In this issue of Blood, Iqbal et al,1 having compiled gene expression profiles from >300 peripheral T-cell lymphomas, expand previous findings on the diagnostic value of molecular signatures that correlate with different histological types of T-cell lymphomas. They report the discovery of 2 molecular subgroups of peripheral T-cell lymphomas, not otherwise specified (PTCL, NOS), characterized by high expression of either GATA-binding protein 3 (GATA-3) or t-box 21 (TBX21) transcription factors and corresponding target genes, with the GATA3 subgroup being associated with distinctly worse prognosis. In an independent study, Wang et al2 also show that GATA3 expression in a subset of PTCL, NOS identifies a subgroup of patients with inferior survival.
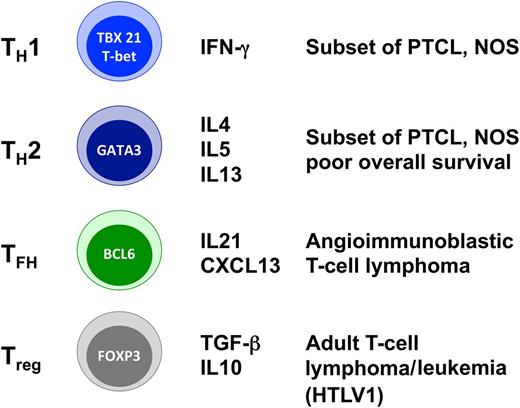
Population subsets of Th cells and corresponding T-cell lymphomas. TH, Th helper cell; TFH, follicular Th cell; Treg, regulatory T cell.
Population subsets of Th cells and corresponding T-cell lymphomas. TH, Th helper cell; TFH, follicular Th cell; Treg, regulatory T cell.
PTCLs—a term collectively designating malignancies derived from mature T cells and natural killer cells—comprise a heterogeneous group of disease entities that are overall rare, accounting for <15% of all non-Hodgkin lymphomas worldwide.3 PTCLs pose challenges in several respects. Clinically, most entities are aggressive diseases with overall poor response to classical treatments and carry a dismal prognosis.4 With respect to classification and diagnosis, in contrast with the main B-cell lymphomas entities that are defined by a combination of morphologic, immunophenotypic, genetic, and clinical features and are linked to a normal cellular counterpart, many T-cell lymphoma entities are pathologically heterogeneous, most of them lack defining genetic aberrations, and their classification relies on less well-characterized diagnostic criteria.5 The paradigm is represented by PTCL, NOS, which represents the largest PTCL entity and is defined “by default” as encompassing cases not fulfilling criteria allowing categorization in a more specific entity. Not unexpectedly, PTCL, NOS is heterogeneous, both pathologically and clinically, and although it is has been repeatedly stated that it may encompass several distinct entities, various attempts to identify biologically relevant subgroups have yet mostly failed.6
The discovery of 2 distinct molecular subgroups of PTCL, NOS identified by unsupervised analysis of genome-wide molecular profiles reported by Iqbal et al represents a major step forward in deciphering the heterogeneity of PTCL, NOS. Interestingly, the 2 subgroups, defined by overexpression of GATA3 or TBX21 (t-bet) and associated target genes, are biologically meaningful. Both GATA3 and TBX21 are transcription factors that are master regulators of gene expression profiles in T helper (Th) cells, skewing Th polarization into Th2 and Th1 differentiation pathways, respectively. The molecular signatures of lymphomas associated with high expression of these transcription factors were also enriched in other Th2- or Th1-associated transcripts, and GATA-3–positive PTCLs are associated with eosinophilia that is typically mediated by Th2 cytokines. These novel findings add to the increasing evidence that cell lineage is a major determinant of PTCL biology and defining factor for the delineation of PTCL entities or subgroups. Considering Th cell subsets (see figure), it is already known that the follicular helper subset represents the cellular origin of angioimmunoblastic T-cell lymphoma,7 whereas the neoplastic cells in human T-lymphotropic virus 1–associated T-cell lymphoma/leukemia generally exhibit a T-regulatory phenotype with expression of FOXP3 transcription factor.8 The new data published by Iqbal and Wang and colleagues now suggest that a large proportion of PTCL, NOS is related to either Th1 or Th2 lineage derivation. There is, however, no perfect overlap between the signatures derived from the lymphoma subgroups and those of the normal Th subsets, likely reflecting the plasticity of the T-cell system. In addition, the TBX21 group might be more heterogeneous as it also comprises a subset of cases with a cytotoxic profile, and using the molecular classifier developed to distinguish between the GATA3 and TBX21 subgroups, there remains a proportion of cases whose gene expression signature is indeterminate and cannot be assigned to one or the other category. Thus, additional studies and analysis of larger cohorts of patients may be necessary to validate the current findings and possibly refine the molecular classification.
The data published by Iqbal et al suggest that the GATA3 signature associates with distinctly less favorable clinical outcome and shorter overall survival than the TBX21 signature. There appears to be a good correlation between the molecular signatures and protein expression; hence, immunohistochemistry for GATA3 and TBX21 is a reliable surrogate to the molecular signatures. Indeed, applying immunohistochemistry for GATA3 on an independent series of cases, Wang et al also showed that GATA3 expression, evidenced in almost half of the cases, identifies a high-risk subset of PTCL, NOS. Although the findings in these 2 independent cohorts are convergent, it will be useful to confirm the clinical correlation of the GATA3 vs TBX21 profiles in another series of PTCL, NOS patients, if possible in the setting of a trial where homogeneous therapy is applied.
Having shown that molecular signatures in PTCL, NOS have a clinical impact, an important question is whether there is a rationale to suggest that these molecular subgroups may benefit from distinct, innovative, and potentially more efficient therapies. Investigating whether the molecular subgroups differ in terms of pathogenesis and oncogenic pathways is a prerequisite to identify novel druggable targets. Evidence for enrichment in intracellular pathways such as the phosphoinositide 3-kinase-associated gene signature in the GATA3 subgroup provides a basis for intervention with specific inhibitors. The experimental data provided in the study by Wang et al point at the role of the tumor microenvironment and how Th2 cytokines may contribute to shaping the phenotype of both the lymphoma cells and tumor-associated macrophages, functionally polarized to promote neoplastic growth, suggesting that tumor microenvironment may represent a therapeutic target. Finally, novel recurrent mutations expected to be discovered by the ongoing high-throughput and deep sequencing studies may segregate with molecular subgroups and possibly provide insights into novel therapies.
Conflict-of-interest disclosure: The authors declare no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal