Key Points
Highly recurring mutations are present in WM, including MYD88 L265P, warts, hypogammaglobulinemia, infection, and myelokathexis-syndrome–like mutations in CXCR4, and ARID1A.
Small, previously undetected CNAs affecting B-cell regulatory genes are highly prevalent in WM.
The genetic basis for Waldenström macroglobulinemia (WM) remains to be clarified. Although 6q losses are commonly present, recurring gene losses in this region remain to be defined. We therefore performed whole genome sequencing (WGS) in 30 WM patients, which included germline/tumor sequencing for 10 patients. Validated somatic mutations occurring in >10% of patients included MYD88, CXCR4, and ARID1A that were present in 90%, 27%, and 17% of patients, respectively, and included the activating mutation L265P in MYD88 and warts, hypogammaglobulinemia, infection, and myelokathexis-syndrome–like mutations in CXCR4 that previously have only been described in the germline. WGS also delineated copy number alterations (CNAs) and structural variants in the 10 paired patients. The CXCR4 and CNA findings were validated in independent expansion cohorts of 147 and 30 WM patients, respectively. Validated gene losses due to CNAs involved PRDM2 (93%), BTG1 (87%), HIVEP2 (77%), MKLN1 (77%), PLEKHG1 (70%), LYN (60%), ARID1B (50%), and FOXP1 (37%). Losses in PLEKHG1, HIVEP2, ARID1B, and BCLAF1 constituted the most common deletions within chromosome 6. Although no recurrent translocations were observed, in 2 patients deletions in 6q corresponded with translocation events. These studies evidence highly recurring somatic events, and provide a genomic basis for understanding the pathogenesis of WM.
Introduction
Waldenström macroglobulinemia (WM) is a distinct indolent B-cell malignancy in the World Health Organization classification. WM shares many clinical and pathological features with other B-cell lymphomas and multiple myeloma, which often complicates the diagnosis of this entity. Immunoglobulin M (IgM) monoclonal gammopathy of unknown significance (MGUS) is a precursor state for WM. Approximately 2% of IgM MGUS patients evolve to a B-cell malignancy per year, with most of these individuals progressing to WM.1,-3 Since the initial description of WM by Jan Waldenström in 1944, the genetic basis for WM has been elusive. We therefore employed whole genome sequencing (WGS) utilizing tumor-derived DNA from 30 patients, which included paired germline/tumor sequencing for 10 patients. The initial findings of this study resulted in the identification of a recurrent somatic mutation, L265P in MYD88, in over 90% of WM patients that distinguished WM from other overlapping entities such as marginal zone lymphoma, chronic lymphocytic leukemia (CLL), and multiple myeloma, wherein MYD88 L265P was either absent or infrequently observed (<10%).4,-6 Importantly, MYD88 L265P was present in IgM MGUS with a frequency of 50% to 80% using allele-specific polymerase chain reaction (AS-PCR), suggesting an early oncogenic event for this mutation and that other genomic events are likely to be responsible for WM disease progression. Activating MYD88 mutations are not unique to WM, and appear in other B-cell lymphomas including primary central nervous system lymphoma (50%) and activated B-cell (ABC) subtype diffuse large B-cell lymphoma (DLBCL) (8% to 29%).7,-9 A multi-hit model would also help explain why no significant differences in treatment response, progression free, and overall survival based solely on MYD88 mutation status determination were observed in a large series of WM patients.10
Little is known about the genetic structural variant (SV) changes such as copy number alterations (CNAs) or translocations in WM. By fluorescence in situ hybridization and array comparative genomic hybridization, deletions in chromosome 6q21-23 have been identified in 40% to 60% of WM patients, with concordant gains in 6p in 41% of those with 6q deletions.11,-13 Gains in chromosomes 3q, 4, 18, 8q, and Xq, as well as losses of 11q23, 13q14, and 17p have also been described in up to 20% of WM cases.14,15 We therefore sought to more fully characterize the molecular events responsible for WM pathogenesis using WGS.
Methods
Sample collection and preparation
Bone marrow (BM) aspirates and peripheral blood (PB) samples were collected in heparinized syringes from 30 patients with the clinicopathological diagnosis of WM, as defined by the Second International Workshop on WM.16 Participants provided informed consent prior to sample collection, in accordance with the Dana-Farber Cancer Institute Institutional Review Board. This study was conducted in accordance with the Declaration of Helsinki. Relevant clinical characteristics and blood work are presented in supplemental Table 1 on the Blood Web site. BM and PB mononuclear cells (PBMCs) were isolated by density gradient centrifugation using Ficoll-Paque (Amersham-Pharmacia Biotech, Piscataway, NJ). WM lymphoplasmacytic cells (LPC) were isolated from the BM mononuclear cells by CD19+ selection using immunomagnetic MACS micro-beads (Miltenyi Biotech, Auburn, CA). To minimize potential contamination from circulating WM LPC, PBMCs were depleted of CD19+ cells using CD19+ MACS micro-beads.
Complete methodology for the WGS analysis and validation is available in the supplemental Methods.
Results
Thirty patients who met the International Consensus Criteria for WM diagnosis were evaluated in this study.16 The clinical characteristics were typical of WM of this cohort and are presented in supplemental Table 1. Tumor DNA from CD19+ BM LPC was isolated from the 30 patients, along with DNA from CD19-depleted PBMCs for use in germline analysis. All 30 tumor and 10 PBMC samples were submitted for WGS with Complete Genomics, as previously described.4
Coding variant analysis
WGS findings were filtered for novel nonsynonymous variants as described in the supplemental Methods. This identified 5835 variants in 4427 genes. The 10-paired genomes were used to identify genes containing at least one somatic variant. Variant data from all 30 WM tumor genomes were then fit to this model wherein each affected gene was categorized as potentially somatic, germline, or unclassified if variants were only observed within the 20 unpaired samples (supplemental Table 2). Genes were further categorized by mutational frequency and validated based on bioinformatic strength and biological relevance, using Sanger sequencing of both tumor and germline DNA (Table 1). These cut-off scores were significantly lower than our previous analysis, allowing for the detection of a number of additional mutations. Primers and representative chromatograms are provided in supplemental Table 3. A full list of somatic and germline mutations affecting genes in the COSMIC cancer gene census are shown in supplemental Tables 4 and 5, respectively.17
Validated somatic variants identified by WGS in WM patients
| Gene . | N . | Chr . | Position . | Reference . | Variant . | Zygosity . | Protein . | Notes . |
|---|---|---|---|---|---|---|---|---|
| ARID1A | 1 | 1 | 27057944 | C | — | hom | Y551Frameshift | Homozygous by aUPD |
| ARID1A | 1 | 1 | 27099946 | C | T | het | R1276Stop | |
| ARID1A | 1 | 1 | 27101665 | C | T | het | Q1650Stop | |
| ARID1A | 1 | 1 | 27106497 | C | T | het | Q2037Stop | Opposite copy loss |
| ARID1A | 1 | 1 | 27107136 | — | A | het | S2249Frameshift | |
| NOTCH2 | 1 | 1 | 120478125 | A | C | het | F1209V | |
| CXCR4 | 1 | 2 | 136872467 | GAAGACTCAG | AC | het | S344Frameshift | |
| CXCR4 | 1 | 2 | 136872481 | AGAT | — | het | S339Frameshift | |
| CXCR4 | 2 | 2 | 136872485 | G | C | het | S338STOP | WHIM associated rs104893626 |
| CXCR4 | 3 | 2 | 136872485 | G | T | het | S338STOP | WHIM associated rs104893626 |
| CXCR4 | 1 | 2 | 136872570 | — | T | het | T311Frameshift | |
| MAP2 | 1 | 2 | 210557805 | G | C | het | W304C | |
| MYD88 | 1 | 3 | 38182032 | C | G | het | S219C | |
| MYD88 | 27 | 3 | 38182640 | T | C | het | L265P | Homozygous by aUPD × 4 |
| MED23 | 1 | 6 | 131915434 | C | T | het | E1013K | |
| SYNE1 | 1 | 6 | 152708319 | C | T | hom | R2792H | Opposite copy loss |
| TRRAP | 1 | 7 | 98530988 | A | T | het | D1326V | |
| TRAF2 | 1 | 9 | 139815501 | G | — | hom | L324Frameshift | Homozygous by aUPD |
| RAG2 | 1 | 11 | 36615549 | A | G | hom | L57P | Opposite copy loss |
| MLL2 | 1 | 12 | 49425598 | A | G | het | S4297L | |
| MLL2 | 1 | 12 | 49448412 | C | — | het | G101Frameshift | |
| TRAF3 | 1 | 14 | 103371839 | — | T | het | L448Frameshift | Opposite copy loss |
| MYBBP1A | 1 | 17 | 4457136 | G | A | het | P177S | |
| MYBBP1A | 1 | 17 | 4457323 | C | T | het | R148Q | |
| TP53 | 1 | 17 | 7577114 | C | T | het | C275Y | |
| TP53 | 1 | 17 | 7577569 | A | C | hom | C238G | Homozygous by aUPD |
| CD79B | 1 | 17 | 62006799 | A | T | het | Y197N | |
| CD79B | 1 | 17 | 62007129 | C | G | het | Exon boundary | |
| MUC16 | 1 | 19 | 9014687 | G | T | het | A12763E |
| Gene . | N . | Chr . | Position . | Reference . | Variant . | Zygosity . | Protein . | Notes . |
|---|---|---|---|---|---|---|---|---|
| ARID1A | 1 | 1 | 27057944 | C | — | hom | Y551Frameshift | Homozygous by aUPD |
| ARID1A | 1 | 1 | 27099946 | C | T | het | R1276Stop | |
| ARID1A | 1 | 1 | 27101665 | C | T | het | Q1650Stop | |
| ARID1A | 1 | 1 | 27106497 | C | T | het | Q2037Stop | Opposite copy loss |
| ARID1A | 1 | 1 | 27107136 | — | A | het | S2249Frameshift | |
| NOTCH2 | 1 | 1 | 120478125 | A | C | het | F1209V | |
| CXCR4 | 1 | 2 | 136872467 | GAAGACTCAG | AC | het | S344Frameshift | |
| CXCR4 | 1 | 2 | 136872481 | AGAT | — | het | S339Frameshift | |
| CXCR4 | 2 | 2 | 136872485 | G | C | het | S338STOP | WHIM associated rs104893626 |
| CXCR4 | 3 | 2 | 136872485 | G | T | het | S338STOP | WHIM associated rs104893626 |
| CXCR4 | 1 | 2 | 136872570 | — | T | het | T311Frameshift | |
| MAP2 | 1 | 2 | 210557805 | G | C | het | W304C | |
| MYD88 | 1 | 3 | 38182032 | C | G | het | S219C | |
| MYD88 | 27 | 3 | 38182640 | T | C | het | L265P | Homozygous by aUPD × 4 |
| MED23 | 1 | 6 | 131915434 | C | T | het | E1013K | |
| SYNE1 | 1 | 6 | 152708319 | C | T | hom | R2792H | Opposite copy loss |
| TRRAP | 1 | 7 | 98530988 | A | T | het | D1326V | |
| TRAF2 | 1 | 9 | 139815501 | G | — | hom | L324Frameshift | Homozygous by aUPD |
| RAG2 | 1 | 11 | 36615549 | A | G | hom | L57P | Opposite copy loss |
| MLL2 | 1 | 12 | 49425598 | A | G | het | S4297L | |
| MLL2 | 1 | 12 | 49448412 | C | — | het | G101Frameshift | |
| TRAF3 | 1 | 14 | 103371839 | — | T | het | L448Frameshift | Opposite copy loss |
| MYBBP1A | 1 | 17 | 4457136 | G | A | het | P177S | |
| MYBBP1A | 1 | 17 | 4457323 | C | T | het | R148Q | |
| TP53 | 1 | 17 | 7577114 | C | T | het | C275Y | |
| TP53 | 1 | 17 | 7577569 | A | C | hom | C238G | Homozygous by aUPD |
| CD79B | 1 | 17 | 62006799 | A | T | het | Y197N | |
| CD79B | 1 | 17 | 62007129 | C | G | het | Exon boundary | |
| MUC16 | 1 | 19 | 9014687 | G | T | het | A12763E |
List of all variants validated by Sanger sequencing ordered by chromosome and position.
aUPD, acquired uniparental disomies; Chr, chromosome; WHIM, warts, hypogammaglobulinemia, infection, and myelokathexis.
As previously described, a somatic T/C mutation in MYD88 resulting in an L265P substitution was observed in 27 of 30 samples. In addition to the L/P-activating substitution in the primary transcript, this mutation resulted in a stop loss in two of the alternative isoforms. The potential functional significance for these shorter transcripts in MYD88 L265P is not known. A subclonal C/G mutation was found in MYD88 resulting in an amino acid substitution (S219C) in 1 patient who also had the L265P mutation.8 No other mutations in MYD88 were observed in the study population.
To better understand the MYD88 wild-type (WT) population, we identified somatic mutations found exclusively in the 3 MYD88 WT patients. Cross-listing these genes with COSMIC, we observed that 2 of the 3 (67%) patients had damaging mutations in MLL2, while mutations in the genes JAK2, PRCC, SUZ12, BCL10, KDM6A, and SETD2 were each observed once. Sanger sequencing confirmed the somatic presence of MLL2 mutations in these patients as previously described.4 Although a larger sample size is needed to determine if the high frequency of MLL2 mutations is typical of the MYD88 WT WM population, it nonetheless represents a gene of great interest since MLL2 mutations are highly recurrent in follicular lymphoma and DLBCL patients.18
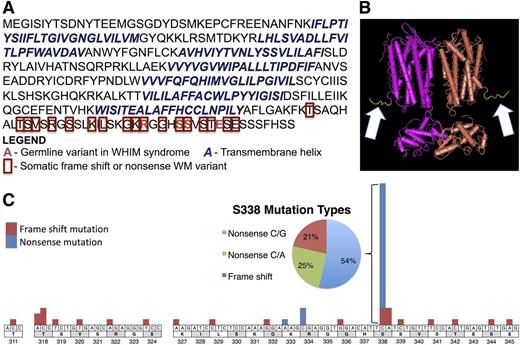
The next most common somatic mutation was observed in the G-protein coupled receptor, CXCR4. CXCR4 is a chemokine receptor that promotes WM survival, migration, and adhesion to the BM stroma through interactions with its ligand, CXCL12.19 Eight of 30 (27%) patients harbored 1 of 5 somatic variants in CXCR4, each of which were identical or functionally similar to mutations associated with WHIM syndrome (Figure 1).20,21 WHIM syndrome is a rare, autosomal dominant genetic disorder that is caused by frameshift or nonsense mutations in the carboxyl-terminal cytoplasmic tail (c-tail) of CXCR4. These mutations occur after the last of the seven transmembrane helices, destroying the c-tail but leaving the regions responsible for ligand binding and downstream signaling via g-proteins intact.22 These c-tail mutations result in the loss of regulatory serines resulting in impaired internalization and prolonged activation.23,-25 To validate this finding, we screened an independent cohort of 147 WM patients for CXCR4 mutations by Sanger sequencing. Combined with the 30 patients in the WGS cohort whose results were validated by Sanger sequencing, 51 of 177 (28.8%) patients had c-terminal mutations in CXCR4. Of these 177 patients, 160 demonstrated the MYD88 L265P mutation. Importantly, 50 of 51 (98%) patients with CXCR4 c-terminal mutations were also MYD88 L265P positive. Conversely, 16 of 17 (94.1%) of the MYD88 WT patients were WT for CXCR4 (P = .026).
Somatic CXCR4 mutations in WM are similar to those found in WHIM syndrome. Somatic CXCR4 nonsense and frameshift mutations found in 177 WM patient samples. (A) Protein sequence for the canonical full-length transcript (NP_003458.1) demonstrates that similar to the variants responsible for WHIM syndrome, these mutations result in a truncation of the cytosolic tail containing the regulatory phospho-serines leaving the seven transmembrane helix region involved in signaling and ligand binding intact. (B) The crystal structure of homodimeric CXCR4 with the carboxyl terminal tail highlighted in yellow and indicated by the white arrows. (C) Precise location and number of WHIM-like somatic mutations in WM at transcript level and detailed mutation-type summary of the most frequently mutated amino acid, S338.
Somatic CXCR4 mutations in WM are similar to those found in WHIM syndrome. Somatic CXCR4 nonsense and frameshift mutations found in 177 WM patient samples. (A) Protein sequence for the canonical full-length transcript (NP_003458.1) demonstrates that similar to the variants responsible for WHIM syndrome, these mutations result in a truncation of the cytosolic tail containing the regulatory phospho-serines leaving the seven transmembrane helix region involved in signaling and ligand binding intact. (B) The crystal structure of homodimeric CXCR4 with the carboxyl terminal tail highlighted in yellow and indicated by the white arrows. (C) Precise location and number of WHIM-like somatic mutations in WM at transcript level and detailed mutation-type summary of the most frequently mutated amino acid, S338.
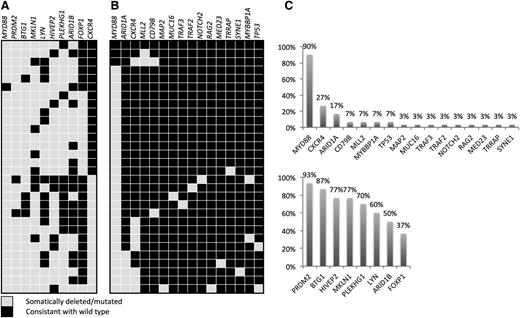
Other genes with validated somatic mutations in this series of patient samples included: ARID1A (5 of 30; 17%), CD79B (2 of 30; 7%), TP53 (2 of 30; 7%), MYBBP1A (2 of 30; 7%), MUC16 (1 of 30; 3%), TRAF2 (1 of 30; 3%), TRAF3 (1 of 30; 3%), RAG2 (1 of 30; 3%), and NOTCH2 (1 of 30; 3%) (Figure 2).
Summary of study results. (A) Copy number results per patient for the independent 30-patient validation cohort as determined by quantitative polymerase chain reaction (qPCR). Somatic MYD88 L265P and WHIM-like CXCR4 mutations were assessed in this population and annotated here for reference. The eight CNA targets selected for validation were chosen based on the 10-paired patient WGS analysis and technical validation studies. (B) Validated mutations in the 30-patient WGS cohort. Somatic status of all findings was confirmed by germline Sanger sequencing. (C) Overall frequency of validated somatic mutation (top) or CNA (bottom) for the independent 30-patient WGS and validation cohorts, respectively.
Summary of study results. (A) Copy number results per patient for the independent 30-patient validation cohort as determined by quantitative polymerase chain reaction (qPCR). Somatic MYD88 L265P and WHIM-like CXCR4 mutations were assessed in this population and annotated here for reference. The eight CNA targets selected for validation were chosen based on the 10-paired patient WGS analysis and technical validation studies. (B) Validated mutations in the 30-patient WGS cohort. Somatic status of all findings was confirmed by germline Sanger sequencing. (C) Overall frequency of validated somatic mutation (top) or CNA (bottom) for the independent 30-patient WGS and validation cohorts, respectively.
SV analysis
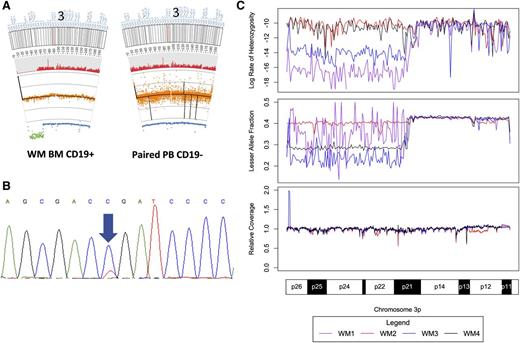
SVs were inferred from high confidence discordant mate-pair and relative coverage data supplied by Complete Genomics. To gain further insight into the SV events underlying WM, we combined individual patient data for lesser allele fraction (LAF), heterozygous call rate, normalized relative coverage, somatic CNAs, and somatic SV calculations with cohort-wide small variant distribution and CNA regions of statistical significance (supplemental Figure 1). These analyses aid in the detection of complex events such as acquired uniparental disomies (aUPD), which can be observed in chromosomes 3p and 21 in supplemental Figure 1. The most common aUPD was at 3p, occurring in 4 of 30 (13%) patients and whose location and size were highly conserved between the samples (Figure 3). This locus included the MYD88 L265P locus, resulting in homozygous somatic mutation in these patients. Additional large aUPDs were observed on chromosomes 1, 2, 5, 9, 17, 21, and X, with many of these affecting the validated small variant findings (Table 1).
aUPD in chromosome 3p in 4 patients with WM. (A) Representative excerpts from paired tumor and germline tissue documenting an aUPD on chromosome 3 for a single WM patient. The red histogram below the ideogram corresponds to the heterozygous call rate. Below that, copy number (black line) and normalized relative coverage (orange) is shown above the LAF measurements (blue). LAF values more than 2 standard deviations from the mean are shown in green. (B) Sanger sequencing of MYD88 L265P for this patient with a near homozygous mutant signal. (C) Heterozygous call rate, LAF, and relative coverage for the 4 patients with aUPDs in chromosome 3p demonstrating the near identical location of the aUPD for the 4 affected patients.
aUPD in chromosome 3p in 4 patients with WM. (A) Representative excerpts from paired tumor and germline tissue documenting an aUPD on chromosome 3 for a single WM patient. The red histogram below the ideogram corresponds to the heterozygous call rate. Below that, copy number (black line) and normalized relative coverage (orange) is shown above the LAF measurements (blue). LAF values more than 2 standard deviations from the mean are shown in green. (B) Sanger sequencing of MYD88 L265P for this patient with a near homozygous mutant signal. (C) Heterozygous call rate, LAF, and relative coverage for the 4 patients with aUPDs in chromosome 3p demonstrating the near identical location of the aUPD for the 4 affected patients.
Translocations were rare events, and we were unable to demonstrate any recurrent translocations in the 10 paired samples (supplemental Table 5). One patient had two distinct t(2;17) translocations disrupting both alleles of RNF213. A t(6;X) translocation disrupting the gene BIA3 was responsible for the chromosome 6q deletion observed in another patient (supplemental Figure 1). A third patient had a chromothriptic event centered on chromosome 6, which made up over half of all translocations observed in this study (supplemental Figure 2), and contributed to 6q deletions observed in the patient’s tumor cells. In this patient, we performed PCR for t(6;7) predicted to disrupt BMP5 and ANKRD7, as well as t(6;11) predicted to disrupt GRIA4 and PKHD1, respectively, and validated both of these findings. For 2 of 5 patients with paired normal/tumor genomes and in whom 6q deletions were identified, translocations were responsible for these regional losses. Macro-level analysis of normalized coverage for all 30 WM samples demonstrated that large deletions in 6q were present in 13 of 30 (43%) cases with corresponding gains in 6p in 3 of 13 (23%) cases. The next most common alteration observed was an amplification of chromosome 4, which occurred in 7 of 30 (23%) patients, confirming previous fluorescence in situ hybridization and array comparative genomic hybridization findings.14,15
Small somatic CNAs
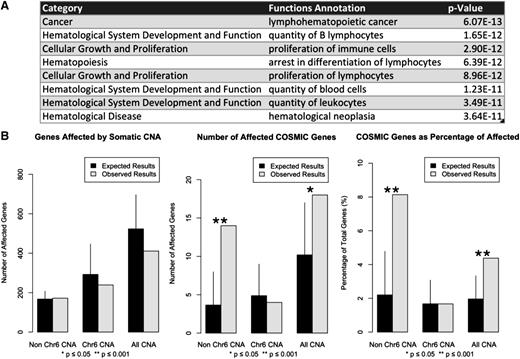
To detect small somatic deletions and amplifications, we calculated 100 kb blocks of the log-transformed ratio of tumor-to-germline relative coverage in each of the paired genomes. Significant CNAs were then compared across all 10 patients in order to identify statistically significant regions. CNAs consisted mostly of isolated 100 kb blocks both within and outside of chromosome 6 (supplemental Table 6 and supplemental Figure 1E). To investigate the biological relevance of these nonchromosome 6 CNAs, we performed functional enrichment analysis on the gene set affected by these alterations. Unique functional annotation results are listed in order of significance in Figure 4A.
Characterization of somatic CNAs in WM. (A) Functional annotation for genes affected by CNA found outside of chromosome 6. The list is ordered by statistical significance and filtered only for duplicated functional annotations matching more than one category. (B) Deletions of matching size were randomly distributed across the genome in 10 000 trials. The number of affected total RefSeq and COSMIC genes was calculated for each group. Results represent mean values with empirical 95% confidence intervals.
Characterization of somatic CNAs in WM. (A) Functional annotation for genes affected by CNA found outside of chromosome 6. The list is ordered by statistical significance and filtered only for duplicated functional annotations matching more than one category. (B) Deletions of matching size were randomly distributed across the genome in 10 000 trials. The number of affected total RefSeq and COSMIC genes was calculated for each group. Results represent mean values with empirical 95% confidence intervals.
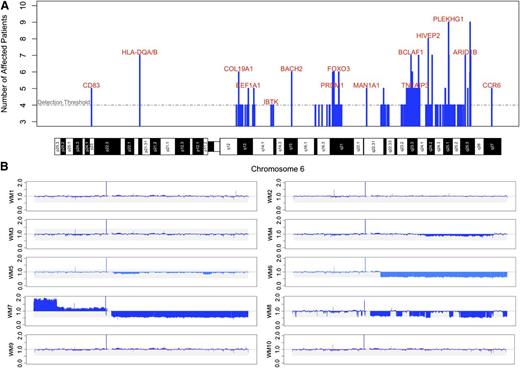
To better characterize these highly recurrent deletions, consecutive blocks of equal significance were merged together to form 194 statistically significant regions with a median length of 100 kb (range = 100 kb to 4900 kb). Of these regions, 134 (69%) were found outside of chromosome 6 with a median length of 100 kb (range = 100 kb to 300 kb), of which 126 of 134 (94%) were restricted to a single 100 kb region. Within chromosome 6, CNAs were larger with only 30 of 60 (50%) CNAs constituting single 100 kb regions (P = 7.709 × 10−12). CNAs targeted COSMIC census genes for 14 of 172 (8.14%) genes outside of chromosome 6 vs 4 of 239 (1.67%) genes within chromosome 6, respectively (P = .0024). To determine the probability of generating these results as a random by-product of genomic instability, we calculated the number of total genes affected by these deletions (N = 411) and the number of these genes found in the COSMIC database (N = 18). We randomly distributed matching deletion sets across the genome, and enumerated the total number of overlapping genes and the number of those genes listed in the COSMIC census (Figure 4B). These simulations revealed a statistically significant increase in targeting of COSMIC genes by CNAs outside of chromosome 6. Affected genes in the COSMIC census were: BTG1 (9 of 10; 90%), FOXP1 (7 of 10; 70%), FNBP1 (7 of 10; 70%), CD74 (7 of 10; 70%), TOP1 (6 of 10; 60%), MYB (5 of 10; 50%), CBLB (5 of 10; 50%), ETV6 (5 of 10; 50%), TNFAIP3 (5 of 10; 50%), FBXW7 (5 of 10; 50%), PRDM1 (5 of 10; 50%), TFE3 (4 of 10; 40%), JAK1 (4 of 10; 40%), MAML2 (4 of 10; 40%), FAM46C (4 of 10; 40%), EBF1 (4 of 10; 40%), STL (4 of 10; 40%), and BIRC3 (4 of 10; 40%). Other affected genes of interest included PRDM2 (8 of 10; 80%), HIVEP2 (8 of 10; 80%), ARID1B (7 of 10; 70%), as well as LYN (7 of 10; 70%).
There were no singular regions of statistical significance in 6q. Some patients had multifocal deletions in this region suggesting multiple target genes (Figure 5). Neither of the previously suspected target genes for 6q loss (ie, PRDM1 and TNFAIP3) were included in the regions of highest statistical significance.11,12 The relative coverage data from all 10-paired patients is shown in Figure 5B, illustrating that only 3 patients had highly clonal deletions in 6q. Two additional patients had subclonal deletions in 6q; therefore, 5 of 10 (50%) paired patients had at least subclonal loss. Deletions in HIVEP2 (8 of 10; 80%), as well as ARID1B (7 of 10; 70%) and BCLAF1 (7 of 10; 70%) constituted the most common deletions in chromosome 6, and were present in patients with and without visible 6q losses.
Somatic deletions identified on chromosome 6 by WGS in WM patients. (A) Frequency of statistically significant chromosome 6 deletions from the 10 paired patients highlighting genes of interest. The positions of the deletions are mapped against chromosome 6 cytogenetic bands. (B) Relative coverage across chromosome 6 for each of the 10 paired samples. Losses in 6q were not always single contiguous deletions, and some patients had deleted segments restricted to a subclone. The frequency of deletions for genes including HIVEP2 and ARID1B were higher than the corresponding number of large block deletions.
Somatic deletions identified on chromosome 6 by WGS in WM patients. (A) Frequency of statistically significant chromosome 6 deletions from the 10 paired patients highlighting genes of interest. The positions of the deletions are mapped against chromosome 6 cytogenetic bands. (B) Relative coverage across chromosome 6 for each of the 10 paired samples. Losses in 6q were not always single contiguous deletions, and some patients had deleted segments restricted to a subclone. The frequency of deletions for genes including HIVEP2 and ARID1B were higher than the corresponding number of large block deletions.
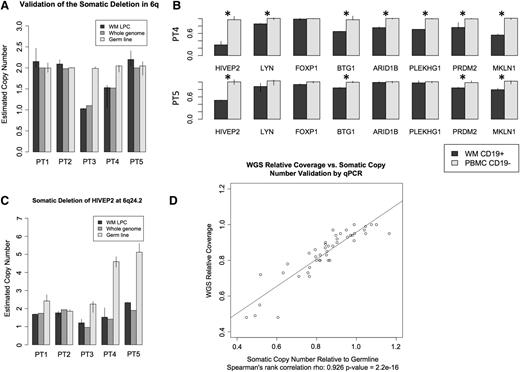
To test our validation system and examine the accuracy of the guanine-cytosine-normalized relative coverage data, we used a PCR-based copy number assay to validate the established 6q22 deletion in a subset of patients (Figure 6A). Having recapitulated the expected results, we conducted additional validation studies for 8 of our top targets in the same 5 patients using commercially available assays. To establish the frequency of these CNAs, we validated these 8 findings in an independent cohort of 30 WM patients revealing the following somatic losses: PDRM2 (28 of 30; 93%) at 1p36.21, BTG1 (26 of 30; 87%) in Chr. 12q21.33, HIVEP2 (23 of 30; 77%) at 6q24.2, MKLN1 (23 of 30; 77%) at 7q32, PLEKHG1 (21 of 30; 70%) at 6q25.1, LYN (18 of 30; 60%) at 8q12.1, ARID1B (15 of 30; 50%) at 6q25.1, and FOXP1 (11 of 30; 37%) at 3p13 (Figure 2). For MKLN1 and HIVEP2, germline CNAs were significant, and therefore, tumor-to-germline relative coverage should be interpreted accordingly (Figure 6C). While many results appeared subclonal, we observed a strong correlation between the PCR-relative copy number and WGS coverage predictions (Figure 6D).
Validation results using qPCR for the most frequent somatic deletions identified by WGS in WM patients. Five patient samples, 3 from the paired and 2 from the unpaired WGS cohorts, were selected for validation studies using qPCR copy number assays. All assays were run in at least triplicate. Results represent median values and ranges. (A) Validation of deletion in the known 6q deletion in 2 patients at HINT3 (6q22.32). (B) Representative validation results normalized to germline as determined by qPCR. Deletions deemed significant by Welch’s t test are denoted by asterisk (*). (C) Somatic relative copy number needs to be interpreted in context. Significant germline copy number variation was noted in both HIVEP2 and MKLN1. (D) Comparison of whole genome and qPCR validation estimates of relative somatic copy number demonstrating similar clonal estimates.
Validation results using qPCR for the most frequent somatic deletions identified by WGS in WM patients. Five patient samples, 3 from the paired and 2 from the unpaired WGS cohorts, were selected for validation studies using qPCR copy number assays. All assays were run in at least triplicate. Results represent median values and ranges. (A) Validation of deletion in the known 6q deletion in 2 patients at HINT3 (6q22.32). (B) Representative validation results normalized to germline as determined by qPCR. Deletions deemed significant by Welch’s t test are denoted by asterisk (*). (C) Somatic relative copy number needs to be interpreted in context. Significant germline copy number variation was noted in both HIVEP2 and MKLN1. (D) Comparison of whole genome and qPCR validation estimates of relative somatic copy number demonstrating similar clonal estimates.
CXCR4 and MYD88 L265P mutation status was determined in BM samples for all 30 patients by Sanger sequencing and AS-PCR, respectively. There were fewer median validated deletions in CXCR4-mutated patients (5) compared with WT (7; P = .002), and with the median total number of 6q CNAs (2) compared with WT (3; P = .007) (Figure 2). This was also true for any combination of two of the three validated 6q genes, thereby ruling out possible biasing by a single gene (P < .023 for all).
Discussion
The most pronounced finding from this study was the discovery of a somatic mutation in MYD88 (L265P), which was present in 90% of patients with WM. The details of this discovery were previously published following the preliminary examination of our WGS results.4 MYD88 serves as an adaptor molecule in Toll-like receptor (TLR) and interleukin-1 receptor signaling, mediating interleukin-1 receptor-associated kinase 4 (IRAK4) and IRAK1 activation, and downstream signaling through nuclear factor κB (NFκB).4,8,26 Mutations in MYD88 are significant given the importance of NFκB signaling in WM cell growth and survival.27 We recently demonstrated that MYD88 L265P can trigger NFκB signaling through an IRAK-independent pathway by direct interaction with Bruton’s tyrosine kinase (BTK) in WM cells, suggesting parallel pathways for NFκB activation.28 In contrast to the findings by Ngo et al who observed multiple MYD88 mutations (L265P, V217F, S219C, M232T, S243N, and T294P) in ABC-subtyped DLBCL patients, our findings were limited to the identification of L265P and S219C as a subclonal event in 1 WM patient with a L265P mutation.8 We also did not observe mutations in CARD11 in WM patients, demonstrating differences in potential oncogenic drivers and dependence on TLR signaling for WM vs ABC-subtyped DLBCL.
The next most common somatic variant after MYD88 L265P was in CXCR4, which was present in 27% of the patients. The mutations identified in WM tumor cells recapitulated those found in the germline of patients with WHIM syndrome. To our knowledge, this is the first time WHIM-like mutations have ever appeared as somatic mutations, and also be associated with a malignant disease. In WHIM syndrome, the loss of regulatory serines in the c-tail of CXCR4 are known to impair receptor internalization, thereby prolonging G-protein and β-arrestin signaling.25 CXCR4 stimulation by its ligand CXCL12 is known to activate AKT1 and mitogen-activated protein kinase family signaling, as well as facilitate cell migration and homing in WM cells.19 The prolonged activation of CXCR4 signaling due to WHIM mutations may therefore exaggerate these effects in WM cells, and deserves further study. Nearly all patients with CXCR4 mutations had also carried the MYD88 L265P mutation, and comprehensive studies will be required to delineate the relative impact of these mutations on WM clinical and treatment response characteristics. The presence of CXCR4 mutations may also offer a targeted approach to therapy of WM by use of CXCR4 antagonists given their successful record in the treatment of WHIM syndrome patients.29 Several antagonists to CXCR4 have been developed and are in clinical trials, including plerixafor, BMS-936564, AMD-070, TG-0054, and others, and warrant investigation alone and/or in combination in WM patients.
Whereas most WM cases in our series did not have mutations in CXCR4, it is possible that CXCR4 signaling may still be critical for many of these patients and offer an opportunity for targeted therapy with CXCR4 antagonists.21 In WM, polymorphisms of the CXCR4 ligand, CXCL12, have been associated with poor posttreatment clinical outcomes.30 Copy loss of RGS17 that was observed in 5 of 10 (50%) patients in our series may also affect this pathway. CXCR4 signals downstream through Giα G-proteins, increasing phosphorylated AKT1 levels and promoting cell survival, while RGS17 inhibits Giα G-proteins by promoting guanosine triphosphate hydrolysis.31,32 In an analogous case, RGS17 inactivation was observed in ovarian cancer affecting Giα G-protein coupled receptor downstream signaling in response to lysophosphatidic acid.31
The other major pathway identified in this study was the loss of chromatin remodeling proteins, ARID1A and ARID1B.33 ARID1A was the third most common single nucleotide variant target in WM, with 5 of 30 (17%) patients having validated nonsense or frameshift mutations. In 1 patient, a mutation in ARID1A (Y551 frameshift) was homozygous as a result of an aUPD, whereas in another patient a nonsense mutation (Q2037*) was opposite CNA loss, resulting in biallelic inactivation (Table 1). Loss of the alternate family member ARID1B was present in 7 of 10 (70%) of the paired patient samples, making it a more frequent 6q target than either PRDM1 or TNFAIP3 (5 of 10; 50% for both).33 One potential target for this pathway was the B-cell development regulator EBF1, which itself was affected by copy number variation deletions in 4 of 10 (40%) patients.34,35 Both ARID1A and ARID1B are members of the switch/sucrose nonfermentable family of proteins that are known to be mutated in other neoplastic malignancies, wherein they are thought to exert their effects via p53 and CDKN1A regulation.36,-38 TP53 itself was mutated in 2 of 30 (7%) of the sequenced genomes, including 1 case of biallelic mutation. Both PRDM2 and TOP1 participated in TP53-related signaling and were deleted in 8 of 10 (80%; 28 of 30 in validation) and 6 of 10 (60%) patients, respectively.39,40 Together, these findings imply that multiple mutations exist in the genome of WM patients which dysregulate the DNA damage response.
The loss of additional cancer-associated genes was observed including ETV6, which was deleted in 6 of 10 (60%) samples analyzed. Deletions of ETV6 have been observed in acute lymphoblastic leukemia (ALL), acute myeloid leukemia, and myelodysplastic syndromes.41,-43 ETV6 is a member of the E-twenty six transcription factor family and acts primarily as a transcriptional repressor involved in hematopoiesis, and functions as a tumor suppressor in myeloid leukemias.44 FOXO3, which was deleted in 6 of 10 (60%) patients, has been shown to negatively regulate growth and survival in mantle cell lymphoma, B-CLL, and natural killer cell neoplasms.45,-47 In B-CLL, phosphorylation of FOXO3 by AKT1 downstream of CXCL12 and CXCR4 prevents FOXO3 nuclear translocation and signaling.46 Upon nuclear translocation, FOXO3 can induce transcription of BTG1, which was deleted in 9 of 10 (90%; 26 of 30 in validation) paired patients.48 While FOXO3 (6q21) was not explicitly validated, 6q deletions were significantly less common in CXCR4-mutated patients and there was a trend for fewer BTG1 deletions as well. BTG1 is a nuclear coactivator that modulates transcription, a member of the antiproliferative TOB/BTG protein family, and was recently shown to be recurrently mutated in several studies of DLBCL.18,49,50 Small deletions affecting BTG1 have been reported in up to 12% of the B-cell precursor subtype of ALL.43,51,52 BTG1 deletions in B-cell precursor subtype of ALL were noted to coincide with deletions in ETV6 and EBF1, both of which were recurrently deleted in our study and suggests that these events may be functionally related.52 Preclinical studies of BTG1 in ALL have shown that the loss of BTG1 is associated with glucocorticoid resistance. These findings may explain the poor responses observed to single-agent steroids in WM, and warrant further investigation of BTG1 loss in treatment outcomes in patients undergoing glucocorticoid-inclusive therapy.53,54 Finally, biallelic loss of RNF213 resulting from two distinct t(2;17)s was observed in 1 of 10 paired patients, and represents an interesting finding since RNF213 is a fusion partner of anaplastic lymphoma kinase and MYC in acute anaplastic large cell lymphoma. In the WM patient with the t(2;17) translocation, RNF213 was translocated to the intergenic space indicating that the loss of RNF213 itself may play an important role in oncogenesis.55,56 It is important to note that most CNAs were consistent with heterozygous loss, and classical tumor suppressors require biallelic loss for oncogenesis. However, these losses could lead to pathway modulation due to altered protein levels or the creation of dominant negatives by partial gene loss.
Many of the mutations identified in this study impact NFκB signaling distal to the TLR4/MYD88 pathway. MYBBP1A, mutated in 2 of 30 (7%) patients, is thought to inhibit NFκB activity by repressing RELA.57 Copy loss of HIVEP2 (8 of 10; 80%; 23 of 30 in validation) and TNFAIP3 in 5 of 10 (50%) of the paired patients is of interest since their loss results in the removal of NFκB-negative regulators in WM.12,58 Chromosome 6q is often deleted in WM patients, and the apparent loss of HIVEP2 in 6q-intact patients is particularly compelling as a role for this gene in the pathogenesis of WM. Likewise, BIRC3 loss in 4 of 10 (40%) patients is associated with splenic marginal zone lymphoma and can increase activation of noncanonical NFκB signaling.59 Interestingly, both BIRC3 partner proteins, TRAF2 and TRAF3, that regulate noncanonical NFκB signaling were each found to be mutated in 1 of 30 (3%) patients. Biallelic loss of TRAF3 has been reported by Braggio et al12 in WM patients. These findings may therefore provide an impetus for studying noncanonical NFκB signaling in WM.
The loss in 7 of 10 (70%; 18 of 30 in validation) paired patients of LYN, a kinase that plays a regulatory role for B-cell receptor signaling, along with mutations in CD79B (2 of 30; 7%), indicates a possible role for B-cell receptor signaling in this disease.60,-62 The loss of IBTK in 4 of 10 (40%) patients and the interaction between activated MYD88 and BTK, raises the possibility for BTK-mediated cross-talk with the TLR/MYD88 pathway wherein BTK plays an important role in NFκB activation.28,63,-65 Moreover, CBLB, a gene disrupted in chronic myelogenous leukemia and lost in 5 of 10 (50%) of the paired patients, has been shown to inhibit the TLR4 response during inflammation by controlling TRL4 and MYD88 association, and subsequent NFκB activation.66,67 However, there is still limited data regarding the structure of the MYD88 L265P mutant protein. Signaling pathways that are well documented in the WT setting will need to be carefully re-examined to clarify the contribution of the L265P mutation to downstream MYD88 signaling.
Our WGS studies in WM patients have therefore identified mutations in genes involving TLR, NFκB, and CXCR4 signaling, chromatin remodeling, and cell-cycle regulation. These findings may denote a multistep process for WM evolution from IgM MGUS to asymptomatic and symptomatic WM, which invariably will require comparative, and even prospective longitudinal sequencing studies. These efforts are particularly warranted since the principal mutation (MYD88 L265P) identified in these studies is also present in 50% to 80% of IgM MGUS patients using AS-PCR, signifying its role as an early oncogenic event.10,68,69 Therefore, other mutations are likely to be acquired in the evolution of MGUS to symptomatic WM, and may offer the opportunity to identify those patients at high risk for disease evolution. Lastly, these studies have also identified novel targets for a rational approach to WM treatment, including potentially, the use of inhibitors targeting TLR and CXCR4 signaling.
Presented in abstract form at the 48th annual meeting of the American Society of Clinical Oncology, Chicago, IL, June 1, 2012.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Yaoyu Wang and John Quackenbush at the Center for Cancer Computational Biology at the Dana-Farber Cancer Institute for their assistance in developing the copy number analysis.
This work was supported by Peter S. Bing, the International Waldenström’s Macroglobulinemia Foundation, the Coyote Fund for Waldenström’s Macroglobulinemia, the D’Amato Family Fund for Genomic Discovery, the Edward and Linda Nelson Fund for Waldenström’s Macroglobulinemia Research, and the WM patients who provided their samples.
Authorship
Contribution: Z.R.H. and S.P.T. designed the study and wrote the manuscript; Z.R.H. performed the data analysis and conducted the copy number validation; Z.R.H. and X.L. designed Sanger sequencing primers; G.Y., Y.Z., Y.C., X.L., and L.X. prepared the samples and performed the Sanger validation studies; S.P.T. and P.S. provided patient care, and obtained consent and samples; and R.J.M., C.T., and C.J.P. selected samples and provided clinical data analysis.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Steven P. Treon, Bing Center for Waldenström’s Macroglobulinemia, Dana-Farber Cancer Institute, M547, 450 Brookline Ave, Boston, MA 02215; e-mail: steven_treon@dfci.harvard.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal