal transduction pathways. (A) NB4 cells were transduced with SLPI-specific or control-RFP–tagged shRNA. RFP+ cells were sorted and treated with ATRA for 2 days. Representative western blot images showing total NF-κB p65 and IκBα proteins. Ponceau was used as a loading control. (B) BM CD34+ cells were transduced with SLPI-specific or control-RFP–tagged shRNA, RFP+ cells were sorted and treated or not with 10 ng/mL of G-CSF for 10 minutes; phospho-ERK1/2, phospho-STAT5, and phospho–LEF-1 proteins were quantified using intracellular staining with appropriate antibody and FACS analysis. Mean fluorescence intensity of phospho-ERK1/2, phospho-STAT5, and phospho–LEF-1 proteins in RFP+ cell population is presented. Data are mean ± SD and are derived from 2 independent experiments, each in duplicate (*P < .05). (C) c-Myc, survivin, and cyclin D1 mRNA expression in NB4 cells transduced with shRNA constructs and treated with ATRA as indicated in panel A. c-Myc, survivin, and cyclin D1 mRNA expression, normalized to that of β-actin and presented in arbitrary units (AUs), was measured by qRT-PCR; data represent mean ± SD of triplicates.
SLPI is involved in signal transduction pathways. (A) NB4 cells were transduced with SLPI-specific or control-RFP–tagged shRNA. RFP+ cells were sorted and treated with ATRA for 2 days. Representative western blot images showing total NF-κB p65 and IκBα proteins. Ponceau was used as a loading control. (B) BM CD34+ cells were transduced with SLPI-specific or control-RFP–tagged shRNA, RFP+ cells were sorted and treated or not with 10 ng/mL of G-CSF for 10 minutes; phospho-ERK1/2, phospho-STAT5, and phospho–LEF-1 proteins were quantified using intracellular staining with appropriate antibody and FACS analysis. Mean fluorescence intensity of phospho-ERK1/2, phospho-STAT5, and phospho–LEF-1 proteins in RFP+ cell population is presented. Data are mean ± SD and are derived from 2 independent experiments, each in duplicate (*P < .05). (C) c-Myc, survivin, and cyclin D1 mRNA expression in NB4 cells transduced with shRNA constructs and treated with ATRA as indicated in panel A. c-Myc, survivin, and cyclin D1 mRNA expression, normalized to that of β-actin and presented in arbitrary units (AUs), was measured by qRT-PCR; data represent mean ± SD of triplicates.
We next evaluated whether there was any functional connection between diminished SLPI levels and the defective LEF-1 expression and abrogated G-CSFR–triggered myeloid differentiation that we observed in CD33+ myeloid progenitor cells of CN patients. G-CSFR activation initiates phosphorylation of a number of proteins, including STAT5 (signal transducer and activator of transcription 5) and ERK1/2. Recently, it was shown that phosphorylated ERK1/2 regulates LEF-1 in hematopoietic cells.41 It is also known that SLPI is involved in the phosphorylation of ERK1/2.33 Therefore, we examined ERK1/2 phosphorylation levels in CD34+ cells transduced with SLPI or control shRNA and treated with G-CSF for 10 minutes. We also measured phospho-STAT5 levels as a classical downstream effector of the G-CSFR activation. As expected, stimulation of control-shRNA–transduced cells with G-CSF resulted in phosphorylation of ERK1/2 and STAT5 proteins. In contrast, G-CSF failed to phosphorylate ERK1/2 and STAT5 after transduction of cells with SLPI-specific shRNA (Figure 5B). Moreover, using MotifScan software, we identified tyrosine residue (Tyr114) in the LEF-1 protein, which could be phosphorylated and generated rabbit polyclonal antibody specifically recognized this phosphotyrosine on the LEF-1 protein. We verified the specificity of this antibody by FACS analysis of NB4 cells treated or not with phosphatase inhibitor cocktail (supplemental Figure 5A). We found that reduced phosphorylated ERK1/2 levels were accompanied by a reduction in G-CSF–induced phosphorylation of LEF1 protein on tyrosine 114 (Figure 5B) and diminished expression of the LEF-1 target genes, c-Myc, survivin, and cyclin D1 (Figure 5C).
Dysregulated G-CSFR–induced intracellular signaling in CD34+ hematopoietic cells transduced with SLPI shRNA
To evaluate the mechanism of defective G-CSFR–induced differentiation of CD34+ hematopoietic cells following downregulation of SLPI, additional to ERK1/2 phosphorylation and LEF-1 regulation, we examined changes in gene expression using Affymetrix GeneChip microarrays. After transducing CD34+ cells with SLPI or control shRNA-RFP constructs and serum-starving for 24 hours, RFP+ cells were obtained by fluorescence-activated cell sorting and treated with G-CSF for 18 hours. Microarray analyses were then performed using RNA isolated from sorted, G-CSF–treated cells (Figure 6A). We found no differences in cell morphology between studied groups 18 hours after G-CSF treatment (data not shown). Experiments were conducted on 3 independent biological replicates. Unsupervised hierarchical clustering confirmed global changes in the gene expression signature of G-CSF–treated CD34+ cells after downregulation of SLPI compared with control-shRNA–transduced CD34+ cells (data not shown).
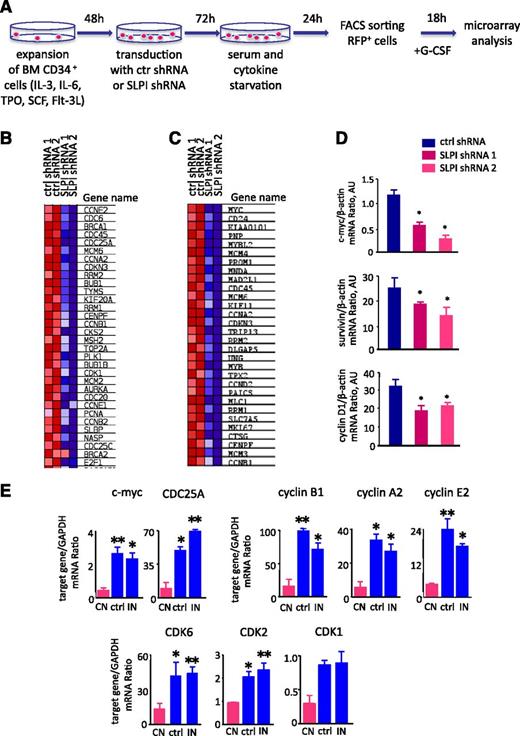
SLPI is involved in a cell-cycle regulatory network. (A) Schematic diagram of the experimental design. BM CD34+ cells were transduced with SLPI-specific or control-RFP–tagged shRNA constructs and serum-starved for 24 hours. RFP+ cells were sorted, treated with G-CSF for 18 hours, and lysed in RNA lysis buffer. RNA from sorted cells was isolated and used for microarray analysis with Affymetrix GeneChip microarrays (Gene Expression Omnibus accession number GSE53353, National Center for Biotechnology Information tracking system number 16934073). (B-C) Microarray data were processed using GSEA software. Heatmaps show differences in mRNA expression of target genes involved in a cell regulatory network (B) and the c-Myc signaling pathway (C) in SLPI-deficient CD34+ cells compared with control-shRNA–transduced cells treated with G-CSF. Upregulation of mRNA expression is shown in red and downregulation in blue. (D) c-Myc, survivin, and cyclin D1 mRNA expression in CD34+ cells transduced with shRNA constructs as indicated in panel A and treated with G-CSF. c-Myc, survivin, and cyclin D1 mRNA expression, normalized to that of β-actin and presented in arbitrary units (AUs), was measured by qRT-PCR; data represent mean ± SD of triplicates. (E) Expression of c-Myc, CDC25, cyclin B, cyclin D1, cyclin E2, and CDK6, CDK3, CDK4 mRNA in BM CD33+ myeloid cells of studied groups. CN, severe congenital neutropenia; ctrl, control; IN, idiopathic neutropenia. mRNA expression of indicated above target genes, normalized to that of GAPDH and presented in AUs, was measured by qRT-PCR; data represent mean ± SD of triplicates (*P < .05; **P < .01).
SLPI is involved in a cell-cycle regulatory network. (A) Schematic diagram of the experimental design. BM CD34+ cells were transduced with SLPI-specific or control-RFP–tagged shRNA constructs and serum-starved for 24 hours. RFP+ cells were sorted, treated with G-CSF for 18 hours, and lysed in RNA lysis buffer. RNA from sorted cells was isolated and used for microarray analysis with Affymetrix GeneChip microarrays (Gene Expression Omnibus accession number GSE53353, National Center for Biotechnology Information tracking system number 16934073). (B-C) Microarray data were processed using GSEA software. Heatmaps show differences in mRNA expression of target genes involved in a cell regulatory network (B) and the c-Myc signaling pathway (C) in SLPI-deficient CD34+ cells compared with control-shRNA–transduced cells treated with G-CSF. Upregulation of mRNA expression is shown in red and downregulation in blue. (D) c-Myc, survivin, and cyclin D1 mRNA expression in CD34+ cells transduced with shRNA constructs as indicated in panel A and treated with G-CSF. c-Myc, survivin, and cyclin D1 mRNA expression, normalized to that of β-actin and presented in arbitrary units (AUs), was measured by qRT-PCR; data represent mean ± SD of triplicates. (E) Expression of c-Myc, CDC25, cyclin B, cyclin D1, cyclin E2, and CDK6, CDK3, CDK4 mRNA in BM CD33+ myeloid cells of studied groups. CN, severe congenital neutropenia; ctrl, control; IN, idiopathic neutropenia. mRNA expression of indicated above target genes, normalized to that of GAPDH and presented in AUs, was measured by qRT-PCR; data represent mean ± SD of triplicates (*P < .05; **P < .01).
Using gene set enrichment analysis (GSEA), we identified pathways affected by inhibition of SLPI in CD34+ primary hematopoietic cells treated with G-CSF. Intriguingly, GSEA analysis revealed that the highest number of pathways that were markedly dysregulated in SLPI-shRNA–transduced cells was related to the c-Myc–triggered signaling (Figure 6B) and cell-cycle signaling (Figure 6C). Notably, c-Myc is a well-known LEF-1 target gene, and we confirmed downregulation of c-Myc mRNA expression in SLPI-shRNA–transduced cells compared with control-shRNA–transduced cells by qRT-PCR (Figure 6D). Interestingly, the same genes that were downregulated in SLPI-shRNA–transduced CD34+ primary hematopoietic cells show diminished expression in CD33+ myeloid progenitor cells of CN patients compared with healthy individuals and patients with idiopathic neutropenia (Figure 6E): cell-cycle regulatory genes (c-Myc, CDC25A, and CDC6), cyclins (CcnA2, CcnB1, and CcnE2), and cyclin-dependent kinases (Cdk1, Cdk2, and Cdkn6).
Using Ingenuity Pathway Analysis software, we found that “hematological system development and function” pathway as well as “hematopoiesis” pathway including 249 and 188 molecules, respectively, were affected in SLPI-shRNA–transduced cells, in comparison with control-shRNA–transduced samples (supplemental Table 1). Moreover, among the top affected associated network functions were Molecular Transport, Nucleic Acid Metabolism, Cell Cycle, Cellular Development, Cellular Growth and Proliferation, and Amino Acid Metabolism (supplemental Table 2). Among the affected molecular and cellular functions pathways were Cellular Development; Molecular Transport; DNA Replication, Recombination, and Repair; Cell Death and Survival; and Cellular Movement (supplemental Table 3).
Discussion
We demonstrate that the levels of SLPI, a natural inhibitor of NE, are severely downregulated in myeloid cells and plasma of CN patients compared with patients with idiopathic neutropenia and healthy individuals (Figure 7). NE is known to stimulate SLPI mRNA synthesis,1,7,8 and we had previously demonstrated that NE mRNA and protein expression are severely downregulated in CN patients.18 Here, we found that transduction of myeloid cells with WT NE induced an increase in SLPI mRNA and protein levels, whereas the 3 mutant forms of NE investigated were not able to induce SLPI expression. Our in vitro studies also showed that inhibition of NE in myeloid cells resulted in diminished SLPI expression. The mechanism by which NE induces SLPI mRNA expression is unclear. NE could be involved in the degradation of protein suppressors of SLPI or histones on regulatory regions of the SLPI gene. In all likelihood, mutated NE would not be capable of degrading SLPI suppressors; however, it is able to activate the UPR and ER stress in myeloid cells.15-17 Indeed, induction of UPR and ER stress by DDT led to diminished expression of SLPI and C/EBPα and to increased mRNA levels of C/EBPβ, caspase-9, and MCL1. Interestingly, similar changes in the expression levels of the above-mentioned factors were detected previously in myeloid cells of CN patients.13,39 It would be interesting to evaluate which signaling components of the UPR are involved in the inhibition of SLPI and C/EBPα. Another possible mechanism of SLPI downregulation in CN patients is that the diminished NE protein levels are not sufficient for induction of SLPI. We previously demonstrated that myeloid progenitors of CN patients exhibit a severely dysregulated myeloid-specific signal-transduction program with diminished expression of myeloid-specific transcription factors LEF-1 and C/EBPα and elevated expression of C/EBPβ.19,39 We demonstrated that downregulation of SLPI is connected to this transcriptional deregulation: C/EBPα binds to the SLPI promoter and inhibition of LEF-1 led to diminished SLPI expression, most probably due to reduced levels of C/EBPα, which is a target gene of LEF-1. Intriguingly, we found diminished SLPI and ELANE levels in CN patients carrying either ELANE or HAX1 mutations. The severely diminished ELANE, LEF-1, and C/EBPα levels in patients with HAX1 mutations19 contribute to the reduced levels of SLPI (Figure 7). Moreover, we found that G-CSF–induced phosphorylation of the HAX1 interaction partner HCLS1 is defective in myeloid cells of CN patients. Diminished HCLS1 expression and phosphorylation due to mutations in HAX1 leads to downregulation of ELANE and therefore to diminished SLPI levels.40
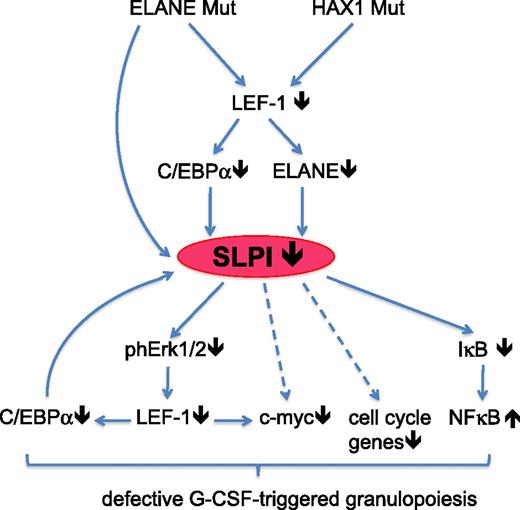
Proposed model of the abrogated granulopoiesis in CN patients due to the diminished SLPI levels. In CN patients harboring ELANE or HAX1 mutations, expression of LEF-1 transcription factor is severely downregulated. This resulted in the diminished expression of LEF-1 target genes, including ELANE and C/EBPα. Both decreased ELANE levels and mutated ELANE failed to induce SLPI, which is normally activated by wild-type ELANE. Low SLPI expression causes abrogated G-CSF–triggered phosphorylation of ERK1/2, phosphorylation and activation of LEF-1 transcription factor, c-Myc–triggered proliferation, and activation of cell-cycle genes. Expression of the myeloid-specific target gene of LEF-1, C/EBPα, is diminished and C/EBPα failed to activate the SLPI promoter. NF-κB signaling is also dysregulated. These pathological intracellular events ultimately lead to the defective granulocytic differentiation of hematopoietic cells in CN patients.
Proposed model of the abrogated granulopoiesis in CN patients due to the diminished SLPI levels. In CN patients harboring ELANE or HAX1 mutations, expression of LEF-1 transcription factor is severely downregulated. This resulted in the diminished expression of LEF-1 target genes, including ELANE and C/EBPα. Both decreased ELANE levels and mutated ELANE failed to induce SLPI, which is normally activated by wild-type ELANE. Low SLPI expression causes abrogated G-CSF–triggered phosphorylation of ERK1/2, phosphorylation and activation of LEF-1 transcription factor, c-Myc–triggered proliferation, and activation of cell-cycle genes. Expression of the myeloid-specific target gene of LEF-1, C/EBPα, is diminished and C/EBPα failed to activate the SLPI promoter. NF-κB signaling is also dysregulated. These pathological intracellular events ultimately lead to the defective granulocytic differentiation of hematopoietic cells in CN patients.
We showed that downregulation of SLPI in hematopoietic progenitor cells and in a myeloid cell line resulted in markedly diminished granulocytic differentiation without any effects on monocytopoiesis and erythropoiesis. Thus, one possible mechanism for the selective maturation arrest of granulopoiesis in CN patients could be defective SLPI synthesis. SLPI mRNA expression is downregulated in both myeloid progenitors and mature granulocytes of CN patients. It has been previously reported that SLPI is maximally expressed in myelocytes/metamyelocytes and that SLPI protein is secreted by granulocytes.4,5,42 We identified a marked reduction in SLPI expression levels in CD33+ cells (corresponding to myeloblasts, promyelocytes, and myelocytes), which are defective in CN patients, compared with SLPI expression in these cells of healthy individuals. We and others have detected intracellular SLPI protein in myeloid progenitors.4 In myeloid cells, intracellular SLPI likely plays a role in regulating the signal transduction program that directs myeloid differentiation. SLPI modulates intracellular signaling pathways, such as NF-κB, ERK, and TGF-β,32,33,43 and prevents synthesis of TNF-α and matrix metalloproteinases.29,44,45 In line with published data, we detected diminished expression of IκBα and elevation of NF-κB p65 after inhibition of SLPI (Figure 7). However, we did not detect any differences in TNF-α synthesis by hematopoietic cells deficient in SLPI. Most probably, SLPI-dependent regulation of TNF-α is cell-type dependent. SLPI also induces activating phosphorylation of ERK1/2 in human ovarian cancer cells.33 Interestingly, we found that phosphorylation of ERK1/2, which is an important player in myeloid differentiation,34,35 was almost completely abolished upon G-CSF–induced myeloid differentiation of SLPI-shRNA–transduced CD34+ cells. It was published that LEF-1 is a target of ERK1/2-mediated phosphorylation in hematopoiesis.39 Consistent with defective phosphorylation of ERK1/2, we found that the levels of phosphorylated LEF-1 and expression of LEF-1 target genes was severely diminished in SLPI-shRNA–transduced cells compared with cells transduced with control shRNA. Therefore, we suggest that, in the absence of SLPI, G-CSF fails to phosphorylate ERK1/2, which in turn leads to the diminished phosphorylation and activation of LEF-1 protein (Figure 7). Recently, we described a novel G-CSFR–triggered intracellular mechanism of granulopoiesis, reporting that G-CSF activates the enzyme nicotinamide phosphoribosyltransferase (Nampt), which is important for NAD+ generation and activation of sirtuins, NAD+-dependent protein deacetylases.39 Microarray analyses in 6 CN patients showed that elevated Nampt levels are associated with severely diminished SLPI levels. The mechanism whether SLPI is directly or indirectly regulated by Nampt has to be investigated. It would be interesting to evaluate whether granulocytic differentiation is affected in SLPI−/− mice. These mice exhibited impaired wound healing46 and a lower survival rate after lipopolysaccharide administration.47 In SLPI-knockout mice, not only high NE activity but also large amounts of TGF-β and IL-6 as well as elevated DNA-binding activities of NF-κB and C/EBPβ were observed.46,47
High levels of SLPI have been demonstrated in cancer cells, including ovarian cancer, gastric cancer, and oral squamous cell carcinoma,46,48 and SLPI is involved in the proliferation48-50 and survival of tumor cells and neutrophils.30,31 Consistent with this, myeloid cells of SLPI-deficient CN patients exhibit cell-cycle arrest and elevated apoptosis.11-14 Some of the activities of SLPI are independent of its protease inhibitory activity.37,50 In line with these observations, our microarray analyses of the intracellular signaling pathways affected by inhibition of SLPI in G-CSF–treated CD34+ hematopoietic cells revealed that SLPI has a strong effect on c-Myc–triggered cell proliferation and the cell-cycle regulatory network (Figure 7). Signaling systems that regulate the cell cycle are also significantly affected by downregulation of c-Myc, CDC25A, CDC6, cyclins (CcnA2, CcnB1, and CcnE2), and cyclin-dependent kinases (Cdk1, Cdk2, and Cdkn6). Intriguingly, genes that were downregulated by shRNA-mediated knockdown of SLPI in hematopoietic cells are similar to those that are decreased in myeloid progenitors of CN patients. Whether SLPI directly affects c-Myc expression or acts via regulation of LEF-1 (a known upstream regulator of c-Myc) remains to be investigated.
Restoration of defective SLPI expression by treatment with recombinant SLPI protein may be a therapeutic option for reversing the maturation arrest of granulopoiesis in CN patients. Fixed doses of recombinant SLPI protein have been already used in in vivo studies. Because of its cationic, arginine-rich structure, SLPI binds predominantly to negatively charged cell membranes and is readily internalized. Accordingly, it has been reported that SLPI binds to annexin II protein on the cell surface of monocytes and macrophages.30
Taken together, by searching for mechanisms of defective granulopoiesis, we identified a previously unrecognized function of SLPI as an essential regulator of myeloid differentiation. SLPI is thus a new-candidate myelopoiesis regulatory factor playing an important role in controlling the proliferation, differentiation, and cell cycle of myeloid cells by regulating intracellular signaling.
The online version of this article contains a data supplement.
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE53353).
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the physicians of the Severe Chronic Neutropenia International Registry for providing patient material and the study subjects for their cooperation. This work was supported by the Madeleine-Schickedanz Kinderkrebsstiftung, DFG-SK-92/4, REBIRTH Cluster of Excellence of the Hannover Medical School, and the Federal Ministry of Education and Research (German Network on Congenital Bone Marrow Failure Syndromes).
Authorship
Contribution: J.S. and K.W. made initial observations, designed the experiments, analyzed the data, supervised experimentation, and wrote the manuscript; O.K. and W.E. performed the main experiments; A.G. and M.Ü. performed WB analysis; M.K. performed ChIP assay; M.Ü., S.K., and J.S. analyzed microarray data; C.Z. provided patient material; and K.H. performed immunohistochemistry of the BM slides.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Karl Welte, Department of Molecular Hematopoiesis, Hannover Medical School, Carl-Neuberg-Strasse 1, 30625 Hannover, Germany; e-mail: welte.karl.h@mh-hannover.de; or Julia Skokowa, Department of Molecular Hematopoiesis, Hannover Medical School, Carl-Neuberg-Strasse 1, 30625 Hannover, Germany; e-mail: skokowa.julia@mh-hannover.de.
References
Author notes
O.K. and W.E. contributed equally to this study.