Key Points
dmPGE2 stabilizes the transcription factor HIF1α in stem and progenitor cells.
Pharmacologic stabilization of HIF1α increases CXCR4 and enhances stem and progenitor homing and engraftment.
Abstract
Hematopoietic stem cell (HSC) transplantation is a lifesaving therapy for a number of immunologic disorders. For effective transplant, HSCs must traffic from the peripheral blood to supportive bone marrow niches. We previously showed that HSC trafficking can be enhanced by ex vivo treatment of hematopoietic grafts with 16-16 dimethyl prostaglandin E2 (dmPGE2). While exploring regulatory molecules involved in dmPGE2 enhancement, we found that transiently increasing the transcription factor hypoxia-inducible factor 1-α (HIF1α) is required for dmPGE2-enhanced CXCR4 upregulation and enhanced migration and homing of stem and progenitor cells and that pharmacologic manipulation of HIF1α is also capable of enhancing homing and engraftment. We also now identify the specific hypoxia response element required for CXCR4 upregulation. These data define a precise mechanism through which ex vivo pulse treatment with dmPGE2 enhances the function of hematopoietic stem and progenitor cells; these data also define a role for hypoxia and HIF1α in enhancement of hematopoietic transplantation.
Introduction
Hematopoietic stem cell (HSC) transplantation is a curative treatment of immunologic malignancies, inherited metabolic diseases, and congenital immunodeficiencies, and is an attractive method for gene therapy. Transplantation success is partly dictated by the quality and number of donor cells transplanted, and is dependent on their ability to home to bone marrow (BM) niches, self-renew and differentiate. Some sources of HSCs display reduced engraftment efficiency because of inadequate number, and/or poor homing. Identifying strategies to enhance homing and expansion of HSCs can improve transplant efficiency, particularly when HSC number is limited. It is known that prostaglandin E2 (PGE2) can stimulate hematopoietic stem and progenitor cell (HSPC) proliferation.1,2 Recently, the long-acting PGE2 analog 16-16 dimethyl PGE2 (dmPGE2) was identified in a zebrafish chemical screen as a regulator of hematopoiesis, and ex vivo exposure to dmPGE2 was shown to increase engraftment in murine and nonhuman primate models.3,4 This strategy has now progressed to a phase 1 clinical trial.5 We previously demonstrated that part of the mechanisms of action for PGE2 were the result of increases in homing, survival, and proliferation of murine and human HSCs.6 PGE2 enhances HSC homing primarily by increasing CXCR4 expression on HSCs; however, the mechanism(s) whereby PGE2 modulates CXCR4 and HSC homing has not been defined.
HSCs have been reported to lie in hypoxic BM niches7-10 that support stabilization of hypoxia-inducible factor 1 α (HIF1α) within HSCs. HSCs that reside in hypoxic niches have greater hematopoietic-repopulating ability,11 although recent evidence suggests that HSCs may inherently maintain hypoxic status independently from their specific BM localization.12 HIF1α dose-dependently regulates HSC activity,9 and intracellular oxygenation status plays a role in HSC quiescence and expansion.9,13,14 Given that HIF1α and hypoxia regulate CXCR4 transcription in some cancer cell lines15-20 and PGE2 can stabilize HIF1α in prostate cancer cells,21-23 we hypothesized that PGE2 may increase CXCR4 and HSC engraftment through effects on HIF1α. Herein, we demonstrate that PGE2 stabilizes HIF1α protein and transcriptional activity, which is required for enhanced HSPC homing, and identify a pharmacologic target for ex vivo enhancement of HSC function.
Materials and methods
Mice
Mice were bred in-house or were purchased from Jackson Laboratory (Bar Harbor, ME) and maintained in the Indiana University School of Medicine animal facility. Conditional HIF1α knockout (KO) mice were generated by breeding HIF1αFlox/Flox and tamoxifen-Cre mice, then crossing hemizygous floxed pups with homozygous HIF1αFlox/Flox mice. Resulting Cre+HIF1αFlox/Flox mice were used. Experiments were approved by the Institutional Animal Care and Use Committee of the Indiana University School of Medicine. Additional materials and methods are provided in the supplemental Methods (available on the Blood Web site).
Results and discussion
We previously demonstrated that short-term exposure to PGE2 upregulates HSPC CXCR4 and enhances their migration to stromal cell-derived factor 1 (SDF-1) and BM homing in vivo. However, the mechanisms whereby PGE2 modulates HSPC CXCR4 and homing are unknown. In prostate cancer cells and renal tubular cells,21,22 PGE2 stabilizes HIF1α protein without affecting messenger RNA (mRNA), and inhibiting PGE2 biosynthesis reduces HIF1α and HIF-responsive genes.23 In HEK cells and in microglial cells, HIF1α upregulates CXCR4 by interacting with hypoxia response elements (HREs) within the CXCR4 promoter.18,19 We hypothesized that the enhancing effect of PGE2 on HSPC CXCR4, migration, and homing could result from HIF1α stabilization and enhanced HIF1α transcriptional activity and that pharmacologic manipulation of HIF1α may be an adjunct or alternative strategy to enhance HSC engraftment. We first determined whether PGE2 stabilizes HIF1α in primary BM cells (BMCs), compared with the prolyl hydroxylase domain inhibitor dimethyloxalyglycine (DMOG), a known HIF1α stabilizer.24 Pulse treatment of lineageneg BMCs with 1 µM of dmPGE2 or 5 µM of DMOG significantly increased HIF1α protein expression by ∼35% (Figure 1A), with no effect on HIF1α mRNA (Figure 1B). Concomitantly, mRNA levels of 2 downstream HIF1 responsive genes adrenomedullin and glucose transporter-1 were significantly increased after PGE2 and DMOG treatment (Figure 1B), confirming that HIF1α stabilization led to increased HIF1α transcriptional activity.
PGE2increases HIF1α protein and downstream responsive genes. (A) (Top) Representative blot of HIF1α protein 4 hours after treatment with vehicle, 1 µM of dmPGE2, or 5 µM of DMOG. (Bottom) Densitometry analysis of HIF1α protein expression in mouse lineageneg BMCs treated with vehicle, dmPGE2, or DMOG. Data are expressed as X ± standard error of the mean (SEM) percent change in protein levels over vehicle control from 3 separate experiments. (B) Expression of HIF1 responsive genes in mouse lineageneg BMCs after treatment with dmPGE2 and DMOG as determined by quantitative reverse-transcription polymerase chain reaction. Data are X ± SEM, N = 3 experiments. *P < .05. (C) (Top) Dose-response blot of HIF1α protein 4 hours after treatment with vehicle or DMOG. (Bottom) Densitometry analysis of HIF1α protein expression in mouse lineageneg BMC. (D) Dose response of CXCR4 on SKL cells treated with vehicle or DMOG. Lineageneg BMCs were treated with DMOG for 2 hours at 37°C, washed, and incubated in RPMI with 10% heat-inactivated fetal calf serum for 16 hours. CXCR4 expression on LSK-gated cells was analyzed by flow cytometry. (E) In vitro transwell migration of murine SKL cells. One million lineageneg BMCs were treated with vehicle, dmPGE2, or DMOG. Cells were assayed for the ability to migrate to 100 ng/mL recombinant murine SDF-1 for 4 hours at 37°C. Data are expressed as X ± SEM, N = 9. (F) BMCs from CD45.1 mice were treated with vehicle, 1 µM of dmPGE2, or 5 µM of DMOG and 1 × 106 treated lineageneg cells transplanted into lethally irradiated CD45.2 mice. At 16 hours later, BM was analyzed for homed SKL cells. Data are represented as X ± SEM from 2 separate experiments (N = 4-5 mice/group/experiment, each assayed individually). (G) Similar homing experiment with or without the addition of AMD3100 10 minutes before transplantation. Data are represented as X ± SEM from 2 separate experiments (N = 4-5 mice/group/experiment, each assayed individually) *P < .05. (H) Percent contribution (chimerism) and (I) frequency analysis for DMOG determined by Poisson statistics using LCALC software. Vehicle P0= 121 319 and DMOG P0= 53 956. (J) Competitive repopulating units of DMOG and vehicle-treated cells in peripheral blood 6 months after transplant. Data are represented as X ± SEM from 2 pooled experiments (N = 5 mice/group/experiment, each assayed individually). (K) Secondary transplant percent chimerism at 6 months after transplant (N = 6 mice/group each assayed individually). *P < .05.
PGE2increases HIF1α protein and downstream responsive genes. (A) (Top) Representative blot of HIF1α protein 4 hours after treatment with vehicle, 1 µM of dmPGE2, or 5 µM of DMOG. (Bottom) Densitometry analysis of HIF1α protein expression in mouse lineageneg BMCs treated with vehicle, dmPGE2, or DMOG. Data are expressed as X ± standard error of the mean (SEM) percent change in protein levels over vehicle control from 3 separate experiments. (B) Expression of HIF1 responsive genes in mouse lineageneg BMCs after treatment with dmPGE2 and DMOG as determined by quantitative reverse-transcription polymerase chain reaction. Data are X ± SEM, N = 3 experiments. *P < .05. (C) (Top) Dose-response blot of HIF1α protein 4 hours after treatment with vehicle or DMOG. (Bottom) Densitometry analysis of HIF1α protein expression in mouse lineageneg BMC. (D) Dose response of CXCR4 on SKL cells treated with vehicle or DMOG. Lineageneg BMCs were treated with DMOG for 2 hours at 37°C, washed, and incubated in RPMI with 10% heat-inactivated fetal calf serum for 16 hours. CXCR4 expression on LSK-gated cells was analyzed by flow cytometry. (E) In vitro transwell migration of murine SKL cells. One million lineageneg BMCs were treated with vehicle, dmPGE2, or DMOG. Cells were assayed for the ability to migrate to 100 ng/mL recombinant murine SDF-1 for 4 hours at 37°C. Data are expressed as X ± SEM, N = 9. (F) BMCs from CD45.1 mice were treated with vehicle, 1 µM of dmPGE2, or 5 µM of DMOG and 1 × 106 treated lineageneg cells transplanted into lethally irradiated CD45.2 mice. At 16 hours later, BM was analyzed for homed SKL cells. Data are represented as X ± SEM from 2 separate experiments (N = 4-5 mice/group/experiment, each assayed individually). (G) Similar homing experiment with or without the addition of AMD3100 10 minutes before transplantation. Data are represented as X ± SEM from 2 separate experiments (N = 4-5 mice/group/experiment, each assayed individually) *P < .05. (H) Percent contribution (chimerism) and (I) frequency analysis for DMOG determined by Poisson statistics using LCALC software. Vehicle P0= 121 319 and DMOG P0= 53 956. (J) Competitive repopulating units of DMOG and vehicle-treated cells in peripheral blood 6 months after transplant. Data are represented as X ± SEM from 2 pooled experiments (N = 5 mice/group/experiment, each assayed individually). (K) Secondary transplant percent chimerism at 6 months after transplant (N = 6 mice/group each assayed individually). *P < .05.
Given the similar effects of dmPGE2 and DMOG on HIF1α, we investigated if DMOG similarly affects HSPC function. Pulse exposure of lineageneg BMC dose-dependently increased HIF1α protein expression (Figure 1C) with a concomitant dose-response increase in CXCR4 levels on Sca-1+ c-kit+ lineageneg (SKL) cells (Figure 1D). Functionally, DMOG pulse exposure also enhanced SKL migration to SDF-1 comparable to dmPGE2 (Figure 1E). In a congenic in vivo homing model, treatment of BMCs with DMOG enhanced SKL cell homing equivalent to dmPGE2 (Figure 1F). As we reported for PGE2,6 enhanced homing of SKL cells by DMOG was completely blocked by the selective CXCR4 antagonist AMD3100 (Figure 1G). In a limiting dilution, head-to-head congenic transplant model, DMOG treatment of BMC before transplantation significantly increased peripheral blood chimerism at 6 months after transplantation compared with vehicle-treated cells (Figure1H), with no differences in lineage reconstitution between recipient, vehicle, and DMOG-treated cells (data not shown). Enhanced chimerism as a result of DMOG treatment stemmed from a twofold increase in HSC frequency (Figure 1I) and competitive repopulating units (Figure 1J). Enhanced chimerism was maintained in secondary transplants (Figure 1K). That the enhancing effects of dmPGE2 on HSPC homing and engraftment can be duplicated by DMOG suggests that PGE2 may mediate its effect on HSPC CXCR4 through stabilization of HIF1α.
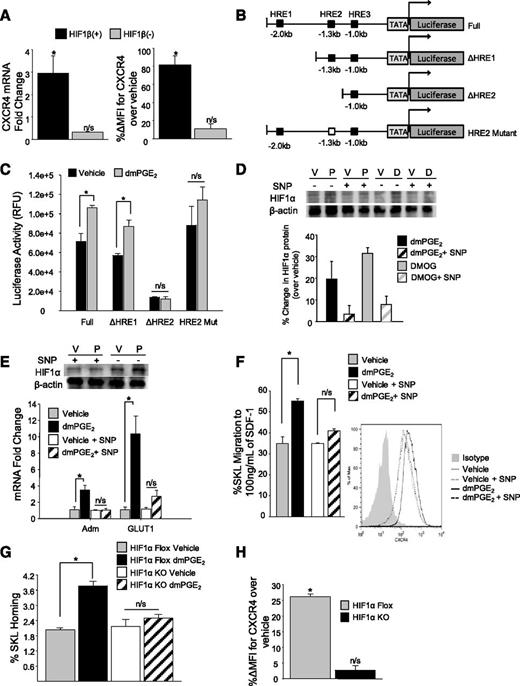
Under normoxic conditions, HIF1α is ubiquitinated and targeted for proteasomal degradation.25-27 In the absence of oxygen, HIF1α is stabilized and translocated to the nucleus by HIF1β, where the translocated HIF complex interacts with HREs within the promoter of responsive genes.28 To further define a HIF1α requirement for PGE2-induced CXCR4 upregulation, we used mutant mouse hepatoma cells lacking or reconstituted with the HIF1β nuclear translocator.29 In mutant hepatoma cells stably transfected with HIF1β, dmPGE2 increased CXCR4 mRNA approximately threefold, and CXCR4 surface expression approximately twofold, whereas dmPGE2 failed to increase CXCR4 in HIF1β (−) mutant cells (Figure 2A). The CXCR4 promoter contains 3 potential HREs, with the HRE located −1.3 kb from the transcriptional start site being necessary for hypoxia-induced CXCR4 upregulation in HEK cells.18 Using a series of luciferase CXCR4 promoter vectors containing specific combinations of HREs (Figure 2B), a significant increase in luciferase activity was observed in dmPGE2-treated cells transfected with the vector containing the −1.3 kb of HRE. However, when the −1.3 kb of HRE was either absent (ΔHRE2) or mutated (HRE2 Mut), no change in luciferase activity (Figure 2C) was observed after dmPGE2 treatment, indicating that the −1.3 kb of HRE is required for CXCR4 regulation by both hypoxia and PGE2. Taken together, these data indicate that HIF1α transcriptional activity is required for PGE2-induced gene transcription of CXCR4 and results from HIF1α nuclear translocation and binding to the −1.3 kb of HRE within the CXCR4 promoter, although we cannot rule out other regulatory elements between HRE1 and HRE2 that may be contributing to CXCR4 regulation in response to hypoxia.
HIF1α transcriptional activity is necessary for PGE2-induced CXCR4 upregulation. (A) (Left) CXCR4 cell surface expression (X ± SEM; N = 3 experiments) on HIF1β (−) and HIF1β (+) cells 24 hours after treatment with dmPGE2. CXCR4 cell surface expression was determined by flow cytometry. Data are expressed as percent change in mean fluorescence intensity of CXCR4 over vehicle. *P < .05. (Right) CXCR4 cell surface expression (X ± SEM; N = 3 experiments) in HIF1β (−) and HIF1β (+) cells 2 hours after treatment with vehicle or dmPGE2 determined by quantitative reverse-transcription polymerase chain reaction. (B) Schematic of pGL2b luciferase reporter constructs containing various regions of the murine CXCR4 promoter. (C) In vitro Luciferase reporter assay. 293T cells were transfected with either full-length CXCR4 promoter constructs containing all HREs (Full), truncated constructs containing 1 HRE (ΔHRE1) or 2 HREs (ΔHRE2), or a mutated 1.3 kb of HRE (HRE 2 Mut). After 24 hours, cells were split equally and treated with either vehicle or dmPGE2 for 16 hours at 37°C. Luciferase activity was measured using the Firefly Luciferase assay kit (Promega). Data are represented as X ± SEM from 2 separate experiments (N = 6). *P < .05. (D) (Top) Representative blot of HIF1α protein 4 hours after treatment with vehicle, dmPGE2, or DMOG, with or without the addition of SNP. (Bottom) Densitometry analysis of HIF1α protein expression in mouse lineageneg BMCs treated with vehicle, dmPGE2, or DMOG, with or without the addition of SNP. Data are expressed as X ± standard deviation percent change in protein levels over vehicle control from 2 separate experiments. (E) Expression of HIF1 responsive genes after treatment with vehicle or dmPGE2 with or without SNP. Data are expressed as X ± SEM, N = 3 experiments. (F) (Left) In vitro transwell migration of murine SKL cells to 100 ng/mL of SDF-1. Lineageneg cells were treated with vehicle or 1 µM of dmPGE2 with or without 100 µM of SNP for 2 hours at 37°C. Data are expressed as mean ± SEM, N = 3 experiments. *P < .05. (Right) Representative fluorescence-activated cell sorter histogram showing CXCR4 expression on SKL cells compared with isotype control. (G) In vivo homing of HIF1α KO cells. BMCs from conditional HIF1α KO or floxed control (CD45.2) mice were treated with vehicle or dmPGE2, and 1 × 106 treated lineageneg cells were transplanted into lethally irradiated BoyJ (CD45.1) mice. At 16 hours later, BM was analyzed for homed SKL cells. Data are represented as X ± SEM from 1 experiment (N = 4-5 mice/group/experiment, each assayed individually). (H) CXCR4 expression (X ± SEM) on HIF1α KO and HIF1α Floxed SKL cells after treatment with dmPGE2. N = 3 mice/group, each assayed individually. *P < .05.
HIF1α transcriptional activity is necessary for PGE2-induced CXCR4 upregulation. (A) (Left) CXCR4 cell surface expression (X ± SEM; N = 3 experiments) on HIF1β (−) and HIF1β (+) cells 24 hours after treatment with dmPGE2. CXCR4 cell surface expression was determined by flow cytometry. Data are expressed as percent change in mean fluorescence intensity of CXCR4 over vehicle. *P < .05. (Right) CXCR4 cell surface expression (X ± SEM; N = 3 experiments) in HIF1β (−) and HIF1β (+) cells 2 hours after treatment with vehicle or dmPGE2 determined by quantitative reverse-transcription polymerase chain reaction. (B) Schematic of pGL2b luciferase reporter constructs containing various regions of the murine CXCR4 promoter. (C) In vitro Luciferase reporter assay. 293T cells were transfected with either full-length CXCR4 promoter constructs containing all HREs (Full), truncated constructs containing 1 HRE (ΔHRE1) or 2 HREs (ΔHRE2), or a mutated 1.3 kb of HRE (HRE 2 Mut). After 24 hours, cells were split equally and treated with either vehicle or dmPGE2 for 16 hours at 37°C. Luciferase activity was measured using the Firefly Luciferase assay kit (Promega). Data are represented as X ± SEM from 2 separate experiments (N = 6). *P < .05. (D) (Top) Representative blot of HIF1α protein 4 hours after treatment with vehicle, dmPGE2, or DMOG, with or without the addition of SNP. (Bottom) Densitometry analysis of HIF1α protein expression in mouse lineageneg BMCs treated with vehicle, dmPGE2, or DMOG, with or without the addition of SNP. Data are expressed as X ± standard deviation percent change in protein levels over vehicle control from 2 separate experiments. (E) Expression of HIF1 responsive genes after treatment with vehicle or dmPGE2 with or without SNP. Data are expressed as X ± SEM, N = 3 experiments. (F) (Left) In vitro transwell migration of murine SKL cells to 100 ng/mL of SDF-1. Lineageneg cells were treated with vehicle or 1 µM of dmPGE2 with or without 100 µM of SNP for 2 hours at 37°C. Data are expressed as mean ± SEM, N = 3 experiments. *P < .05. (Right) Representative fluorescence-activated cell sorter histogram showing CXCR4 expression on SKL cells compared with isotype control. (G) In vivo homing of HIF1α KO cells. BMCs from conditional HIF1α KO or floxed control (CD45.2) mice were treated with vehicle or dmPGE2, and 1 × 106 treated lineageneg cells were transplanted into lethally irradiated BoyJ (CD45.1) mice. At 16 hours later, BM was analyzed for homed SKL cells. Data are represented as X ± SEM from 1 experiment (N = 4-5 mice/group/experiment, each assayed individually). (H) CXCR4 expression (X ± SEM) on HIF1α KO and HIF1α Floxed SKL cells after treatment with dmPGE2. N = 3 mice/group, each assayed individually. *P < .05.
To support the hypothesis that HIF1α is required for transcriptional upregulation of CXCR4 by PGE2, we used pharmacologic and genetic approaches to determine whether the effects of PGE2 on increasing CXCR4 expression and migration to SDF-1 in primary HSPCs were mediated by HIF1α stabilization. Using sodium nitroprusside (SNP), a compound that inhibits HIF1α protein accumulation,30,31 we saw an inhibition in both DMOG and dmPGE2-mediated HIF1α stabilization (Figure 2D), as well as dmPGE2-mediated upregulation of HIF-responsive genes (Figure 2E). SNP also blocked the increase in migration to SDF-1 and CXCR4 expression, normally seen with PGE2 (Figure 2F). Conditional deletion of HIF1α resulted in loss of PGE2-induced enhanced HSPC homing in HIF1α KO cells (Figure 2G), and no increase in CXCR4 expression after dmPGE2 treatment was observed (Figure 2H), further supporting the observation that HIF1α is required for PGE2-induced CXCR4 upregulation and enhanced homing.
In summary, we provide new mechanistic insight into the effects of PGE2 on HSC homing and engraftment and define a new ex vivo pharmacologic strategy to enhance hematopoietic transplantation. This finding that HIF1α stabilization by DMOG improves HSC homing and engraftment, along with recent evidence that human CD34+ cells cultured in hypoxic conditions exhibit elevated levels of CXCR432 and that in vivo administration of DMOG enhances HSC recovery after total body irradiation,33 suggests several potential therapeutic strategies targeting HIF1α to improve HSC engraftment and repopulation.
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors would like to thank Dr Mircea Ivan for providing HIF1β mutant cell lines, Dr Wilhelm Krek for providing the Luciferase Reporter vectors, as well as Anthony Sinn and Kacie Peterman from the Melvin and Bren Simon Cancer Center in Vivo Therapeutics Core for irradiation assistance and mouse transplants.
This study was supported by National Institutes of Health (NIH) grant HL096305 (L.M.P.). J.H. was supported by NIH training grants DK07519, HL07910, and HL087735. J.M.S. was supported by NIH training grant DK07519. Flow cytometry was performed in the Flow Cytometry Resource Facility of the Indiana University Simon Cancer Center (NCI P30 CA082709). Additional core support was provided by a Center of Excellence in Hematology grant P01 DK090948.
Authorship
Contribution: J.M.S. designed and performed experiments, performed data analysis and interpretation, and wrote the manuscript; J.H. designed research and wrote the manuscript; P.S. performed experiments; and L.M.P. designed experiments, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: J.H. and L.M.P. have received consulting fees from Fate Therapeutics. The remaining authors declare no competing financial interests.
Correspondence: Louis M. Pelus, Department of Microbiology and Immunology, Indiana University School of Medicine, 950 West Walnut St, Indianapolis, IN 46202; e-mail: lpelus@iupui.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal