Key Points
DPPIV (CD26) is a new specific marker of CML LSC that aids CML diagnostics and the measurement, characterization, and purification of LSC.
DPPIV on CML LSC degrades SDF-1 and thereby promotes the niche-escape of LSC, which may contribute to extramedullary myeloproliferation in CML.
Abstract
Chronic myeloid leukemia (CML) is a stem cell (SC) neoplasm characterized by the BCR/ABL1 oncogene. Although mechanisms of BCR/ABL1-induced transformation are well-defined, little is known about effector-molecules contributing to malignant expansion and the extramedullary spread of leukemic SC (LSC) in CML. We have identified the cytokine-targeting surface enzyme dipeptidylpeptidase-IV (DPPIV/CD26) as a novel, specific and pathogenetically relevant biomarker of CD34+/CD38─ CML LSC. In functional assays, CD26 was identified as target enzyme disrupting the SDF-1-CXCR4-axis by cleaving SDF-1, a chemotaxin recruiting CXCR4+ SC. CD26 was not detected on normal SC or LSC in other hematopoietic malignancies. Correspondingly, CD26+ LSC decreased to low or undetectable levels during successful treatment with imatinib. CD26+ CML LSC engrafted NOD-SCID-IL-2Rγ−/− (NSG) mice with BCR/ABL1+ cells, whereas CD26─ SC from the same patients produced multilineage BCR/ABL1– engraftment. Finally, targeting of CD26 by gliptins suppressed the expansion of BCR/ABL1+ cells. Together, CD26 is a new biomarker and target of CML LSC. CD26 expression may explain the abnormal extramedullary spread of CML LSC, and inhibition of CD26 may revert abnormal LSC function and support curative treatment approaches in this malignancy.
Introduction
Chronic myeloid leukemia (CML) is a stem cell (SC) neoplasm characterized by the reciprocal translocation t(9;22).1-3 The resulting oncoprotein, BCR/ABL1, is considered essential for the initiation and manifestation of the disease.2,3 In line with this assumption, the BCR/ABL1 kinase inhibitor imatinib produces major and stable cytogenetic responses in a majority of patients with chronic phase (CP) CML.4,5 Nevertheless, in most patients, imatinib is unable to eradicate the disease, a phenomenon that has been explained by intrinsic and acquired drug resistance in residual leukemic SC (LSC).6-11 Whereas intrinsic resistance is common to most or all LSC, acquired resistance results from additional (subclone-specific) lesions, such as BCR/ABL1 mutations.7-13
The concept of LSC is based on the assumption that only a subset of leukemic cells display long-term self-renewal and thus leukemia-propagating activity.12-16 This hypothesis has major implications for the development of new curative treatment concepts.16-21 In fact, current research focuses on LSC-specific drug targets and novel drugs that can attack and eliminate LSC.17-21 However, the issue of targeting LSC is complex.21 One important aspect is that LSC interact with the SC-related bone marrow (BM) microenvironment, the so called “SC niche” and thereby may escape natural control mechanisms or drug effects.21-24
In CML, LSC supposedly reside within the CD34+/CD38─/Lin─ fraction of the leukemic clone.7-10,25-27 However, normal hematopoietic SC also exhibit this phenotype so that additional markers are required to discriminate CML LSC from normal SC. Recent data suggest that CML LSC aberrantly express IL-1RAP.28 Moreover, it has been described that CML LSC express higher levels of CD33 and CD123 compared with normal SC.29,30 Other studies have shown that LSC in CML express several surface antigens that are relevant for SC-niche interactions, including CD44 and CXCR4.31,32 The latter antigen is of considerable importance because SC are usually attracted into the SC niche by the chemotactic effect of stroma-derived factor-1 (SDF-1), a ligand of CXCR4. However, although CML LSC express CXCR4, their migratory response to SDF-1 is poor compared with normal SC.31-33
In the present article, we show that CD34+/CD38─/Lin─ CML LSC specifically co-express dipeptidylpeptidase IV (DPPIV = CD26) and that this enzyme disrupts LSC-niche interactions by degrading SDF-1. Moreover, our data show that CD26 is a robust biomarker for the quantification and isolation of CML LSC.
Materials and methods
Patients and cell sampling
Eighty-eight patients with CML (36 females, 52 males) were examined. The median age was 56 years (range 18-86). The patients’ characteristics are shown in supplemental Table 1. Peripheral blood (PB) and/or BM cells were collected at diagnosis and in the follow-up. Control cohorts are shown in supplemental Table 2 (normal BM), supplemental Table 3 (idiopathic cytopenia), supplemental Table 4 (other hematologic disorders), and supplemental Table 5 (CD26+ AML cases). All donors gave written informed consent. All studies, including the examination of LSC and their growth in a xenotransplantation mouse model, were approved by the local ethics committee of the Medical University of Vienna. Studies were conducted in accordance with the Declaration of Helsinki.
Isolation of cells, phenotyping, and purification of LSC
Phenotyping of CD34+/CD38─/Lin─ SC and CD34+/CD38+ progenitor cells was performed by multicolor flow cytometry as described29,30 using CML samples (n = 132) or control samples and antibodies shown in supplemental Table 6. The gating algorithm is shown in supplemental Figure 1A. In follow-up examinations, the percentage of CD26+ SC was determined in patients treated with imatinib or nilotinib. In 9 CML patients, CD34+/CD38─ SC and CD34+/CD38+ progenitor cells were purified from mononuclear cells (MNC) by cell sorting as described,30 and in 14 CML patients, the CD26+ LSC fraction and CD26─ SC fraction of CD34+/Lin─ cells were purified by cell sorting. A detailed description is provided in the supplemental material.
Xenotransplantation assay
In xenotransplantation experiments, various subfractions of CML leukocytes were injected intravenously into irradiated nonobese diabetic SCID-IL-2Rγ−/− (NSG) mice. A detailed description of mouse experiments is provided in the supplemental Appendix.
In vitro culture experiments and chemotaxis assay
CML MNC, SC subfractions (CD34+/CD38─/CD26+, CD34+/CD38─/CD26─), and CD34+/CD38+ progenitor cells were cultured in a long-term culture-initiating cell (LTC-IC) assay as described.30 CML MNC were cultured in the absence or presence of vildagliptin (0.1-10 µM) or sitagliptin (0.1-10 µM). CML MNC or purified LSC were also cultured in short-term suspension culture or in short-term methylcellulose cultures in the absence or presence of vildagliptin or sitagliptin (0.1-10 µM). Colonies were counted under an inverted microscope and examined for BCR/ABL1 mRNA expression by quantitative polymerase chain reaction (qPCR). Migration of CXCR4+ U937 cells or primary (sorted) CD34+ CML cells against SDF-1 was determined in a double-chamber chemotaxis assay as reported.31,32 A detailed description of bioassays used in this study is provided in the supplemental Methods.
Immunohistochemistry experiments
Immunohistochemistry was performed on formalin-fixed and paraffin-embedded BM sections obtained from patients with newly diagnosed CML, and BM sections obtained from NSG mice injected with CD26+ CML LSC and CD26─ SC obtained from (the same) CML patients. The antibodies used and the staining protocols are provided in supplemental Table 7. The distribution of CML (progenitor) cells was investigated by microscopy and documented by photography.
Gene array analysis, qPCR, and cytogenetics
Highly purified (sorted) CD34+/CD38─ and CD34+/CD38+ cells from CML CP or cord blood (CB) samples were subjected to RNA isolation and gene array analysis using Affymetrix technology. Technical details are provided in the supplemental material. A complete set of mRNA data are available at Gene Expression Omnibus (accession #GSE40721). mRNA expression levels (selected genes) were confirmed by qPCR using primers shown in supplemental Table 8. BCR/ABL1 and CD26 mRNA levels were quantified in sorted CML LSC, LTC-IC-derived or short-term culture-derived colonies, and NSG mouse-engrafting cells. RNA isolation and PCR assays are described in the supplemental material. ABL served as a reference gene. In clinical follow-up samples, BCR/ABL1 mRNA levels were adjusted according to the international scale (IS).34 Conventional cytogenetics and fluorescence in situ hybridization (FISH) were performed according to published protocols.35
Statistical analysis
Differences in marker expression or the percentage of LSC in various sample series were determined by statistical tests described in the supplemental material.
Results
CD34+/CD38─/Lin─ CML LSC express a unique phenotype
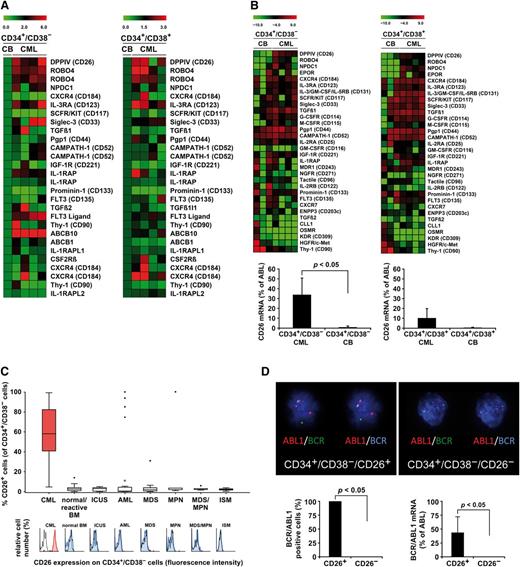
To identify novel robust markers and targets of LSC, we performed gene array studies (Figure 1A) and qPCR analyses (Figure 1B) on highly purified (>98% pure) CD34+/CD38─ SC derived from patients with CP CML. In addition, flow cytometry was performed (Figure 1C and Table 1). Antigens that were clearly detectable not only by gene array and qPCR, but also on the surface of LSC, were CD9, CD25, CD26, CD33, CD36, CD44, CD52, CD90, CD105, CD114, CD117, CD123, CD164, CD184, IL-1RAP, ROBO4, and NPDC1. Several of these antigens were found to be expressed at higher levels on the surface of CML LSC compared with normal SC (supplemental Table 9). Three of these molecules, CD25, CD26, and IL-1RAP, were found to be rather specifically (aberrantly) expressed on CML LSC but were not or were only weakly expressed on normal SC (supplemental Figure 2A and Table 1). The most specific marker of CML LSC appeared to be CD26 (supplemental Figure 2A). Overall, there was a good correlation between expression of CD26 and CD25 and a less significant correlation between expression of CD26 and IL-1RAP on CML LSC, which appeared to be caused by the fact that in several patients, LSC did not express IL-1RAP. Supplemental Figure 2B shows the reactivity of LSC with 3 different CD26 mAb, and supplemental Tables 9-11 show an overview of upregulated genes in CML LSC.
LSCs in CML express CD26. (A) Heat map analysis of gene array data of RNA isolated from sorted CD34+/CD38─ SCs (left) or CD34+/CD38+ progenitor cells (right) obtained from patients with chronic phase (CP) CML (n = 4) and cord blood (CB) SC. Gene array analyses were performed as described in the supplemental material available on the Blood Web site. Expression levels were normalized to CB control. Top-regulated genes of interest are shown. (B) Heat map analysis of pPCR data (upper panel). mRNA analyses were performed on sorted CD34+/CD38─ SC (upper left) and CD34+/CD38+ cells (upper right) obtained from 3 CB samples and 4 patients with CML. Top-regulated genes of interest are shown. Lower panel: qPCR analysis of expression of CD26 mRNA in sorted CD34+/CD38─ SC (lower left) and CD34+/CD38+ cells (lower right) obtained from 7 patients with CML and 3 CB samples. CD26 mRNA levels are expressed as percent of ABL. Results represent the mean ± SD (CML, n = 7; CB, n = 3). (C) Multicolor flow cytometry analysis of CD26 expression on CD34+/CD38─ SC in patients with CML (CP, n = 33; AP, n = 3; BP, n = 2), normal/reactive BM (n = 33), and BM of patients with idiopathic cytopenia of unknown significance (ICUS) (n = 15), AML (n = 39), myelodysplastic syndrome (MDS) (n = 12), JAK2 V617F+ myeloproliferative neoplasms (MPN) (n = 6), MDS/MPN overlap disorders (n = 5), and indolent systemic mastocytosis (ISM) (n = 7). The upper panel shows the percentage of CD26+ SC. Boxes represent the 25th to 75th percentile. The lower subpanel shows representative staining examples: red indicates a positive staining-reaction, blue indicates a negative stain. The isotype-matched control antibody is also shown (black open histograms). (D) The upper panel shows a FISH analysis of purified CD34+/CD38−/CD26+ CML LSC (left) and CD34+/CD38−/CD26− SC (right) in one CML patient. FISH was performed on cytospin slides using a triple color–staining approach, with 1 probe specific for ABL1 (red) and 2 (green and blue) specific for BCR. Red-green and red-blue fusion-spots indicate BCR/ABL1. Quantitative FISH data are shown in supplemental Table 13. Lower panel: Percentage of BCR/ABL1+ cells by FISH (left panel) and percent BCR/ABL1 mRNA relative to ABL mRNA (right panel) in sorted CD34+/CD38−/CD26+ LSC and CD34+/CD38−/CD26− SC from the same patients. FISH data are expressed as mean ± SD from 3 patients, and BCR/ABL1 mRNA levels from sorted cells as mean ± SD from 4 different CML patients.
LSCs in CML express CD26. (A) Heat map analysis of gene array data of RNA isolated from sorted CD34+/CD38─ SCs (left) or CD34+/CD38+ progenitor cells (right) obtained from patients with chronic phase (CP) CML (n = 4) and cord blood (CB) SC. Gene array analyses were performed as described in the supplemental material available on the Blood Web site. Expression levels were normalized to CB control. Top-regulated genes of interest are shown. (B) Heat map analysis of pPCR data (upper panel). mRNA analyses were performed on sorted CD34+/CD38─ SC (upper left) and CD34+/CD38+ cells (upper right) obtained from 3 CB samples and 4 patients with CML. Top-regulated genes of interest are shown. Lower panel: qPCR analysis of expression of CD26 mRNA in sorted CD34+/CD38─ SC (lower left) and CD34+/CD38+ cells (lower right) obtained from 7 patients with CML and 3 CB samples. CD26 mRNA levels are expressed as percent of ABL. Results represent the mean ± SD (CML, n = 7; CB, n = 3). (C) Multicolor flow cytometry analysis of CD26 expression on CD34+/CD38─ SC in patients with CML (CP, n = 33; AP, n = 3; BP, n = 2), normal/reactive BM (n = 33), and BM of patients with idiopathic cytopenia of unknown significance (ICUS) (n = 15), AML (n = 39), myelodysplastic syndrome (MDS) (n = 12), JAK2 V617F+ myeloproliferative neoplasms (MPN) (n = 6), MDS/MPN overlap disorders (n = 5), and indolent systemic mastocytosis (ISM) (n = 7). The upper panel shows the percentage of CD26+ SC. Boxes represent the 25th to 75th percentile. The lower subpanel shows representative staining examples: red indicates a positive staining-reaction, blue indicates a negative stain. The isotype-matched control antibody is also shown (black open histograms). (D) The upper panel shows a FISH analysis of purified CD34+/CD38−/CD26+ CML LSC (left) and CD34+/CD38−/CD26− SC (right) in one CML patient. FISH was performed on cytospin slides using a triple color–staining approach, with 1 probe specific for ABL1 (red) and 2 (green and blue) specific for BCR. Red-green and red-blue fusion-spots indicate BCR/ABL1. Quantitative FISH data are shown in supplemental Table 13. Lower panel: Percentage of BCR/ABL1+ cells by FISH (left panel) and percent BCR/ABL1 mRNA relative to ABL mRNA (right panel) in sorted CD34+/CD38−/CD26+ LSC and CD34+/CD38−/CD26− SC from the same patients. FISH data are expressed as mean ± SD from 3 patients, and BCR/ABL1 mRNA levels from sorted cells as mean ± SD from 4 different CML patients.
Expression of surface antigens on CD34+/CD38− and CD34+/CD38+ cells in patients with CML and in normal/reactive BM
| . | . | Expression on CP CML cells . | Expression on AP CML cells . | Expression on normal BM cells . | |||
|---|---|---|---|---|---|---|---|
| CD . | Antigen . | CD34+/CD38− . | CD34+/CD38+ . | CD34+/CD38− . | CD34+/CD38+ . | CD34+/CD38− . | CD34+/CD38+ . |
| CD9 | Tetraspanin 29 | + | + | n.t. | n.t. | +/− | +/− |
| CD25 | IL-2RA | ++ | +/– | ++ | + | − | +/− |
| CD26 | DPPIV | ++ | +/− | ++ | +/− | − | − |
| CD33 | Siglec-3 | ++ | ++ | ++ | ++ | + | ++ |
| CD36 | TSPR | +/− | + | n.t. | n.t. | − | +/− |
| CD41 | ITGA2B | + | + | +/− | +/− | + | + |
| CD42b | GP1BA | − | − | − | − | − | − |
| CD44 | Pgp1 | ++ | ++ | ++ | ++ | ++ | ++ |
| CD46 | MCP | ++ | ++ | ++ | ++ | ++ | ++ |
| CD49f | ITGA6B | + | + | + | +/− | + | + |
| CD52 | CAMPATH | + | +/− | n.t. | n.t. | + | + |
| CD62 | SELP | − | − | − | − | − | − |
| CD90 | Thy-1 | + | +/− | n.t. | n.t. | + | +/− |
| CD96 | Tactile | − | − | − | − | − | − |
| CD105 | Endoglin | ++ | ++ | ++ | ++ | + | + |
| CD114 | G-CSFR | + | +/− | + | +/− | + | + |
| CD117 | SCFR/KIT | ++ | ++ | ++ | ++ | ++ | ++ |
| CD123 | IL-3RA | + | + | + | + | +/− | + |
| CD130 | IL6ST | − | − | − | − | +/− | +/− |
| CD133 | Prominin 1 | ++ | +/− | ++ | +/− | ++ | ++ |
| CD135 | FLT3 | + | +/− | + | +/− | + | + |
| CD151 | RAPH | + | + | + | + | + | + |
| CD164 | Sialomucin | + | + | + | + | ++ | ++ |
| CD184 | CXCR4 | + | + | + | + | + | + |
| CD200 | OX2 | ++ | + | + | + | ++ | ++ |
| CD203c | ENPP3 | − | − | − | − | − | − |
| CD243 | MDR1 | − | − | − | − | − | − |
| CD271 | NGF-R | − | − | n.t. | n.t. | − | − |
| n.c. | CLL1 | +/− | +/− | n.t. | n.t. | − | +/− |
| n.c. | IL-1RAP | + | + | n.t. | n.t. | − | +/− |
| n.c. | NPDC1 | + | + | n.t. | n.t. | + | + |
| n.c. | ROBO4 | + | +/− | n.t. | n.t. | + | +/− |
| . | . | Expression on CP CML cells . | Expression on AP CML cells . | Expression on normal BM cells . | |||
|---|---|---|---|---|---|---|---|
| CD . | Antigen . | CD34+/CD38− . | CD34+/CD38+ . | CD34+/CD38− . | CD34+/CD38+ . | CD34+/CD38− . | CD34+/CD38+ . |
| CD9 | Tetraspanin 29 | + | + | n.t. | n.t. | +/− | +/− |
| CD25 | IL-2RA | ++ | +/– | ++ | + | − | +/− |
| CD26 | DPPIV | ++ | +/− | ++ | +/− | − | − |
| CD33 | Siglec-3 | ++ | ++ | ++ | ++ | + | ++ |
| CD36 | TSPR | +/− | + | n.t. | n.t. | − | +/− |
| CD41 | ITGA2B | + | + | +/− | +/− | + | + |
| CD42b | GP1BA | − | − | − | − | − | − |
| CD44 | Pgp1 | ++ | ++ | ++ | ++ | ++ | ++ |
| CD46 | MCP | ++ | ++ | ++ | ++ | ++ | ++ |
| CD49f | ITGA6B | + | + | + | +/− | + | + |
| CD52 | CAMPATH | + | +/− | n.t. | n.t. | + | + |
| CD62 | SELP | − | − | − | − | − | − |
| CD90 | Thy-1 | + | +/− | n.t. | n.t. | + | +/− |
| CD96 | Tactile | − | − | − | − | − | − |
| CD105 | Endoglin | ++ | ++ | ++ | ++ | + | + |
| CD114 | G-CSFR | + | +/− | + | +/− | + | + |
| CD117 | SCFR/KIT | ++ | ++ | ++ | ++ | ++ | ++ |
| CD123 | IL-3RA | + | + | + | + | +/− | + |
| CD130 | IL6ST | − | − | − | − | +/− | +/− |
| CD133 | Prominin 1 | ++ | +/− | ++ | +/− | ++ | ++ |
| CD135 | FLT3 | + | +/− | + | +/− | + | + |
| CD151 | RAPH | + | + | + | + | + | + |
| CD164 | Sialomucin | + | + | + | + | ++ | ++ |
| CD184 | CXCR4 | + | + | + | + | + | + |
| CD200 | OX2 | ++ | + | + | + | ++ | ++ |
| CD203c | ENPP3 | − | − | − | − | − | − |
| CD243 | MDR1 | − | − | − | − | − | − |
| CD271 | NGF-R | − | − | n.t. | n.t. | − | − |
| n.c. | CLL1 | +/− | +/− | n.t. | n.t. | − | +/− |
| n.c. | IL-1RAP | + | + | n.t. | n.t. | − | +/− |
| n.c. | NPDC1 | + | + | n.t. | n.t. | + | + |
| n.c. | ROBO4 | + | +/− | n.t. | n.t. | + | +/− |
CP, chronic phase; AP, accelerated phase; CLL1, C type lectin-like molecule 1; CML, chronic myeloid leukemia; DPPIV, dipeptidylpeptidase IV; G-CSF, granulocyte colony-stimulating factor; IL-2, interleukin-2; n.c., not yet clustered; NGF, nerve growth factor; n.t., not tested; SCF, SC factor; TSPR, thrombospondin receptor; ++, strongly expressed in a majority of cases; +, clear expression in majority of cases; +/−, expression in minority of cases or weak expression in majority of cases; −, no expression in a vast majority of cases.
Identification of DPPIV (CD26) as a specific biomarker of CML LSC
In consecutive validation experiments, only CD26 was found to meet all requirements for a specific marker of CML LSC. In fact, whereas CD25 and IL-1RAP are expressed on different types of cells in the CML clone, expression of CD26 was found to be restricted essentially to LSC and is otherwise only expressed on basophils and T cells (supplemental Table 12). Second, whereas IL-1RAP and CD25 are frequently expressed on LSC in acute myeloid leukemia (AML) and other types of leukemias, CD26 was only detectable on LSC in CML but not on SC in other myeloid neoplasms, with the exception of a very few patients with AML (Figure 1C and supplemental Figure 2A). As was determined by FISH, nearly 100% of the purified CD26+ LSC expressed BCR/ABL1, whereas the CD26─ SC from the same patients were all BCR/ABL1– (Figure 1D and supplemental Table 13). Expression of BCR/ABL1 in CD26+ CML LSC was also confirmed by qPCR (Figure 1D).
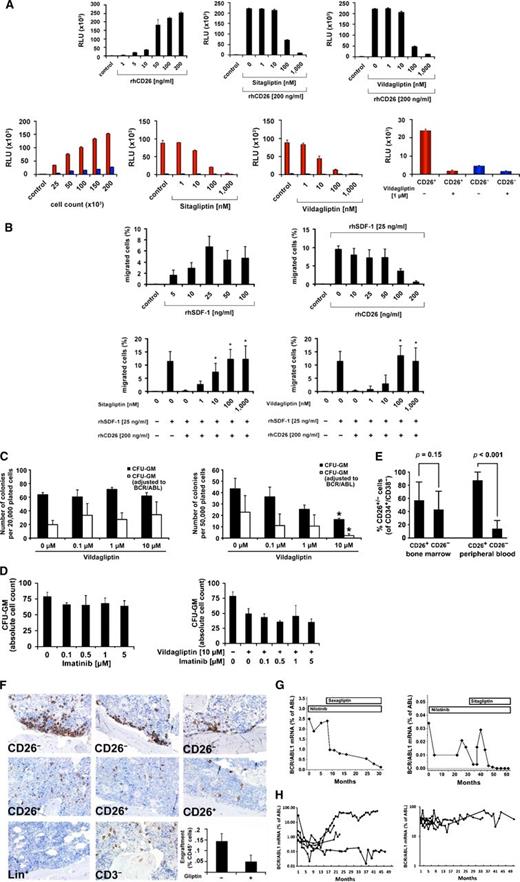
CD26+ LSC exhibit long-term proliferation- and NSG repopulation activity
In short-term colony assays and in LTC-ICs, colony cells derived from CD26+ LSC (obtained from CP CML patients) contained BCR/ABL1 mRNA, whereas cells of colonies derived from CD26─ SC (from the same patients) did not contain BCR/ABL1 (Figure 2A). In NSG mice, CD26+ LSC obtained from CP CML patients were found to produce a myeloid (granulo-monocytic) BCR/ABL1+ engraftment after 16 and 28 weeks, whereas CD26─ SC isolated from the same patients (CP CML) produced BCR/ABL1– multilineage (myeloid and B lineage) engraftment (Figure 2B-D). As expected, the more mature CD34─ myeloid cells (CD14+, CD15+) in CP CML patients did not engraft or survive in NSG mice after 32 weeks (Figure 3).
Clonogenic growth and in vivo repopulation of CML SCs. (A) Left: Sorted (>98% pure) CD34+/CD38−/CD26− SC, CD34+/CD38−/CD26+ SC, and CD34+/CD38+ sorted progenitor cells (CP CML) were plated in methylcellulose and incubated at 37°C for 16 days. Then CFU-GM–derived colonies were counted under an inverted microscope (black bars). Thereafter, colonies were picked and BCR/ABL1 mRNA expression was determined by qPCR (white bars). Results represent the mean ± SD of triplicates. Right: Purified CD34+/CD38−/CD26−, CD34+/CD38−/CD26+, and CD34+/CD38+ cells were incubated on irradiated M2-10B4 feeder cells for 3 weeks. Then cells were harvested and cultured in a CFU-GM assay. CFU-GM colonies (black bars) were counted by microscopy and BCR/ABL1 mRNA levels were determined by qPCR (white bars). (B) Experimental setup of xenograft experiments with purified CML cells. First, CML cells (CP) were depleted from lineage+ cells and separated into CD26+ cells and CD26− cells by cell sorting (purity >98%). The 2 fractions were separately injected intravenously into irradiated NSG mice (0.1-0.8 × 106 CD26+ cells per mouse and 0.5-1.5 × 106 CD26− cells per mouse). (C) After 16 weeks or 28 weeks, flushed BM cells of NSG mice injected with CD26− cells (blue symbols) or CD26+ cells (red symbols) were analyzed for the presence of CD33+ myeloid cells (circles) and CD19+ B cells (triangles) by flow cytometry. (D) Flow cytometry patterns of engrafted cells in 2 representative mice, one injected with CD26− SC, where multilineage BCR/ABL1– engraftment was found (upper panel) and one with CD26+ SC, where engraftment with BCR/ABL1+ myeloid cells was seen (lower subpanel). The table shows a summary of engraftment results with percentages of human CD45+ cells, CD33+ cells, and CD19+ cells, and the detection of BCR/ABL1 by qPCR. n.d., not determined.
Clonogenic growth and in vivo repopulation of CML SCs. (A) Left: Sorted (>98% pure) CD34+/CD38−/CD26− SC, CD34+/CD38−/CD26+ SC, and CD34+/CD38+ sorted progenitor cells (CP CML) were plated in methylcellulose and incubated at 37°C for 16 days. Then CFU-GM–derived colonies were counted under an inverted microscope (black bars). Thereafter, colonies were picked and BCR/ABL1 mRNA expression was determined by qPCR (white bars). Results represent the mean ± SD of triplicates. Right: Purified CD34+/CD38−/CD26−, CD34+/CD38−/CD26+, and CD34+/CD38+ cells were incubated on irradiated M2-10B4 feeder cells for 3 weeks. Then cells were harvested and cultured in a CFU-GM assay. CFU-GM colonies (black bars) were counted by microscopy and BCR/ABL1 mRNA levels were determined by qPCR (white bars). (B) Experimental setup of xenograft experiments with purified CML cells. First, CML cells (CP) were depleted from lineage+ cells and separated into CD26+ cells and CD26− cells by cell sorting (purity >98%). The 2 fractions were separately injected intravenously into irradiated NSG mice (0.1-0.8 × 106 CD26+ cells per mouse and 0.5-1.5 × 106 CD26− cells per mouse). (C) After 16 weeks or 28 weeks, flushed BM cells of NSG mice injected with CD26− cells (blue symbols) or CD26+ cells (red symbols) were analyzed for the presence of CD33+ myeloid cells (circles) and CD19+ B cells (triangles) by flow cytometry. (D) Flow cytometry patterns of engrafted cells in 2 representative mice, one injected with CD26− SC, where multilineage BCR/ABL1– engraftment was found (upper panel) and one with CD26+ SC, where engraftment with BCR/ABL1+ myeloid cells was seen (lower subpanel). The table shows a summary of engraftment results with percentages of human CD45+ cells, CD33+ cells, and CD19+ cells, and the detection of BCR/ABL1 by qPCR. n.d., not determined.
CML SC express functional DPPIV activity. (A) DPPIV enzyme activity of recombinant human (rh) DPPIV/CD26 (upper panel) and cell-derived CD26 (lower panel) measured as described in the supplemental material. Upper right: rhCD26 (200 ng/mL) was preincubated with various concentrations of sitagliptin or vildagliptin (as indicated) at 37°C for 30 minutes. Lower left: DPPIV activity in lysates of CD26-transfected KU812 cells (red bars) and control-vector–transfected CD26− KU812 cells (blue bars) (each 50 × 103). Before being analyzed, cells were incubated with various concentrations of sitagliptin or vildagliptin for 30 minutes. Lower right: Primary sorted CD26+ SC (red bars) and CD26− SC (blue bars) (each 25 × 103 cells) from a patient with CP CML were incubated with or without vildagliptin at 37°C for 30 minutes. DPPIV enzyme activity was expressed as relative luminescence units (RLU) per 50 × 103 cells. Results represent the mean ± SD of triplicates. (B) For chemotactic migration, CXCR4+ U937 cells (1.5 × 105/well) were applied in a double-chamber chemotaxis assay. Cells were allowed to migrate through 5-µm filters against various concentrations of SDF-1 (upper left) at 37°C for 4 hours. Transmigrated cells were counted by flow cytometry. In a next step, SDF-1 (25 ng/mL) was incubated with various concentrations of rhCD26 overnight and then used to induce migration of U937 cells (upper right subpanel). Finally, 200 ng/mL of rhCD26 were preincubated with various concentrations of sitagliptin (lower left) or vildagliptin (lower right) for 30 minutes, before SDF-1 (25 ng/mL) was added (overnight). Results represent the mean ± SD from 4 experiments. *P < .05 compared with untreated cells. (C) Left: CML MNC were plated in methylcellulose supplemented with cytokines in the absence (control) or presence of various concentrations of vildagliptin at 37°C for 16 days. Then CFU-GM colonies (black bars) were counted. Thereafter, colonies were picked and BCR/ABL1 mRNA expression was determined by qPCR (white bars). Right: CML MNC were incubated on irradiated M2-10B4 feeder cells for 3 weeks in the absence (control) or presence of various concentrations of vildagliptin. Then cells were harvested and plated in a CFU assay for 16 days. CFU-GM colonies (black bars) and BCR/ABL1+ colonies (white bars) are shown. *P < .05 compared with untreated cells. (D) CML MNC were plated on a feeder layer of irradiated M2-10B4 cells in various concentrations of imatinib (left panel) or imatinib in combination with 10 µM vildagliptin (right panel) for 48 hours. Thereafter, nonadherent cells of each well were harvested and plated in a CFU assay. After 2 weeks, colonies were counted. (E) The percentage of CD26+ and CD26− SC in the BM (left bars) and PB (right bars) of patients with CML determined by flow cytometry. (F) NSG mouse BM sections stained with an antibody against human CD45. NSG mice were injected with CD26+ SC, CD26− SC, lineage-positive (Lin+; CD14+, CD15+ sorted) cells (negative control), or CD3–depleted MNC (positive control) obtained from patients with CP CML. CD26+ LSC produced diffuse engraftment in the NSG BM, whereas normal CD26− SC produced myeloid engraftment at the endosteal surface. Original magnification ×40/0.85. (F) Lower right: Engraftment of CD26+ CML LSC after preincubation with control medium (−) or vildagliptin, 10 µM (+) at 37°C for 30 minutes. Engraftment in NSG mice (3 mice per group) was determined by measuring the percentage of CD45+ cells in the BM (after 18 weeks) by flow cytometry. Results represent the mean ± SD of 3 mice (each group). (G) BCR/ABL1 transcript levels, quantified by qPCR, in 2 patients with imatinib-resistant CML who received nilotinib (left panel patient: 200 mg/day; right panel patient: 800 mg/day), and later, because of uncontrolled diabetes mellitus, saxagliptin, 5 mg daily per os (left panel) or sitagliptin, 50 mg daily per os (right panel). (H) BCR/ABL1 mRNA levels in 8 CML patients (4 with high BCR/ABL1 burden and 4 with low BCR/ABL1 burden) who lost MMR or CMR during therapy with nilotinib and did not receive a gliptin. BCR/ABL1 transcript levels were quantified by qPCR according to the international scale.
CML SC express functional DPPIV activity. (A) DPPIV enzyme activity of recombinant human (rh) DPPIV/CD26 (upper panel) and cell-derived CD26 (lower panel) measured as described in the supplemental material. Upper right: rhCD26 (200 ng/mL) was preincubated with various concentrations of sitagliptin or vildagliptin (as indicated) at 37°C for 30 minutes. Lower left: DPPIV activity in lysates of CD26-transfected KU812 cells (red bars) and control-vector–transfected CD26− KU812 cells (blue bars) (each 50 × 103). Before being analyzed, cells were incubated with various concentrations of sitagliptin or vildagliptin for 30 minutes. Lower right: Primary sorted CD26+ SC (red bars) and CD26− SC (blue bars) (each 25 × 103 cells) from a patient with CP CML were incubated with or without vildagliptin at 37°C for 30 minutes. DPPIV enzyme activity was expressed as relative luminescence units (RLU) per 50 × 103 cells. Results represent the mean ± SD of triplicates. (B) For chemotactic migration, CXCR4+ U937 cells (1.5 × 105/well) were applied in a double-chamber chemotaxis assay. Cells were allowed to migrate through 5-µm filters against various concentrations of SDF-1 (upper left) at 37°C for 4 hours. Transmigrated cells were counted by flow cytometry. In a next step, SDF-1 (25 ng/mL) was incubated with various concentrations of rhCD26 overnight and then used to induce migration of U937 cells (upper right subpanel). Finally, 200 ng/mL of rhCD26 were preincubated with various concentrations of sitagliptin (lower left) or vildagliptin (lower right) for 30 minutes, before SDF-1 (25 ng/mL) was added (overnight). Results represent the mean ± SD from 4 experiments. *P < .05 compared with untreated cells. (C) Left: CML MNC were plated in methylcellulose supplemented with cytokines in the absence (control) or presence of various concentrations of vildagliptin at 37°C for 16 days. Then CFU-GM colonies (black bars) were counted. Thereafter, colonies were picked and BCR/ABL1 mRNA expression was determined by qPCR (white bars). Right: CML MNC were incubated on irradiated M2-10B4 feeder cells for 3 weeks in the absence (control) or presence of various concentrations of vildagliptin. Then cells were harvested and plated in a CFU assay for 16 days. CFU-GM colonies (black bars) and BCR/ABL1+ colonies (white bars) are shown. *P < .05 compared with untreated cells. (D) CML MNC were plated on a feeder layer of irradiated M2-10B4 cells in various concentrations of imatinib (left panel) or imatinib in combination with 10 µM vildagliptin (right panel) for 48 hours. Thereafter, nonadherent cells of each well were harvested and plated in a CFU assay. After 2 weeks, colonies were counted. (E) The percentage of CD26+ and CD26− SC in the BM (left bars) and PB (right bars) of patients with CML determined by flow cytometry. (F) NSG mouse BM sections stained with an antibody against human CD45. NSG mice were injected with CD26+ SC, CD26− SC, lineage-positive (Lin+; CD14+, CD15+ sorted) cells (negative control), or CD3–depleted MNC (positive control) obtained from patients with CP CML. CD26+ LSC produced diffuse engraftment in the NSG BM, whereas normal CD26− SC produced myeloid engraftment at the endosteal surface. Original magnification ×40/0.85. (F) Lower right: Engraftment of CD26+ CML LSC after preincubation with control medium (−) or vildagliptin, 10 µM (+) at 37°C for 30 minutes. Engraftment in NSG mice (3 mice per group) was determined by measuring the percentage of CD45+ cells in the BM (after 18 weeks) by flow cytometry. Results represent the mean ± SD of 3 mice (each group). (G) BCR/ABL1 transcript levels, quantified by qPCR, in 2 patients with imatinib-resistant CML who received nilotinib (left panel patient: 200 mg/day; right panel patient: 800 mg/day), and later, because of uncontrolled diabetes mellitus, saxagliptin, 5 mg daily per os (left panel) or sitagliptin, 50 mg daily per os (right panel). (H) BCR/ABL1 mRNA levels in 8 CML patients (4 with high BCR/ABL1 burden and 4 with low BCR/ABL1 burden) who lost MMR or CMR during therapy with nilotinib and did not receive a gliptin. BCR/ABL1 transcript levels were quantified by qPCR according to the international scale.
LSC-derived DPPIV (CD26) cleaves SDF-1 and facilitates a SC escape from the BM niche, resulting in niche-independent spread of leukemic cells
Highly purified CD34+/CD38─/CD26+ CML LSC exhibited DPPIV (CD26) enzymatic activity, whereas CD34+/CD38─/CD26─ SC from the same patients did not express DPPIV activity (Figure 3A). The DPPIV-targeting drugs sitagliptin and vildagliptin were found to block the activity of recombinant DPPIV, DPPIV transfected into CML cells (KU812), and DPPIV obtained from primary CML LSC (Figure 3A). In addition, recombinant DPPIV blocked the SDF-1–induced migration of CXCR4+ U937 cells, and this DPPIV effect was reverted by sitagliptin and vildagliptin (Figure 3B). The same result was obtained with primary CML LSC (supplemental Figure 3). Furthermore, we found that although gliptins did not affect colony formation per se, gliptin exposure of CML LSC is associated with reduced numbers of BCR/ABL1+ colony-forming unit–granulocyte macrophage (CFU-GM) in LTC-IC (Figure 3C). In addition, when plated on M2-10B4 cells, short-term exposure of CML cells to vildagliptin resulted in CFU depletion, presumably through (better) attachment of SC to the stroma cell layer, and imatinib augmented this gliptin-effect (Figure 3D). These drug effects were not seen with normal BM SC (supplemental Figure 4A-B). Collectively, these data suggest that CD26+ CML LSC escape from niche cells because of disrupted SDF-1-CXCR4 interactions. Correspondingly, relative to normal SC, the numbers of circulating CD26+ CML LSC in our CML patients were higher than BM LSC numbers (Figure 3E). The escape hypothesis was further supported by data obtained in NSG mice. In fact, whereas normal CD26─ SC isolated from our CML patients engrafted near or at the endosteal layer (SC niche) in NSG mice, CD26+ LSC from the same patients showed a diffuse engraftment pattern (Figure 3F). Such diffuse pattern of infiltration of CD34+ cells and CD26+ cells was also found in our CML patients (supplemental Figure 5).
Targeting of CD26 with gliptins suppresses the leukemic spread in CML
To test the hypothesis that gliptin exposure redistributes CML LSC back into the SC niche to control their subsequent spread, we incubated primary CP CML LSC with vildagliptin before injecting these cells into NSG mice. As is shown in Figure 3F, preincubation of CML cells with vildagliptin resulted in reduced engraftment of leukemic cells. We also screened our CML cohort for patients who were treated with nilotinib but remained BCR/ABL1+ and had received a gliptin because of uncontrolled diabetes mellitus. We identified 2 such patients. In both of them, BCR/ABL1 transcript levels decreased during treatment with saxagliptin or sitagliptin (Figure 3G). In other CML patients who had lost or did not achieve major molecular remission (MMR) during nilotinib and did not receive a gliptin, BCR/ABL1 continued to increase or remained stable, but did not decrease (Figure 3H).
Regulation of expression of CD25, CD26, and CXCR4 on LSC in CML
Although CD26 is specifically expressed on BCR/ABL1+ LSC, BCR/ABL1 did not induce expression of CD26 in Mo7e cells (supplemental Figure 6A) or Ba/F3 cells (not shown), and only a marginal increase in CD25 mRNA was seen (supplemental Figure 6A). Moreover, imatinib did not block expression of CD25 or CD26 in CML LSC (supplemental Figure 6B-C). Nilotinib was found to slightly downregulate expression of CD25 and CD26 on CML LSC (supplemental Figure 6B). We also found that CD25 and CD26 are expressed on CML LSC independent of IL-3 and granulocyte colony-stimulating factor (G-CSF) activity. In particular, no significant change in expression of CD25 and CD26 was found when CML LSC were exposed to a blocking anti–G-CSF antibody, a blocking anti–IL-3 antibody, or recombinant IL-3 (supplemental Figure 7A-D). We also examined the effects of gliptins, imatinib, and rhCD26 on expression of CXCR4 on CML cells. However, we were not able to detect any change in CXCR4 expression on KU812 cells or CD34+/CD38− CML LSC after exposure to these compounds (supplemental Figure 8). Whereas freshly isolated CB SC (CD34+/CD38─) failed to express CD26, these cells started to express low amounts of CD26 after short-term culture (supplemental Figure 9A-B). However, exposure of CB SC to G-CSF did not result in upregulation of CD26 (supplemental Figure 9A-B).
Successful treatment with imatinib eliminates circulating CML LSC
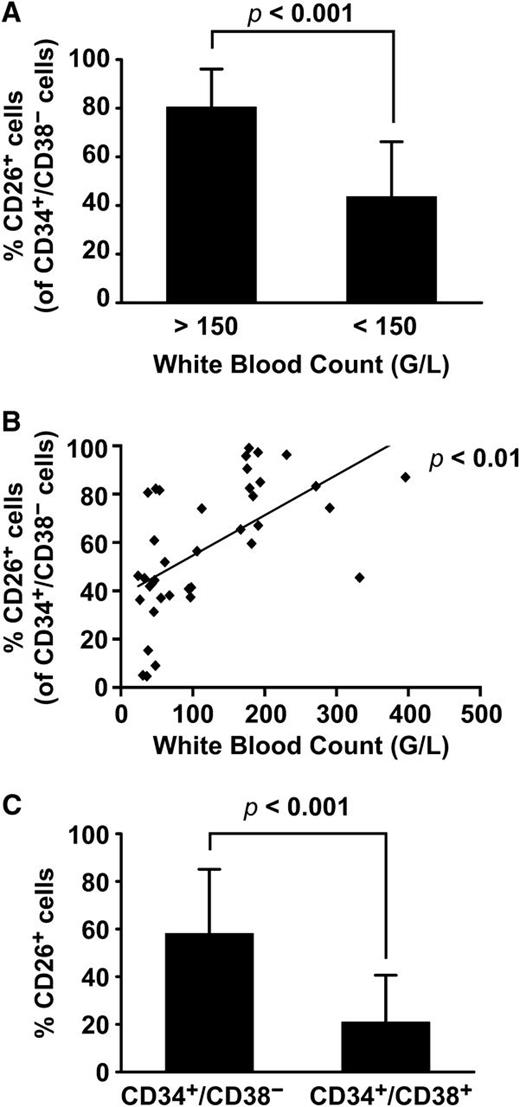
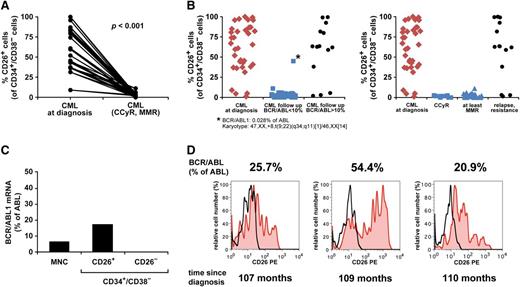
Although BM and PB SC were found to express CD26 in all CML patients at diagnosis, the expression levels and percentage of CD26+ cells varied from donor to donor (Figure 1C). The percentage of CD26+ SC was found to correlate with white blood counts (Figure 4) but did not correlate with other prognostic variables or scores (Sokal, Hasford, EUTOS) (Figure 5). During successful therapy with imatinib, CD26+ LSC decreased substantially, and in most cases, CD26+ LSC were no longer detectable in the PB (Figure 6A-B). However, during the initial phase of successful therapy (at month 3) residual CD26+ LSC were detected and we were able to sort out these cells in one patient. As expected, BCR/ABL1 mRNA was detectable in the residual CD26+ LSC, whereas the sorted CD26─ SC did not express BCR/ABL1 (Figure 6C). In most patients with resistant or relapsed CML, CD26+ LSC were detectable in the PB and BM (Figure 6B,D). We also examined the expression of CD26 on LSC in CML BP. In most of these patients, including lymphoid BP, LSC expressed CD25, and CD26 at the protein and mRNA level (supplemental Figure 10A-B). However, in a few patients with BP, LSC did not express CD26 (supplemental Figure 10A). IL-1RAP was also found to be expressed on LSC in most BP patients (supplemental Figure 10C).
Correlation between leukemic burden and CD26+ SC in patients with CML. (A) The comparison of the percentage of CD26+ BM SC (of all CD34+/CD38− cells) in patients with high white blood counts (WBC) at diagnosis (>150 G/L, n = 15, left bar) with that in patients in whom lower WBC were measured (<150, G/L, n = 23). (B) The correlation between WBC and the percentage of CD26+ SC in CML at diagnosis in all patients (n = 38) are shown. (C) The comparison of the percentage of CD26+ SC with the percentage of CD26+ progenitor cells (CD34+/CD38+) in patients with freshly diagnosed CML are shown. As is visible, CD26 is primarily expressed in the CD34+/CD38− fraction of the leukemic clone.
Correlation between leukemic burden and CD26+ SC in patients with CML. (A) The comparison of the percentage of CD26+ BM SC (of all CD34+/CD38− cells) in patients with high white blood counts (WBC) at diagnosis (>150 G/L, n = 15, left bar) with that in patients in whom lower WBC were measured (<150, G/L, n = 23). (B) The correlation between WBC and the percentage of CD26+ SC in CML at diagnosis in all patients (n = 38) are shown. (C) The comparison of the percentage of CD26+ SC with the percentage of CD26+ progenitor cells (CD34+/CD38+) in patients with freshly diagnosed CML are shown. As is visible, CD26 is primarily expressed in the CD34+/CD38− fraction of the leukemic clone.
Correlation between the numbers of CD26+ SCs and CML scores. (A-B) In 27 patients with CML (BM samples, n = 26; PB, n = 1), the percentage of CD26+ SC (of all CD34+/CD38− cells) and CD26+ progenitor cells (of all CD34+/CD38+ cells) was determined by flow cytometry. Percentage of CD26+ SC (left) and CD26+ progenitor cells (right subpanel) in various categories of the Sokal score (A) and Hasford score (B). In high-risk patients, phenotypically more mature progenitor cells tend to acquire CD26. Sokal scoring: high (>1.2), intermediate (int) (0.8-1.2), and low (<0.8). Hasford scoring: high (>1480), int (781-1480), low (≤780). (C) Correlations between the percentages of CD26+ SC and the 3 scoring systems. As shown, no substantial correlation was found.
Correlation between the numbers of CD26+ SCs and CML scores. (A-B) In 27 patients with CML (BM samples, n = 26; PB, n = 1), the percentage of CD26+ SC (of all CD34+/CD38− cells) and CD26+ progenitor cells (of all CD34+/CD38+ cells) was determined by flow cytometry. Percentage of CD26+ SC (left) and CD26+ progenitor cells (right subpanel) in various categories of the Sokal score (A) and Hasford score (B). In high-risk patients, phenotypically more mature progenitor cells tend to acquire CD26. Sokal scoring: high (>1.2), intermediate (int) (0.8-1.2), and low (<0.8). Hasford scoring: high (>1480), int (781-1480), low (≤780). (C) Correlations between the percentages of CD26+ SC and the 3 scoring systems. As shown, no substantial correlation was found.
The decrease in CD26+ SCs correlates with clinical responses in CML. (A) The percentage of CD26+ SC (CD34+/CD38− cells) in patients with CML at diagnosis (n = 21) and at the time of complete cytogenetic response (CCyR) and/or MMR after treatment (imatinib n = 17, nilotinib n = 2, dasatinib n = 2) are shown. (B) Left panel: The percentage of CD26+ SC (CD34+/CD38− cells) in patients with CML at diagnosis (n = 35), those who had <10% BCR/ABL1 during the first year of therapy (n = 46; imatinib n = 38, nilotinib n = 7, dasatinib n = 1), and patients with >10% BCR/ABL1 in the follow-up (n = 14; imatinib n = 4, nilotinib n = 2, dasatinib n = 1, multiple therapies n = 7) are shown. (B) Right panel: The percentage of CD26+ SC (CD34+/CD38− cells) in patients with CML at diagnosis (n = 35), patients with CCyR without MMR (n = 10), patients with MMR (n = 36), and patients with fully relapsed or resistant CML (n = 14) are shown. (C) BCR/ABL1 mRNA levels in sorted CD34+/CD38−/CD26+ and CD34+/CD38−/CD26− SC in a patient with CML after 3 months of treatment with imatinib (400 mg/day). (D) Percentage of CD26+ SC (CD34+/CD38− cells) and BCR/ABL1 mRNA levels (according to IS) in a patient with CML who had developed resistance against imatinib and nilotinib. The figure shows 3 different time points.
The decrease in CD26+ SCs correlates with clinical responses in CML. (A) The percentage of CD26+ SC (CD34+/CD38− cells) in patients with CML at diagnosis (n = 21) and at the time of complete cytogenetic response (CCyR) and/or MMR after treatment (imatinib n = 17, nilotinib n = 2, dasatinib n = 2) are shown. (B) Left panel: The percentage of CD26+ SC (CD34+/CD38− cells) in patients with CML at diagnosis (n = 35), those who had <10% BCR/ABL1 during the first year of therapy (n = 46; imatinib n = 38, nilotinib n = 7, dasatinib n = 1), and patients with >10% BCR/ABL1 in the follow-up (n = 14; imatinib n = 4, nilotinib n = 2, dasatinib n = 1, multiple therapies n = 7) are shown. (B) Right panel: The percentage of CD26+ SC (CD34+/CD38− cells) in patients with CML at diagnosis (n = 35), patients with CCyR without MMR (n = 10), patients with MMR (n = 36), and patients with fully relapsed or resistant CML (n = 14) are shown. (C) BCR/ABL1 mRNA levels in sorted CD34+/CD38−/CD26+ and CD34+/CD38−/CD26− SC in a patient with CML after 3 months of treatment with imatinib (400 mg/day). (D) Percentage of CD26+ SC (CD34+/CD38− cells) and BCR/ABL1 mRNA levels (according to IS) in a patient with CML who had developed resistance against imatinib and nilotinib. The figure shows 3 different time points.
Discussion
The concept of LSC has been developed with the intention to explain the biology and functional hierarchy of myeloid leukemias and to develop novel curative drug therapies.6-11 CML LSC supposedly reside within a CD34+/CD38─ fraction of the leukemic clone.7-10,25-27 However, this phenotype is shared also by normal BM SCs. We performed gene array, PCR, and flow cytometry studies to identify novel markers of CML LSC. Of all surface antigens examined, the surface enzyme CD26 (DPPIV) was identified as a specific, reliable, and functionally important marker of LSC in CML. In fact, in contrast to other antigens, only CD26 was found to be expressed invariably on LSC in all patients but was not detectable on CD34+/CD38─ SC in normal BM samples or on LSC in other myeloid neoplasms. Moreover, in most patients with CML, CD26 was expressed only on CD34+/CD38─ LSC but not on more mature CD34+/CD38+ progenitors.
Several different markers are considered to be expressed more or less specifically on LSC in CML.28-30 In the present study, we were able to confirm expression of these antigens, including Siglec-3 (CD33), Pgp1 (CD44), IL-3RA (CD123), and IL-1RAP. In particular, CD34+/CD38─/CD26+ LSC in CML coexpressed these antigens in most donors. In addition, we were able to show that CD26+ CML LSC coexpress IL-2RA (CD25). Whereas CD33 and CD123 were also detectable on normal BM SC, albeit at lower levels, CD25 and IL-1RAP were only detectable on CML LSC. However, unlike CD26, CD25 and IL-1RAP were also found to be expressed quite frequently on LSC in other leukemias, especially AML. With regard to CD25, these data confirm previous studies.36 As expected, we found a good correlation between expression of CD25 and CD26 on CML LSC, but only a rough correlation between expression of CD26 and IL-1RAP, which is best explained by the fact that in several patients, CML LSC did not express substantial amounts of IL-1RAP. Most cell-surface antigens detected on CML LSC were also identified by gene array and PCR analyses in our experiments, thereby confirming previous studies.28,37,38 Altogether, our data show that CD26 is a reliable and robust biomarker of CML LSC.
To confirm that CD26 is specifically expressed in CML LSC, we purified CD26+ and CD26─ SC from CP-CML samples to near homogeneity (>98% purity) by cell sorting. As expected, only the CD26+ LSC, but not the CD26─ SC, were found to express BCR/ABL1 by FISH and qPCR. As assessed by FISH, virtually all CD26+ SC were found to be clonal cells expressing BCR/ABL1. Moreover, only the CD26+ LSC gave rise to BCR/ABL1+ colonies in vitro in short-term and LTC-IC cultures. Finally, only the CD26+ SC were found to repopulate the NSG marrow with BCR/ABL1+ cells, whereas CD26─ SC from the same patients produced BCR/ABL1–, nonleukemic, engraftment. These data show that CD26 is a specific marker for CML LSC. However, we were not able to demonstrate reproducible CML engraftment in secondary recipient mice. This may be explained by the fact that the development of a chronic leukemia exceeds the observation period or even the lifetime of a mouse, which is in turn explained by the premalignant nature and thus low proliferation rate of CML LSC.19,39 Alternatively, the homing defect of CML LSC was responsible for the low rate and slow time of engraftment. The possibility that only more mature cells engrafted or even mimicked engraftment by maintenance could be excluded in control experiments. In fact, isolated Lin+/CD34─ CML cells (SC-depleted fractions) did not produce a stable cell population or detectable engraftment in NSG mice. Finally, our NSG mice engrafted AML cells as full-blown leukemias in the same NSG assay.40
In CML, the BCR/ABL1 mutation is considered an early and decisive driver mutation that triggers various signaling pathways and expression of diverse effector molecules in CML cells.2-4 However, not all CML-related antigens and aberrantly expressed markers may be triggered by BCR/ABL1.41 We found that expression of CD26 on CML LSC is not dependent solely on BCR/ABL1 activity. Notably, BCR/ABL1 did not induce expression of CD26 in Mo7e or Ba/F3 cells, and expression of CD26 on CML LSC was not inhibited by the addition of imatinib. These observations suggest that expression of CD26 depends on other, as yet unknown, mechanisms that may be relevant in an early phase of CML development. Alternatively, the turnover of CD26 on the surface of CML LSC is very low, so that inhibition of BCR/ABL1 was ineffective. We also asked whether expression of CD26 on CML LSC is dependent on autocrine cytokine stimulation. Notably, it has been described that CML LSC produce IL-3, G-CSF, or other cytokines in an autocrine fashion.33,42-44 However, exposure to IL-3 or a neutralizing anti–IL-3R antibody did not modulate expression of CD25 or CD26 on CML LSC, and the same was found with a neutralizing anti–G-CSF antibody. Therefore, the most likely explanation is that CD26 expression is an early BCR/ABL1-independent event in leukemogenesis that may be critical for disease initiation and/or manifestation. The fact that CD26 is detectable on LSC in all patients would favor this hypothesis. Moreover, even in primary lymphoid BP of CML (BCR/ABL1+), LSC expressed CD26. However, in one patient with BP, CD26 was no longer detectable on LSC, and also the CML cell lines examined were found to be CD26–. This observation suggests that in a terminal stage of disease, CD26 may no longer be required to trigger the spread and expansion of LSC.
In CML, one of the key features is the extramedullary spread of stem and progenitor cells, resulting in overwhelming extramedullary myelopoiesis.2,3 However, although CML LSC are known to exhibit a homing defect and altered response to niche-related chemokines such as SDF-1,31-33 the exact mechanism of LSC mobilization in CML remains unknown. In the current article, we show that CML LSC specifically express the SDF-1–degrading surface enzyme DPPIV (CD26). These data suggest that LSC-derived CD26 may degrade SDF-1 and thereby blocks SDF-1–induced migration of CML LSC. Indeed, LSC-derived CD26 was found to exhibit DPPIV activity. In addition, we were able to show that CML-derived CD26 as well as recombinant CD26 inhibit SDF-1–induced migration of CXCR4+ leukemic cells. As expected, these effects of CD26 were neutralized by gliptins. Together, these data show that CD26 is a functionally active SDF-1–degrading surface enzyme expressed on CML LSC. Expression of CD26 may thereby also explain the migration and homing defect of CML LSC. This hypothesis was further supported by the observation that CD26+ LSC produced a diffuse engraftment in the BM of NSG mice, whereas the CD26─ SC from the same patients engrafted at the endosteal region. We also asked whether normal SC express CD26. However, although we studied it extensively, we were unable to detect CD26 on SC obtained from normal or reactive BM. In freshly isolated CB samples, we were also unable to detect CD26 on CD34+/CD38─ SC. However, after short-term culture, CB SC expressed low amounts of CD26. These observations suggest that under certain (in vitro) conditions, normal (CB-derived) SC may express some CD26, which would confirm previous data.45-48 However, in contrast to a previous study,47 we were unable to show that G-CSF promotes the expression of CD26 on CB SC.
To validate CD26 as a new marker of CML LSC, we asked whether CD26 expression would correlate with disease activity. To address this question, we examined follow-up samples of our CML patients treated with imatinib or nilotinib. At the time of complete cytogenetic remission (CCyR) and/or MMR, CD26 was no longer detectable on CD34+/CD38─ SC in these patients, suggesting that most CML LSC were suppressed, had redistributed, or were depleted in these patients. In contrast, in patients with relapsing CML or resistant disease, at least a subfraction of CD34+/CD38─ cells co-expressed CD26. In addition, we were able to show that the residual CD26+ SC that can be detected shortly after the start of imatinib therapy (month 3) are BCR/ABL1+ cells, whereas the CD26─ SC isolated from the same samples are BCR/ABL1–. These data suggest that CD26 can be used as a follow-up parameter to monitor the presence and numbers (percentage) of CML LSC in tyrosine kinase inhibitor–treated patients.
Finally, we addressed the question of whether CD26 may serve as a novel therapeutic target in CML. To test this hypothesis, we first incubated CML LSC with vildagliptin before injecting these cells into NSG mice. Indeed, gliptin exposure resulted in a decreased engraftment of CML cells. In a second step, we examined nilotinib-treated CML patients receiving a gliptin because of uncontrolled diabetes mellitus. We identified 2 such patients, and in both, gliptin treatment was followed by a substantial decrease of BCR/ABL1 to undetectable or near undetectable levels. Although these data need to be confirmed in a clinical trial, they are encouraging and may point to the important role of CD26 as an LSC target in CML.
In summary, our data show that CML LSC specifically express CD26, and that CD26 expression is functionally important and may disrupt SDF-1-CXCR4 interactions in the SC niche. Whether CD26 can also be used as a new SC target in CML is currently under investigation.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Günther Hofbauer and Andreas Spittler (both at the Cell Sorting Core Unit of the Medical University of Vienna), Verena Suppan, Gerlinde Mitterbauer-Hohendanner, and Tina Bernthaler for their excellent technical assistance.
This work was made possible using patient samples donated as part of the United Kingdom (UK) SPIRIT2 Trial.
This study was supported by The Austrian Science Fund (Special Research Programs F4704-B20) and the Glasgow Experimental Cancer Medicine Centre, which is funded by Cancer Research UK and the Chief Scientist’s Office (Scotland); and a programme grant from Cancer Research UK (C11074/A11008) (T.L.H.).
Authorship
Contribution: H.H. and I.S. performed key laboratory experiments; S.C.-R. and G.S. provided qPCR data, immunocytochemistry, and immunohistochemistry stains; T.R. and M.W. provided xenotransplant experiments; G.H. and M.M. provided vital cell line models and experiments with transfected cell lines; M.B. provided microarray experiments; K.B. analyzed surface marker expression of patients by flow cytometry; S.H. provided vital logistic support; B.S. provided FISH experiments; W.R.S. provided clinical data and patients; T.L.H. provided sorting experiments and CD34+ CML samples; C.M. provided molecular biology data; and P.V. designed the study and wrote the paper.
Conflict-of-interest disclosure: P.V. received research funding and honoraria from Novartis and BMS. T.L.H. holds research funding and serves on the advisory board of Novartis and BMS. The remaining authors declare no competing financial interests.
Correspondence: Peter Valent, Department of Internal Medicine I, Division of Hematology and Hemostaseology, Medical University of Vienna, Austria, Währinger Gürtel 18-20, A-1090 Vienna, Austria; e-mail: peter.valent@meduniwien.ac.at.
References
Author notes
H.H. and I.S. contributed equally to this study.