Key Points
Diagnostic signatures for PTCL subtypes and 2 novel subgroups with distinct oncogenic pathway and prognostic importance in PTCL-NOS were identified.
Demonstrated that ALK(–) ALCL is a distinct molecular entity and the tumor microenvironment has prognostic significance in AITL patients.
Abstract
Peripheral T-cell lymphoma (PTCL) encompasses a heterogeneous group of neoplasms with generally poor clinical outcome. Currently 50% of PTCL cases are not classifiable: PTCL-not otherwise specified (NOS). Gene-expression profiles on 372 PTCL cases were analyzed and robust molecular classifiers and oncogenic pathways that reflect the pathobiology of tumor cells and their microenvironment were identified for major PTCL-entities, including 114 angioimmunoblastic T-cell lymphoma (AITL), 31 anaplastic lymphoma kinase (ALK)-positive and 48 ALK-negative anaplastic large cell lymphoma, 14 adult T-cell leukemia/lymphoma and 44 extranodal NK/T-cell lymphoma that were further separated into NK-cell and gdT-cell lymphomas. Thirty-seven percent of morphologically diagnosed PTCL-NOS cases were reclassified into other specific subtypes by molecular signatures. Reexamination, immunohistochemistry, and IDH2 mutation analysis in reclassified cases supported the validity of the reclassification. Two major molecular subgroups can be identified in the remaining PTCL-NOS cases characterized by high expression of either GATA3 (33%; 40/121) or TBX21 (49%; 59/121). The GATA3 subgroup was significantly associated with poor overall survival (P = .01). High expression of cytotoxic gene-signature within the TBX21 subgroup also showed poor clinical outcome (P = .05). In AITL, high expression of several signatures associated with the tumor microenvironment was significantly associated with outcome. A combined prognostic score was predictive of survival in an independent cohort (P = .004).
Introduction
Malignancies derived from the T-cell and natural killer (NK) cell lineages comprise ∼10% of all non-Hodgkin lymphomas (NHLs) in Western countries and are more prevalent in Asia.1,2 The World Health Organization (WHO) classification recognizes a number of distinctive subtypes of peripheral T-cell lymphoma (PTCL), including angioimmunoblastic T-cell lymphoma (AITL), anaplastic large cell lymphoma (ALCL), adult T-cell leukemia/lymphoma (ATLL), and entities derived mostly from NK cells including extranodal NK/T-cell lymphoma of the nasal type (ENKTL).2 There are additional rare PTCLs that are mostly extranodal tumors.
The varied morphology and lack of definitive diagnostic markers for most of the subtypes make the diagnosis and classification of these diseases challenging and impede their investigation. With current immunophenotypic and molecular markers, about 30% to 50% of PTCL cases are not classifiable and are categorized as PTCL not otherwise specified (PTCL-NOS).2 Although a few recurrent genetic abnormalities have been reported in PTCL,3-5 no specific genetic abnormality is diagnostic of particular PTCL subtypes, with the exception of t(2;5)(p23;q35) in ALK(+)ALCL.
Gene expression profiling (GEP) has been performed to improve the diagnosis of PTCL and to better understand its pathobiology.6-11 AITL has been shown to have a unique molecular profile, which suggests its derivation from follicular T-helper cells.6-11 Approximately 20% of PTCL-NOS cases would be classified as AITL using a GEP signature.12 There are also similarities in the molecular profiles of ALK(+)ALCL and ALK(–) ALCL, suggesting shared pathogenetic mechanisms.13,14 However, in-depth molecular analysis of PTCL has often been limited by the small sample size and/or the lack of corresponding clinical data. In the current study, we performed GEP analysis of a large series of PTCL and ENKTCL cases (n = 372) to derive robust molecular classifiers of PTCL subtypes, identify previously unrecognized subtypes, and develop prognostic models.
Materials and methods
Study populations
GEP was performed on cryopreserved pretreatment biopsy specimens from newly diagnosed PTCL patients from institutes in the Lymphoma Leukemia Molecular Profiling Project (LLMPP), Singapore, and International (I)-PTCL group consortiums. The diagnoses were made by a panel of expert hematopathologists in accordance with the WHO classification.2 To increase the sample size, and to include patients of Asian origin, clinical and GEP data from 3 large studies (http://www.ncbi.nlm.nih.gov/geo/: GSE6338, GSE14879, GSE19069)8,10,12,15,16 were included. This study has been approved by the respective institutional review boards of the participating institutes. This study was conducted in accordance with the Declaration of Helsinki.
Gene-expression profiling
The methods for isolation and processing of RNA and acquisition of GEP raw data have been described previously.12,16 GEP was performed using the HG-U133-plus2.0 arrays (Affymetrix Inc., Santa Clara, CA). The GEP data from published PTCL cohorts were compared with newly generated data sets for quality control. We excluded 53 of 425 PTCL cases from further analysis because of (1) failure to meet GEP data quality control and (2) rare PTCL entities with insufficient cases for analysis.
Diagnostic modeling and statistical analysis
The detailed statistical and experimental methods for the identification of molecular diagnostic signatures and prognostic signatures for PTCL subgroups are presented in supplemental Methods, available on the Blood Web site. The gene signature enrichment analysis was performed as described previously.17
Immunohistochemistry and in situ hybridization
A standard panel of T-cell immunomarkers (CD2, CD3, CD4, CD5, CD8, CD43, and PD1), cytotoxic markers (TIA-1, granzyme B, perforin), B-cell immunomarkers (CD20, CD79a, and PAX5), and other immunomarkers (CD10, CD21, CD30, and CD56) were used in various combinations on corresponding FFPE tissue sections to confirm the diagnosis using standard IHC procedures.1 Immunostaining for ALK expression and/or a fluorescence in-situ hybridization assay for ALK translocation were performed for suspected cases of ALCL. The presence of EBV was confirmed by in situ hybridization for EBERs.16 Other immunostains, polymerase chain reaction analyses, and fluorescence in situ hybridization studies were performed as needed, and all cases were diagnosed according to criteria of the WHO classification.2 The majority of PTCL cases from LLMPP and iPTCL1 series were reviewed on a multiheaded microscope. A consensus diagnosis was reached by the expert hematopathologists in a conference, if at least 3 of the 4 experts agreed on the diagnosis.
In addition to the standard panel of T-cell markers, we performed immunohistochemical staining on deparaffinized tissues sections with GATA3 (mouse monoclonal antibody, clone HG3-35, 1:25; Santa Cruz Biotechnology, Santa Cruz, CA) or TBX21 (mouse monoclonal antibody, clone 4B10, 1:100; BD Biosciences, San Jose, CA) overnight at 4°C, after peroxidase and protein blocking. The microwave epitope retrieval in 10 mM Tris/HCl pH9 containing 1 mM ethylenediamine tetraacetic acid was used for both antibodies, and appropriate negative (no primary antibody) and positive controls (tonsil or breast tumor sections) were stained in parallel with each set of tumors studied.
IDH2 mutation analysis
Clinical outcome analysis
The details on the creation of a prognostic model for AITL are described in supplemental section 1 (supplemental Figure 6). The clinical outcome of PTCL entities was performed as described earlier.1 The Fisher exact test for categorical data and Student t tests for continuous data were used for analysis between groups. False discovery rates were calculated using the Benjamini Hochberg method.
Results
Patient characteristics
Of the 372 PTCL cases included in this study, 193 PTCL cases are new cases from the LLMPP and Singapore, whereas the other cases have been reported earlier. The various PTCL entities showed similar clinical outcomes in different cohorts, with a superior clinical outcome in patients with ALK(+)ALCL (supplemental Figure 1A-B) who also had a younger median age (27 years) (supplemental Table 1A).
Refinement of molecular diagnostic signatures for major PTCL entities
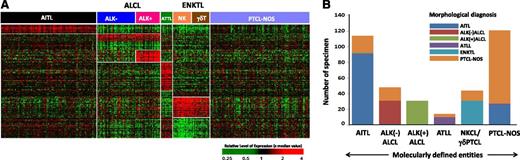
Each pathologically diagnosed entity was examined by the molecular classifiers to determine the molecular diagnosis. A compound covariate classifier19 was used to identify specimens diagnosed as PTCL-NOS by pathology that had molecular characteristics of other subtypes. The analysis was performed with a leave-one-out cross-validated method, with the subtype of cases reassigned accordingly (details in supplemental Methods). Genes in the signatures and enriched pathways in each molecular subtype as analyzed by Gene Set Enrichment Analysis17 are shown in Table 1 and supplemental Table 1B. These were largely consistent with our previous observations.12,16
Functional annotation* of the representative genes in the molecular signatures of the common PTCL entities
| PTCL subgroups . | Unique genes . | Major functional category . | Gene symbols . |
|---|---|---|---|
| AITL | 21 | Cell morphology/Intracellular signaling | EFNB2,ROBO1,S1PR3,ANK2,LPAR1,SNAP91,SOX8 |
| Cell migration/Vascularization | LPAR1,RAMP3,S1PR3,ROBO1,EFNB2,TUBB2B, SOX8 | ||
| Cell cycle | SOX8, ARHGEF10 | ||
| ATLL | 75 | T-cell homeostasis/activation/differentiation | NOTCH1, ZBTB17,CCNE1,FGF18,MYCN,PTHLH,SMARCA2,WNK1, NKX2-1, CYP26A1, HPSE,CTLA4, MYCN, PELI1,PRKCB, SPAST, ALS2, KIF3B, ZFYVE27 |
| Cell cycle/Proliferation | FGF18,MYCN,NKX2-1,NOTCH1,PTHLH,SMARCA2,CCNE1, GF18, WNK1, CTLA4, PELI1, PRKCB, ZBTB17, HPSE, FNTB, REL-1 | ||
| ALCL | 40 | Cell morphology/Intracellular signaling and interaction | DMRT1,SLC19A21,STK3, PERP, TNFRSF8, TMOD1, BATF3 |
| P53-induced genes | CDC14B, PERP,WDFEY3,TMOD1 | ||
| ALK+ vs ALK– | 29 | Cell cycle/Proliferation | AGT, ALK, ANXA3, BTBD11, CCNA1, DNER, GAS1, HS6ST2, IL1RAP, PCOLCE2, PDE4DIP, SLC16A3, TIAM2, TUBB6, WNT7B, SMOX, TMEM158, NLRP7, ADRB2, GALNT2 |
| Cell morphology/intracellular signaling and interaction | DNER, ADRB2, AGT, TIAM2, HS6ST2, GAS1, IL1RAP, WNT7B, ARHGEF10, HRASLS | ||
| ENKTL | 80 | NK-cell activation/survival | CD244,FASLG,KIR2DL4/LOC100287534,KLRD1,SH2D1B, FASLG |
| NK cell markers | CD244,FASLG,KLRC2,KLRD1,FASLG,NCAM1 | ||
| TBX21 vs GATA3 | 100 | Cell growth/Proliferation | ATP6V0D1,AXL,CD59,CHI3L1,CLTC,COL6A1,CREG1,CTSB,CTSC,CTSS,CYBB,FABP3,FPR1,FTL,GUCA2A,HCK,IFI30,IL13RA1,JAK2,LILRB1,NR1H3,PDXK,PITPNA,PLSCR1,PRDX3,PRKG1PSAP, SLC7A7,SOD2, TCN2,THY1,TYR, UBE2L6, WARS, AXL, FTL, SIRPA, STAT1 |
| TBX21 | T-cell activation/Th1 differentiation | CSF2,IFNG,CD28, STAT1, AXL, CD28, CD40, CD59, CSF2, FTL,IFNG,LILRB1, SIRPA,TBX21 | |
| GATA3 | Cell cycle /proliferation | MSH6, EGR1, CAT, EGR1, CAT, SEPT6, GATA3 |
| PTCL subgroups . | Unique genes . | Major functional category . | Gene symbols . |
|---|---|---|---|
| AITL | 21 | Cell morphology/Intracellular signaling | EFNB2,ROBO1,S1PR3,ANK2,LPAR1,SNAP91,SOX8 |
| Cell migration/Vascularization | LPAR1,RAMP3,S1PR3,ROBO1,EFNB2,TUBB2B, SOX8 | ||
| Cell cycle | SOX8, ARHGEF10 | ||
| ATLL | 75 | T-cell homeostasis/activation/differentiation | NOTCH1, ZBTB17,CCNE1,FGF18,MYCN,PTHLH,SMARCA2,WNK1, NKX2-1, CYP26A1, HPSE,CTLA4, MYCN, PELI1,PRKCB, SPAST, ALS2, KIF3B, ZFYVE27 |
| Cell cycle/Proliferation | FGF18,MYCN,NKX2-1,NOTCH1,PTHLH,SMARCA2,CCNE1, GF18, WNK1, CTLA4, PELI1, PRKCB, ZBTB17, HPSE, FNTB, REL-1 | ||
| ALCL | 40 | Cell morphology/Intracellular signaling and interaction | DMRT1,SLC19A21,STK3, PERP, TNFRSF8, TMOD1, BATF3 |
| P53-induced genes | CDC14B, PERP,WDFEY3,TMOD1 | ||
| ALK+ vs ALK– | 29 | Cell cycle/Proliferation | AGT, ALK, ANXA3, BTBD11, CCNA1, DNER, GAS1, HS6ST2, IL1RAP, PCOLCE2, PDE4DIP, SLC16A3, TIAM2, TUBB6, WNT7B, SMOX, TMEM158, NLRP7, ADRB2, GALNT2 |
| Cell morphology/intracellular signaling and interaction | DNER, ADRB2, AGT, TIAM2, HS6ST2, GAS1, IL1RAP, WNT7B, ARHGEF10, HRASLS | ||
| ENKTL | 80 | NK-cell activation/survival | CD244,FASLG,KIR2DL4/LOC100287534,KLRD1,SH2D1B, FASLG |
| NK cell markers | CD244,FASLG,KLRC2,KLRD1,FASLG,NCAM1 | ||
| TBX21 vs GATA3 | 100 | Cell growth/Proliferation | ATP6V0D1,AXL,CD59,CHI3L1,CLTC,COL6A1,CREG1,CTSB,CTSC,CTSS,CYBB,FABP3,FPR1,FTL,GUCA2A,HCK,IFI30,IL13RA1,JAK2,LILRB1,NR1H3,PDXK,PITPNA,PLSCR1,PRDX3,PRKG1PSAP, SLC7A7,SOD2, TCN2,THY1,TYR, UBE2L6, WARS, AXL, FTL, SIRPA, STAT1 |
| TBX21 | T-cell activation/Th1 differentiation | CSF2,IFNG,CD28, STAT1, AXL, CD28, CD40, CD59, CSF2, FTL,IFNG,LILRB1, SIRPA,TBX21 | |
| GATA3 | Cell cycle /proliferation | MSH6, EGR1, CAT, EGR1, CAT, SEPT6, GATA3 |
Ingenuity software (Ingenuity Systems, Inc.) was used to classify genes into functional categories.
Angioimmunoblastic T-cell lymphoma.
The genes in the signature reflected associations with angiogenesis and vascular endothelial function or cell migration (Table 1). The molecular diagnostic signature for AITL predicted 77% (90/117) of the pathologically diagnosed AITL as such, whereas 21 of 150 (14%) pathologically diagnosed PTCL-NOS cases were reclassified as AITL, similar to our previous observations20 (Figure 1A-B). An IDH2 mutation (R172K/S/T/G/M) was identified in 37% (17/46) of concordantly diagnosed AITL cases, and in 36% (4/11) of the cases of PTCL-NOS that were reclassified as AITL, but IDH2 was mutated in only 7% (1/14) AITL cases reclassified as PTCL-NOS (supplemental Figure 2).
Molecular diagnostic signatures of PTCL subgroups. (A) Unique gene expression signatures were identified for major PTCL entities using compound covariate prediction model (see Materials and methods for details), and the predictor score from top ranking genes for each subtype was used to classify a PTCL patient. ALCL and ENKTL groups are further differentiated into ALK(+)ALCL and ALK(–)ALCL, and NK and γδ T-cell subgroups, respectively. Each column represents a PTCL patient and each row represents a unique gene of the classifier. The relative gene expression scale is indicated below. (B) Pathological vs molecular diagnosis comparison. Substantial number of cases from PTCL-NOS were molecularly classified into WHO recognized PTCL subgroups: (i) AITL (n = 21, 14%); (ii) ALK(–)ALCL (n = 17, 11%); (iii) ATLL (n = 4, 3%); (iv) γδ-PTCL (n = 13, 9%). However, 26 AITL cases (22%) were not molecularly classifiable and changed to PTCL-NOS.
Molecular diagnostic signatures of PTCL subgroups. (A) Unique gene expression signatures were identified for major PTCL entities using compound covariate prediction model (see Materials and methods for details), and the predictor score from top ranking genes for each subtype was used to classify a PTCL patient. ALCL and ENKTL groups are further differentiated into ALK(+)ALCL and ALK(–)ALCL, and NK and γδ T-cell subgroups, respectively. Each column represents a PTCL patient and each row represents a unique gene of the classifier. The relative gene expression scale is indicated below. (B) Pathological vs molecular diagnosis comparison. Substantial number of cases from PTCL-NOS were molecularly classified into WHO recognized PTCL subgroups: (i) AITL (n = 21, 14%); (ii) ALK(–)ALCL (n = 17, 11%); (iii) ATLL (n = 4, 3%); (iv) γδ-PTCL (n = 13, 9%). However, 26 AITL cases (22%) were not molecularly classifiable and changed to PTCL-NOS.
Adult T-cell leukemia/lymphoma.
The diagnostic signature included MYC, REL-1, and NOTCH-1 known to be induced by the HTLV1-encoded TAX oncogene.20,21 The molecular diagnosis of ATLL identified 10 of 11 pathologically diagnosed cases, and 4 (∼3%) PTCL-NOS cases were reclassified as ATLL, similar to our previous observations12 (Figure 1A-B and Table 1). The reclassified cases also had a poor clinical outcome (median overall survival [OS] <0.6 years) and HTLV1 seropositivity.16
Extranodal NK/T-cell lymphoma, nasal type.
The molecular classifier representing the distinctive phenotypic and functional characteristics of NK cells16 was able to identify 97% (31/32) ENKTL cases and reclassified 9% (13/150) of PTCL-NOS as ENKTL (Figure 1A-B and Table 1). These cases could then be classified as NKCL or γδ-PTCL, the latter subgroup showing a high expression of CD3γ and CD3δ mRNA as well as transcripts encoding TCRγ chain16 (supplemental Figure 3).
Anaplastic large cell lymphoma.
The ALCL molecular signature identified 97% (58/60) pathologically defined ALCL cases and reclassified 11% (17/150) of PTCL-NOS (as ALCL, which were further classified as ALK(-)ALCL) (Figure 1A-B). The ALCL signature included some previously defined genes (eg, TNFRSF8 [CD30], BATF3, TMOD1),22 as well as a subset of p53 transcriptional targets. As expected, genes associated with TCR signaling (LCK, FYB, CSK1) were significantly downregulated (Table 1).
ALK(+)ALCL and ALK(–)ALCL subtypes.
We derived a distinct gene signature (Figure 2A) that was able to differentiate ALK(+)ALCL from ALK(–)ALCL, which showed a concordance with the pathological diagnosis of 100% (31/31) for ALK(+)ALCL and 94% (30/32) for ALK(–)ALCL. Interestingly, all 17 of molecularly reclassified ALCL cases from PTCL-NOS were classified as ALK(–)ALCL (Figure 2B), with a gene expression profile that was remarkably similar to other ALK(–)ALCL. They all expressed three genes (TNSFR8 [CD30], BATF3, and TMOD1) that were recently reported for ALK(–) ALCL.12 ALK(–)ALCL were significantly enriched for expression of MYC, IRF4 target gene signatures, as well as for proliferation and mTOR pathway signatures, compared with PTCL-NOS (supplemental Table 1B). In contrast, ALK(+)ALCL cases were enriched for the expression of signatures of HIF1-α target genes, IL10-induced genes, and H-ras/K-ras–induced genes compared with ALK(–)ALCL, which showed marginal enrichment of PI3K pathway–regulated genes (Figure 2B).
ALK(–)ALCL is molecularly distinct from PTCL-NOS and ALK(+)ALCL. (A) A unique gene expression signature for ALCL was identified and then ALK(+)ALCL cases were separated from ALK(–)ALCL (reclassified ALCL cases are indicated at the top). (B) Representative gene signatures/pathway enriched in ALK(+)ALCL compared with ALK(–)ALCL. (C) A representative re-classified ALCL case (hematoxylin and eosin staining [H&E]) with a pathological diagnosis of PTCL-NOS showing expression of CD30, GZMB, and TIA, but no expression of ALK by immunostaining.
ALK(–)ALCL is molecularly distinct from PTCL-NOS and ALK(+)ALCL. (A) A unique gene expression signature for ALCL was identified and then ALK(+)ALCL cases were separated from ALK(–)ALCL (reclassified ALCL cases are indicated at the top). (B) Representative gene signatures/pathway enriched in ALK(+)ALCL compared with ALK(–)ALCL. (C) A representative re-classified ALCL case (hematoxylin and eosin staining [H&E]) with a pathological diagnosis of PTCL-NOS showing expression of CD30, GZMB, and TIA, but no expression of ALK by immunostaining.
Reevaluation of the histology of these reclassified cases showed that most cases were morphologically consistent with ALCL, and indeed at least 1 or 2 pathologists in the initial review panel considered ALCL as the most likely diagnosis. In particular, these cases expressed CD30 protein strongly (Figure 2C and supplemental Table 1C). During the initial review of these cases, the cytotoxic T-cell proteins TIA1 and GZMB were either not detected by immunohistochemistry (IHC) or the assays were not performed. Reevaluation of these cases revealed that they consistently expressed these cytotoxic markers at both the mRNA and protein levels (Figure 2C), supporting their classification as ALCL.
PTCL-NOS can be divided into 2 major subgroups with significantly different outcomes
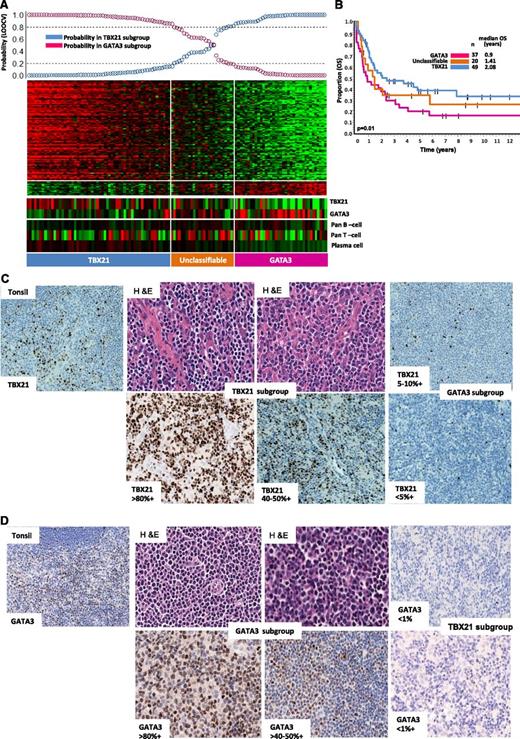
After excluding molecularly reclassified cases, 121 cases remained in the PTCL-NOS subgroup (supplemental Figure 4). Unsupervised hierarchical clustering showed at least two major subclusters with significant difference in OS (P = .03) (supplemental Figure 4A-B). One of the groups (cluster 1) showed high expression of GATA3 and its known target genes (CCR4, IL18RA, CXCR7, IK). The other cluster showed significantly higher expression of TBX21 (T-bet) and EOMES and their known target genes (CXCR3, IL2RB, CCL3, IFNγ). Overall, GATA3 and TBX21 mRNA showed an inverse expression pattern and concordance with GATA3 and TBX21 protein expression (Figure 3A,C-D). We then defined a gene signature (Figure 3A) that can distinguish the GATA3 (33%, 40/121) from the TBX21 subgroup (49%, 59/121) with >80% confidence, and the remaining 22 intermediate cases were termed “Unclassified.” We again observed a significant (P = .01) survival difference (5-year OS of 38%; [95% CI; 25%-56%]) for the TBX21 group compared with (19% [95% CI; 9%-38%]) the GATA3 subgroup. Differences in expression of TBX21 and GATA3 mRNA were also significantly associated with OS (P = .004). No significant differences in immunophenotypic characteristic with regards to CD4 and CD8 positivity and in clinical characteristics at presentation were observed among these patients (supplemental Table 1D).
Two major molecular subgroups within PTCL-NOS with biological and overall survival differences. (A) Bayesian predictor for the GATA3 and TBX21 subgroups was derived using cases from hierarchical cluster 1 and cluster 2. LOOCV was used for classification precision. (B) Overall survival (OS) analysis of molecularly-defined GATA3 and TBX21 subgroups showed significant difference in clinical outcome (P = .01). A subset of cases mentioned in the text lacked clinical outcome data and are not included in OS analysis. (C) Representative cases in the TBX21 subgroup (H&E) with TBX21 immunostain showed positivity ranging from 40% to 80% of cells, whereas positivity in the GATA3 subgroup was <10%. (D) Representative cases in GATA3 subgroup immunostained with GATA3 showing positivity ranging from 50% to 80% of cells, and were negative for TBX21.
Two major molecular subgroups within PTCL-NOS with biological and overall survival differences. (A) Bayesian predictor for the GATA3 and TBX21 subgroups was derived using cases from hierarchical cluster 1 and cluster 2. LOOCV was used for classification precision. (B) Overall survival (OS) analysis of molecularly-defined GATA3 and TBX21 subgroups showed significant difference in clinical outcome (P = .01). A subset of cases mentioned in the text lacked clinical outcome data and are not included in OS analysis. (C) Representative cases in the TBX21 subgroup (H&E) with TBX21 immunostain showed positivity ranging from 40% to 80% of cells, whereas positivity in the GATA3 subgroup was <10%. (D) Representative cases in GATA3 subgroup immunostained with GATA3 showing positivity ranging from 50% to 80% of cells, and were negative for TBX21.
Within the TBX21 group, the expression of cytotoxic (GNLY, PRF, GZM-K,-H-M, LYZ)/cytokine (CXCR3, CXCL12, and CCL-2,-3,-6,-11) transcripts associated with cytotoxic CD8+ T cells was inversely associated with the expression of B-cell/plasma cell and immunoglobulin (Ig) transcripts. These signatures were correlated with protein expression demonstrated by IHC (supplemental Figure 5A-C). Interestingly, patients who showed high expression of the former signatures showed poorer OS (P = .05) (supplemental Figure 5D). Significant enrichment of IFN α/β/γ-regulated gene signatures, a CD8+ T- cell profile and NF-κB pathway signatures were observed in the TBX21 subgroup (FDR <0.025, P < .01) compared with the GATA3 subgroup that showed marginal enrichment for mTOR- and MYC-related gene signatures and significant enrichment of PI3Kinase-induced gene signatures.
Derivation of prognostic model for AITL
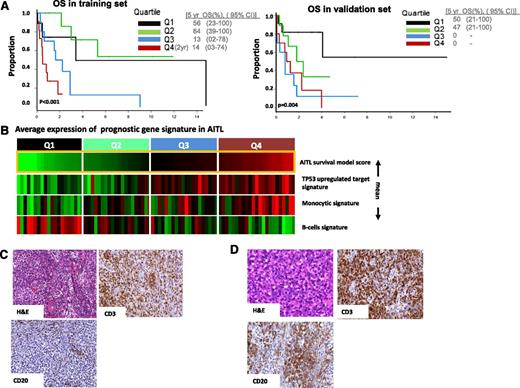
Of the 312 gene signatures in the lymphoid signature database23 evaluated for clinical outcome correlation, 34 gene signatures showed significant correlation (P < .05) in the training set (supplemental Figure 6). Highly correlated signatures were merged, resulting in 4 distinct genetic functional scores with biological plausibility including a B-cell signature, a monocytic signature, cytotoxic signature associated with CD8+ T-cells, and p53-induced target gene signature. The B-cell–associated signatures predicted a favorable outcome, whereas the other three were associated with poorer outcome (Table 2). Correlation of the signature scores resulted in an AIC optimal model24 containing only the B-cell-, monocytic-, and P53-induced target score. In the validation set, we observed that all of the individual scores and the model as a whole were validated (P < .04 and P < .004, respectively) (Figure 4A). The IHC indicated a correlation between the B-cell signature and the abundance of CD20+ B cells in corresponding tumors (Figure 4B). We did not observe any significant association of these prognostic scores with IPI (data not shown).
Gene signatures predictive of survival in the training set and in the independent validation set
| Signature cluster . | Effect of high expression . | Training P value . | Validation P value . |
|---|---|---|---|
| P53-induced gene signature | Poor prognosis | .001 | .01 |
| Cytotoxic T-cell signature | Poor prognosis | .005 | .04 |
| Monocytic/dendritic signature | Poor prognosis | .011 | .01 |
| B cell (GCB cell signature) | Good prognosis | .002 | .01 |
| Miscellaneous gene signatures | Good Prognosis | .009 | .07 |
| Signature cluster . | Effect of high expression . | Training P value . | Validation P value . |
|---|---|---|---|
| P53-induced gene signature | Poor prognosis | .001 | .01 |
| Cytotoxic T-cell signature | Poor prognosis | .005 | .04 |
| Monocytic/dendritic signature | Poor prognosis | .011 | .01 |
| B cell (GCB cell signature) | Good prognosis | .002 | .01 |
| Miscellaneous gene signatures | Good Prognosis | .009 | .07 |
Survival prediction in AITL. (A) Prognostic model in a training set and validated on an independent validation set. The optimal model represents that cases with high B-cell signature and low monocytic signature show favorable outcome compared with other cases (P = .004). (B) AITL cases are arranged according to model score, which represents differences between averages expression of B-cell signature and monocytic/P53 signature (mean). (C) Representative case from the upper (4th) quartile of the model score showing fewer CD20+ cells, whereas in (D) cases in lower(2nd) quartile showed a higher number of CD20+ B cells.
Survival prediction in AITL. (A) Prognostic model in a training set and validated on an independent validation set. The optimal model represents that cases with high B-cell signature and low monocytic signature show favorable outcome compared with other cases (P = .004). (B) AITL cases are arranged according to model score, which represents differences between averages expression of B-cell signature and monocytic/P53 signature (mean). (C) Representative case from the upper (4th) quartile of the model score showing fewer CD20+ cells, whereas in (D) cases in lower(2nd) quartile showed a higher number of CD20+ B cells.
Discussion
The outcome of PTCL patients is generally dismal with the existing treatment regimens and even novel drugs have not improved long-term survival.25 Therefore, a better understanding of the pathogenesis and biology of these tumors is needed to improve treatment and patient outcome. Studies on PTCL have been hindered by the difficulty of obtaining adequate samples for a meaningful analysis. With a large series of PTCL cases for analysis, we have validated and refined previously reported molecular diagnostic and prognostic signatures and oncogenic pathways. Overall, the classifiers for ATLL and ENKTL are highly specific, with little overlap with other entities.22 The signature for ENKTL is largely shared by tumors of the innate immune system: NK cells and γδ T cells, and 9% (13/150) of PTCL-NOS are γδ T-cell lymphomas by molecular classification,16 with clinical outcome similar to other ENKTLs.
The molecular classifiers and pathway signatures in other PTCL entities often reflected contributions from the microenvironment. This is prominently illustrated in AITL, which showed significant enrichment of the TFH gene signature as well as B-cell, follicular dendritic cell, and angiogenesis-related gene signatures from the microenvironment.16 Similarly, the cytokine milieu in AITL included potent interleukins that are angiogenic (IL8), proinflammatory (IL6, IL18), immunosuppressive (IL10), and proliferation-inducing (IL21), and have a vast repertoire of chemokines/receptors (eg, CXCL-2,-9,-13, 14 and CXCR-4,-5,-6), thus indicating a complex immunologic network. The high expression of IL4, IL6, and IL2116 are likely responsible for the expansion of B cells and plasma cells, and the hypergammaglobulinemia and autoimmune phenomenon often seen in this disease. Alternatively, the high expression of transcripts encoding IDO1 (INDO),26 PTGER2, IL10, and transforming growth factor-β indicates the presence of an immunosuppressive microenvironment, which is reflected by the frequent increase in EBV-positive B-cells.
The prognosticator in AITL reflected the importance of the microenvironment as observed in other lymphomas of B-cell lineage.27,28 The association of B cells with good prognosis is quite intriguing and has been reported in several solid tumors.29 It is possible the B-cell and/ plasma cell component is a reflection of the cytokine environment and/or retention of the function of the T cells to provide help for B cell. It is also possible that it is just a reflection of earlier disease with more B and plasma cells. Further studies are required to decipher this relationship. The immunosuppressive signature in AITL is represented by that of the tumor-associated macrophages (TAM). The accumulation of the TAMs is linked to high expression of IL-10 and vascular endothelial growth factor, which promotes the growth of TAMs and prevents maturation of monocytes to dendritic cells.27,28 Reversing this immunosuppressive microenvironment may promote antitumor immunity and the suppression of the proliferation of EBV-transformed B cells.
The molecular classifier for AITL allowed us to reclassify 14% of PTCL-NOS as AITL. These cases were not pathologically diagnosed as AITL because of the absence of certain key IHCs or morphologic features. We observed that IDH2 mutations are mostly confined to AITL vs PTCL-NOS.30 The presence of IDH2 mutations in these reclassified AITL cases at a similar frequency as consensus cases supports the validity of the molecular classification. Approximately 13% of morphologically diagnosed AITL cases were not molecularly classifiable as such. It is likely that some of these cases may represent true AITL cases with low tumor content or with nonrepresentative tissue submitted for GEP studies. However, it is also possible that some of these cases were misdiagnosed as AITL pathologically. We performed IDH2 mutation analysis on 14 available cases, and only 1 mutated case (7%) was detected, suggesting that the molecular classifier rarely missed true AITL cases.
The diagnostic signature for ALK(+)ALCL correlates highly with the pathological diagnosis, and gene signatures and oncogenic pathways are concordant with previous studies.31 We are now able to derive an ALK(–)ALCL signature that interestingly included a previously reported 3-gene discriminator (CD30, BATF3, TMOD1),32 as well as oncogenic pathways and signatures that are distinct from PTCL-NOS and ALK(+)ALCL. We reexamined the 17 reclassified ALK(–)ALCL cases (from PTCL-NOS) and found that these cases were compatible with ALCL but were not diagnosed as such because some key immunostains were not available or were negative. Molecularly, these cases showed GEP features as indistinguishable from the group that had a concordant pathological diagnosis. There are 2 recent reports on CD30+PTCL-NOS,33,34 and some of these cases may overlap with ALK(–)ALCL defined by our study. Interestingly, the clinical outcome of the molecularly defined ALK(–)ALCL patients was poorer compared with the other PTCL-NOS cases (P = .01, data not shown); however, a larger number of cases need to be studied prospectively to confirm this finding. With a robust signature that can clearly define ALK(–)ALCL, we can investigate this entity further without the ambiguities that compromise previous studies.
After excluding all of the cases with defined molecular classifiers, ∼32% of PTCLs (121/372) remained in the nonspecified category (supplemental Table 2). Most of these cases can now be classified into at least 2 meaningful biological subgroups with distinct clinical outcomes. The GATA3 high subgroup was associated with poor survival. This group showed enrichment of gene signatures related to proliferation (MYC), mTOR (P13K), and marginal enrichment of β-catenin gene signature (data not shown). Interestingly, Wnt/β-catenin signaling is required for GATA3 expression.35 The TBX21 high subgroup showed significant enrichment of IFNγ-induced gene signatures and NF-κB gene signatures and more favorable clinical outcome. A subset of cases within this group has high expression of transcripts associated with cytotoxic T cells and probably represents the cytotoxic T-cell lymphoma identified previously12 and, similarly, these patients had poorer outcome compared with cases with low cytotoxic signature. The current finding is biologically relevant, because TBX21 and its paralogue EOMES are important not only for Th2 cell but also for cytotoxic T-cell (CTL) differentiation,36,37 and there could be a dichotomy in cellular polarization in the TBX21 group with CTL at one end of the spectrum. The identification of these novel entities and their major biological and clinical characteristics in a previously undefined, large group of PTCL provides a rational framework and exciting opportunities for further investigations.
Translational relevance
We have defined robust molecular diagnostic and prognostic signatures for the more common subtypes of PTCL and segregated PTCL-NOS into meaningful biological and prognostic subtypes. We have identified enriched oncogenic pathways associated with the different PTCL entities, and the biological insight gained provides possible novel therapeutic targets for intervention. For example, AITL may benefit from FDA-approved drugs that target the NF-κB pathway via bortezomib or carfilzomib or specific inhibitors against IDH2 mutation that are being developed. Our findings also suggest that ALK(–) ALCL patients may benefit from drugs that target mitotic cells (eg, aurora kinase inhibitors) in combination with drugs that target PI3-kinase/AKT pathway. Similarly, a subset of patients in the GATA3 subgroup may benefit from drugs that target mTOR (eg, rapamycin, temsirolimus). Our studies also strongly indicate that the tumor microenvironment significantly contributed to the prognosis of AITL, and reversing the immunosuppressive microenvironment may promote antitumor immunity and the suppression of the proliferation of EBV-transformed B cells.
Our report provides a new perspective on the classification for PTCL and biological rationale for novel therapeutic intervention in specific entities of PTCL that ultimately can be translated in clinical practice both diagnostically and therapeutically to significantly benefit patients with PTCL.
There is an Inside Blood Commentary on this article in this issue.
Presented in part at the 53rd American Society of Hematology (ASH) Annual Meeting, New Orleans, LA, December 5-8, 2012.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Cynthia Lachel, Zhongfeng Liu and Genomic Core facility of University of Nebraska Medical Center for technical assistance.
The study was supported by the National Cancer Institute, National Institutes of Health (1U01CA157581-01). J.I. is a recipient of the young scientist award from the Lymphoma Foundation of America, a fellowship from the Lymphoma Research Foundation, and the Translational Research Program award from the Leukemia & Lymphoma Society, and has also received a University of Nebraska Medical Center-Clinical and Translational Research mentored Scholars program pilot grant. C.W. is a recipient of a fellowship from the Chinese Scholarship Council.
Authorship
Contribution: J.I., W.C.C., and L.M.S. designed research; J.I. and C.W. performed research; J.I., G.W., and L.S. analyzed data and performed statistical analyses; S.G., R A.W., A.R., R.D.G., D.D.W., T.C.G., B.T.T., S.T.L., S.Y.T., L.M.R., E.S.J., E.C., A.M., J.D., R.M.B., J.R.C., R.R.T., G.O., E.G., P.G., P.P.P., S.A.P., W.Y.A., S.N., M.S., F.B., L.d.L., J.M.C., J.A., J.V., and W.C.C. provided materials, reviewed pathology results and immunostaining data, or contributed GEP and clinical data; and J.I., W.C.C., and L.M.S. wrote and finalized the manuscript.
Conflict–of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Javeed Iqbal, Department of Pathology and Microbiology, 983135 Nebraska Medical Center, Omaha, NE 68198; e-mail: jiqbal@unmc.edu; Wing C. (John) Chan, Department of Pathology, City of Hope National Medical Center, Duarte, CA 91010; e-mail:jochan@coh.org; and Louis M. Staudt, Metabolism Branch, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892; e-mail: lstaudt@mail.nih.gov.

![Figure 2. ALK(–)ALCL is molecularly distinct from PTCL-NOS and ALK(+)ALCL. (A) A unique gene expression signature for ALCL was identified and then ALK(+)ALCL cases were separated from ALK(–)ALCL (reclassified ALCL cases are indicated at the top). (B) Representative gene signatures/pathway enriched in ALK(+)ALCL compared with ALK(–)ALCL. (C) A representative re-classified ALCL case (hematoxylin and eosin staining [H&E]) with a pathological diagnosis of PTCL-NOS showing expression of CD30, GZMB, and TIA, but no expression of ALK by immunostaining.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/123/19/10.1182_blood-2013-11-536359/4/m_2915f2.jpeg?Expires=1769128113&Signature=cmIFLqngg0WxxSvGN2kqvCNCu-ZPcGmcAmFfS~R1DjaTYhX79Jwi97xaOSKWeJIxHIZELd~wAns6~fJJvgXIyCGcPsD4YFaBKMXmluh~NWFreHjlG4e~kp7KwZGyBviWbpMi7Vs2oszptKIyFF3BTSs65hDcMGQ4IXcGGq134UVSeHM7CNHiIa-Wgkeke5aoMzgIJ8NZmFegqpQAPKUC~IOxTO0ISVqUYUVuva61x6i-gPs9OrKuaF2udF5WaxQ5iOo2HuchH79ZdJZclA2YdxSuYCECw2ZfSpXUzBRIH1e86EgS352rRFaVETHMKZzZtj8SU-IELGWqBFrekMthJw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal