Abstract
The Myeloproliferative Neoplasms (MPNs), including Polycythemia Vera (PV), Essential Thrombocythemia (ET), and Primary Myelofibrosis (PMF) are clonal hematopoietic disorders. JAK2V617F mutations are observed in approximately 90-95% of PV cases, but only 40-50% of ET and PMF cases. Although JAK2 exon 12 and LNK mutations are observed in the majority of JAK2V617F-negative PV patients, candidate gene and exome sequencing studies to date have not identified activating oncogenes in the majority of JAK2V617F-negative ET/PMF patients. Thus, further genetic investigations are needed to define the mutational architecture of these JAK2 wildtype MPNs in order to gain insight into the biology of these diseases, the clinical implications of genetic events that do occur, and the elucidation of potential therapeutic targets.
To characterize the spectrum of genetic alterations in JAK2 wildtype chronic-phase myeloproliferative neoplasms.
We identified 32 patients with a confirmed diagnosis of an MPN (per 2008 WHO criteria), including MF, PV and ET, who were negative for JAK2V617F using a CLIA-certified allele specific assay for the JAK2 disease allele. Genomic DNA and total RNA was isolated from formalin fixed paraffin embedded (FFPE) tissue, blood and bone marrow aspirates. Adaptor ligated sequencing libraries were captured by solution hybridization using two custom baitsets targeting 374 cancer-related genes and 24 genes frequently rearranged for DNA-seq, and 258 genes frequently rearranged for RNA-seq. All captured libraries were sequenced to high depth (Illumina HiSeq), averaging >590X for DNA and >20,000,000 total pairs for RNA, to enable the sensitive and specific detection of genomic alterations.
High coverage sequencing allowed us to identify JAK2V617F mutations in two patients (allele burden 3-5%) that were below the limit of detection of the CLIA assay. The most common mutations observed in JAK2V617F-negative MPN were in ASXL1 (22% of patients) and in TET2 (9%). Taken together, mutations in known epigenetic modifiers (ASXL1, TET2, DNMT3A, EZH2, MLL) were observed in 43% of samples, including a MLL-PTD mutation in one patient with PMF. We identified mutations in spliceosome components (SRSF2, U2AF1), in a subset of patients, consistent with previous reports, and in each case mutations in spliceosome components were mutually exclusive.
We identified mutations in the JAK-STAT pathway (MPL, TYK2) and the RAS pathway components (KRAS, NF1) in 9% of this patient cohort, suggesting that there are alternate disease alleles that activate signaling in JAK2V617F-negative MPN. RNA-sequencing identified a ETV6-ABL1 fusion in one patient, and we identified amplification of PIK3CA in one patient in our cohort; these data suggest fusion genes and amplifications activate signaling in a subset of patients with JAK2V617F-negative MPN. We also identified novel mutations in MPN patients which have not been reported to date, including mutations in DNA repair genes (ATM and BRCA) in 25% of cases and mutations in the Notch signaling pathway (NOTCH1-4) in 31% of cases. The functional implications of these novel mutations remain to be elucidated.
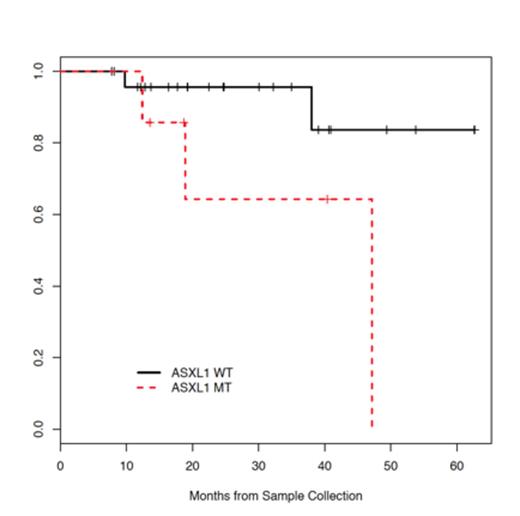
In univariate analysis, ASXL1 mutations were found to associate with impaired overall survival (Figure 1, p=0.049). These findings are consistent with data demonstrating an impaired survival in patients with MDS and PMF, and suggest that ASXL1 mutations represent an important biomarker for adverse outcome in JAK2V617F-negative MPN.
ASXL1 mutations result in impaired survival in patients with JAK2 wildtype MPNs.
ASXL1 mutations result in impaired survival in patients with JAK2 wildtype MPNs.
These data demonstrate that the mutational spectra of JAK2V617F-negative MPN includes genes implicated in epigenetic regulation, novel mutations which activate gene signaling, and fusion genes/copy number alterations which provide a novel mechanism of oncogenic activation not previously reported in MPN. ASXL1 mutations occur frequently in JAK2 wildtype Philadelphia-Chromosome negative MPNs, and are associated with impaired overall survival. Collectively, these findings support the importance of ASXL1 mutations in predicting outcome in JAK2V617F-negative MPN, demonstrate that mutations in signaling effectors and in epigenetic regulators are common in MPN, and illustrate the genetic heterogeneity of JAK2V617F-negative MPN.
Rampal:Foundation Medicine: Consultancy. Levine:Foundation Medicine, Inc: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal