Key Points
TLR-activated FANCA- and FANCC-deficient macrophages overproduce IL-1β.
IL-1β suppresses in vitro expansion of Fancc-deficient multipotent hematopoietic progenitor cells.
Abstract
Hematopoietic stem and progenitor cells with inactivated Fanconi anemia (FA) genes, FANCA and FANCC, are hypersensitive to inflammatory cytokines. One of these, tumor necrosis factor α (TNF-α), is also overproduced by FA mononuclear phagocytes in response to certain Toll-like receptor (TLR) agonists, creating an autoinhibitory loop that may contribute to the pathogenesis of progressive bone marrow (BM) failure and selection of TNF-α–resistant leukemic stem cell clones. In macrophages, the TNF-α overproduction phenotype depends on p38 mitogen-activated protein kinase (MAPK), an enzyme also known to induce expression of other inflammatory cytokines, including interleukin 1β (IL-1β). Reasoning that IL-1β might be involved in a like autoinhibitory loop, we determined that (1) TLR activation of FANCA- and FANCC-deficient macrophages induced overproduction of both TNF-α and IL-1β in a p38-dependent manner; (2) exposure of Fancc-deficient BM progenitors to IL-1β potently suppressed the expansion of multipotent progenitor cells in vitro; and (3) although TNF-α overexpression in FA cells is controlled posttranscriptionally by the p38 substrate MAPKAPK-2, p38-dependent overproduction of IL-1β is controlled transcriptionally. We suggest that multiple inflammatory cytokines overproduced by FANCA- and FANCC-deficient mononuclear phagocytes may contribute to the progressive BM failure that characterizes FA, and that to achieve suppression of this proinflammatory state, p38 is a more promising molecular therapeutic target than either IL-1β or TNF-α alone.
Introduction
Fanconi anemia (FA) is an inherited disease caused by biallelic or X chromosome–linked inactivation of any 1 of 15 different genes, including FANCA and FANCC.1-4 Most FA patients develop progressive bone marrow (BM) failure and have a high relative risk of clonal evolution to acute myelogenous leukemia.5-7 Additionally, FA patients have an equally high risk of developing squamous cell carcinomas and other epithelial malignancies.8 The FA gene products participate in DNA damage response pathways and may protect hematopoietic cells from damage induced by the production of endogenous aldehydes.9,10 There is also evidence that at least some of the FA proteins function in hematopoietic cells to modulate responses to suppressive cytokines in progenitor cells,11-15 and Toll-like receptor (TLR) responses in macrophages,16,17 even in the absence of DNA damage.18-20 Thus, deficiency of either FANCA or FANCC in macrophages results in the overproduction of at least 1 growth-suppressive cytokine, tumor necrosis factor α (TNF-α), to which FA BM progenitors are inherently hypersensitive. This may contribute directly to stem cell dysfunction and BM failure and facilitate the selection and outgrowth of TNF-α–resistant neoplastic hematopoietic cells,21 a notion that has been formally confirmed in Fancc-deficient mice.22
Activation of TLR pathways in macrophages induces production of other inflammatory cytokines, including interleukin 1β (IL-1β),23-25 which is known to promote tumor progression and metastasis.26,27 Expression of IL-1β, like TNF-α, is known to be dependent on the activity of the global stress responsive p38 mitogen-activated protein kinase (MAPK) signal transduction pathway.28 Therefore, in light of our prior studies and the work of others confirming that TNF-α is a potent endogenous suppressor of normal hematopoietic progenitor and stem cell activity in mice,11-15 we reasoned that FA marrow progenitors and stem cells might also be hypersensitive to IL-1β and that TLR activation might induce the overproduction of both TNF-α and IL-1β by FA mononuclear phagocytes. We show here the results of studies that confirm this hypothesis, revealing a second cytokine autoinhibitory loop in aberrant FA hematopoiesis and suggesting that multiple inflammatory cytokines may contribute to suppression of hematopoiesis in this disease.
Materials and methods
Antibodies and reagents
Anti-phospho-p38 MAPK (Thr180/Tyr182; #9216), anti-p38 MAPK (#9212), anti-phospho-MAPKAPK-2 (Thr334; #3007), anti-MAPKAPK-2 (#3042), anti-IL-1β (#2022), anti-A20 (#5630), anti-ASK1 (#3762), anti-HSP90 (#4877), anti-C/EBPβ (#3087), anti-phospho-C/EBPβ (Thr235; #3084), anti-MKK3 (#9232), and anti-MKK6 (#9264), and anti-caspase-1 (#3866) were purchased from Cell Signaling Technology (Danvers, MA). Anti-FANCA (sc-18664), anti-HSP70 (sc-24), anti-MAPKAPK-2 (sc-100393), anti-TAK1 (sc-7162), anti-TRAF6 (sc-8409), and anti-RIP1 (sc-7881 and sc-133102) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-FANCC (#2) was obtained from the Fanconi Anemia Research Fund Antibody Project (Portland, OR). Anti-caspase-1 (ALX-210-804) and R848 (ALX-420-038) were purchased from Enzo Life Sciences (Farmingdale, NY). Anti-GAPDH (ab9485) was purchased from Abcam (Cambridge, MA). The following fluorescently conjugated anti-mouse antibodies for flow cytometry were purchased from BD Pharmingen (San Jose, CA): PE-TER119 (#553873), PE-CD45R (#553080), PE-CD11b (#553311), PE-CD4 (#553730), PE-CD8a (#553033), PE-CD5 (#553023), PE-Ly6G/Ly6C (#553128), PE-CD3e (#553064), PE-CF594-CD117 (c-Kit) (#562417), PE-Cy7-Sca-1 (#558162), FITC-CD34 (#560238), APC-CD135 (Flt3) (#560718), FITC-Ly6A/E (Sca-1) (#557405), and APC-CD117 (c-Kit) (#553356). Anti-TNF-α (MAB610) and anti-IL-1β (MAB201) neutralizing antibodies were purchased from R&D Systems (Minneapolis, MN). Recombinant murine IL-1β (#401-ML), IL-6 (#406-ML), IL-11 (#418-ML), stem cell factor (SCF; #455-ML), and FLT3L (#427-FL) were purchased from R&D Systems. BIRB 796 (doramapimod; #1358) was obtained from Axon Medchem (Groningen, The Netherlands). Glyburide (glibenclamide; G0639) and BIO (6-bromoindirubin-3′-oxime; B1686) were purchased from Sigma-Aldrich (St. Louis, MO).
Cell lines and primary cells
THP-1 human acute monocytic leukemia cells were purchased from the American Type Culture Collection (Manassas, VA), and THP1-XBlue cells (thpx-sp) were purchased from InvivoGen (San Diego, CA). The development and culture conditions of the derivative cell lines we established (T-shNT, T-shFA, and T-shFC) were described previously.16,17
BM-derived macrophages (BMDM) from wild-type and Fancc−/− mice were prepared as described previously.17 Murine macrophages were cultured at a concentration of 50 000 cells per mL for 24 hours in RPMI 1640 medium (Life Technologies, Carlsbad, CA) with 10% fetal bovine serum (FBS; Thermo Scientific Hyclone, Logan, UT) in the presence of the indicated doses of R848. All murine studies were approved by the Portland Veteran’s Administration Institutional Animal Care and Use Committee.
In 4 separate experiments, BM mononuclear cells were prepared from 2 wild-type and 2 Fancc−/− mice using Ficoll-Paque Plus gradient separation (GE Healthcare, Piscataway, NJ) and then pooled according to genotype. Cells were stained with the following anti-mouse antibodies: phycoerythrin (PE)–conjugated lineage markers (B220, CD3e, CD4, CD5, CD8a, CD11b, Gr-1, and Ter119), PE-CF594-CD117 (c-Kit), PE-Cy7-Sca-1, fluorescein isothiocyanate–CD34, and allophycocyanin-CD135 (Flt3) in a solution of Hank’s buffered salt solution/0.5% bovine serum albumin/10 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid. c-Kit+Lineage–Sca-1+ (KLS) cells were isolated on an Influx cell sorter (BD Biosciences) and, based on percent of the KLS cells also bearing the CD34–Flt3– signature (defined herein as long-term hematopoietic stem cells, or LT-HSCs29 ), were plated at 1000 LT-HSCs per well in 500 μL of RPMI, containing penicillin and streptomycin, 10% FBS, mIL-6 (20 ng/mL), mIL-11 (10 ng/mL), mSCF (10 ng/mL), and mFLT3L (50 ng/mL). Half of the wells for each genotype were also treated with murine IL-1β (30 pg/mL) for 7 days. Cells were then counted and stained with the same cocktail of antibodies as described previously and analyzed by LSRII flow cytometer (BD Biosciences).
Primary human CD14+ cells were prepared from peripheral blood mononuclear leukocytes (isolated using Ficoll-Paque Plus; GE Healthcare, Little Chalfont, United Kingdom) using magnetic microbeads (EasySep human CD14 positive selection kit; StemCell Technologies, Vancouver, BC, Canada). Cells were cultured at a concentration of 50 000 cells per mL for 24 hours in RPMI 1640 medium with 10% FBS in the presence of the indicated doses of R848. Conditioned medium was harvested for TNF-α and IL-1β quantification using the Quantikine ELISA (enzyme-linked immunosorbent assay) Kit (R&D Systems). Blood from healthy volunteers and from patients was collected after having obtained informed consent from either the subject or the subject’s parents in accordance with the Declaration of Helsinki and enrolled in a clinical study approved by the Institutional Review Board of Oregon Health & Science University.
Stable suppression of FANCC and FANCA with lentiviral short hairpin RNA (shRNA)
THP1-XBlue cells were transduced with lentiviral particles expressing shRNA targeting FANCA and FANCC, as described previously.16,17 shRNA-expressing cells were selected with 0.6 μg/mL puromycin for 2 to 3 weeks, after which time the cells were maintained in medium containing 0.3 μg/mL puromycin.16 Knockdown of FANCA or FANCC was confirmed by immunoblotting for FANCA, FANCC, and FANCD2 and by cytogenetic confirmation of mitomycin-C–induced chromosomal breakage.16 Immunoblot analyses were performed as described previously.11,16
Immunoassays
A total of 200 000 cells per 200 μL (T-shNT, T-shFA, and T-shFC) or 10 000 cells per 200 μL (primary human CD14+ cells or mouse BMDM) was cultured in 96-well plates and treated with R848 for 24 hours, after which culture supernatants were assayed for IL-1β or TNF-α using Quantikine ELISA Kits (R&D Systems). In some experiments, cultures were pretreated with BIRB 796 for 6 hours, glyburide for 2 hours, or anti-TNF-α or anti-IL-1β neutralizing antibodies for 2 hours before the addition of R848 for 24 hours followed by medium collection.
Real-time quantitative reverse-transcription polymerase chain reaction (qRT-PCR)
Total RNA was prepared from 1 × 106 to 5 × 106 cells using the RNeasy Mini kit (Qiagen, Valencia, CA). Complementary DNA synthesis and real-time PCR were performed as described previously.16,17 Primer and probe sets for human IL-1β (Hs01555410_m1), human TNF-α (Hs00174128_m1), and human 18S ribosomal RNA (rRNA; 4319413E) were purchased as TaqMan Gene Expression Assays (Life Technologies).
Small interfering RNA (siRNA) knockdown
SMARTpool siRNA targeting MK2 and control siRNA were purchased from Thermo Scientific Dharmacon (Lafayette, CO). The pool consisted of 4 distinct sequences: GAACCACCCUUGGAUCAUG, GAAUGACCAUCACCGAGUU, CGAAUGGGCCAGUAUGAAU, and UGAUUGUCAUGGAAUGUUU. Cells were transfected with siRNA using the Amaxa Nucleofector II (Lonza, Basel, Switzerland), as described previously.16 After transfection, the cells were cultured for 24 hours, transfected a second time, and then cultured for 48 hours before stimulation with R848. Knockdown of the protein of interest was confirmed by immunoblotting. Band densitometry was performed using ImageJ software (National Institutes of Health).
Ectopic expression of MAPKAPK-2 constitutively active mutant (MK2-CA)
As previously described, ectopic expression of an MK2-CA containing only the catalytic domain (amino acids [aa] 49-338) in macrophages is sufficient to induce phosphorylation of the MK2 substrate HSP25 and to enhance TLR-induced TNF-α production.30 A PCR fragment containing complementary DNA coding for aa 49 to 338 of MK2 was subcloned into the vector pLXSN (Clontech Laboratories, Mountain View, CA). THP-1 cells expressing pLXSN or pLXSN-MK2-CA were selected using 200 μg/mL G418 (Invitrogen) for 2 to 3 weeks. MK2-CA overexpression was confirmed by immunoblot analysis.
Results
TLR-induced IL-1β overproduction in FANCA- and FANCC-deficient mononuclear phagocyte cell lines and primary cells requires activation of the inflammasome
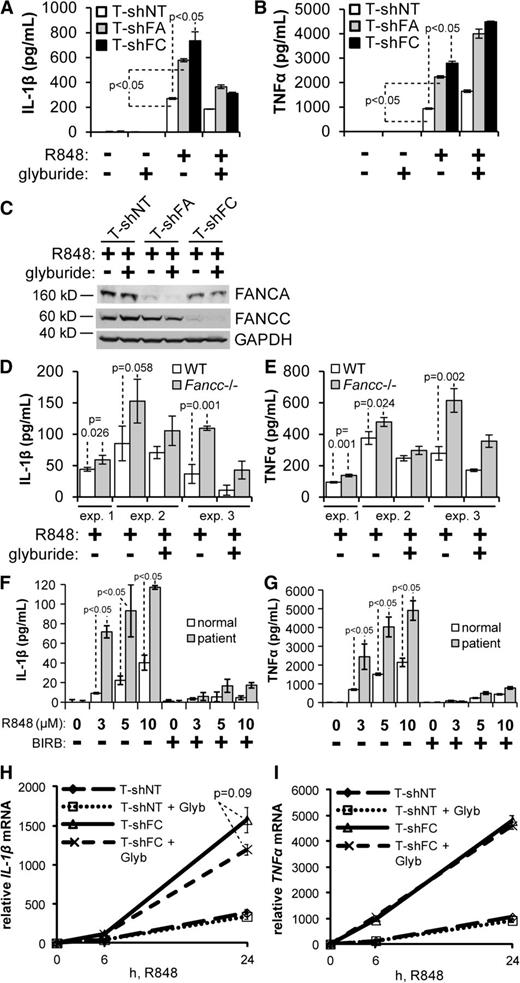
THP-1 human monocytic leukemia cells stably expressing shRNA directed against FANCA (T-shFA) or FANCC (T-shFC) produced 2 and 2.5 times more IL-1β, respectively, in response to R848 (a TLR7/8 ligand) than did cells expressing a nontargeted shRNA (T-shNT) (Figure 1A). R848 treatment did not affect the viability of either cell type. In monocytes, production of IL-1β (but not TNF-α) is known to be controlled by the inflammasome, a multimeric complex activated by a wide range of stress signals.24,31,32 To determine whether TLR-induced IL-1β hypersecretion in T-shFA and T-shFC cells requires activation of the inflammasome, we used 2 inhibitors of functional molecules of the inflammasome complex: glyburide, an inhibitor of the NLRP3 inflammasome,33 and YVAD, an inhibitor of caspase-1–dependent IL-1β secretion. Both glyburide (Figure 1A) and YVAD (supplemental Figure 1A; see the Blood Web site) significantly suppressed IL-1β production by T-shFA, T-shFC, and control cells. As expected, neither glyburide (Figure 1B) nor YVAD suppressed TLR-induced secretion of TNF-α by T-shNT and T-shFC cells (supplemental Figure 1B). Silencing of FANCA and FANCC in these cell lines was confirmed by western blotting and was not affected by the addition of glyburide (Figure 1C).
TLR-induced overproduction of IL-1β by FANCA- and FANCC-deficient cells. (A-C) THP-1 cells expressing shRNA directed against FANCA (T-shFA), FANCC (T-shFC), or a nontargeted shRNA (T-shNT) were plated at a concentration of 106 per mL, pretreated with glyburide (50 μM) for 2 hours, and stimulated with R848 (30 μM) for 24 hours. Secreted IL-1β (A) and TNF-α (B) were measured in the conditioned media by ELISA. One representative experiment of 3 is shown. Each consisted of 3 technical replicates for each condition. (C) Western blotting of indicated proteins from whole cell extracts of the same cells was performed, and molecular weight markers are indicated. (D-E) Wild-type and Fancc−/− BMDMs were plated at a concentration of 50 000 per mL, pretreated with glyburide (50 μM) for 2 hours, and stimulated with R848 (3 μM) for 24 hours. Secreted IL-1β (D) and TNF-α (E) were measured in the culture media by ELISA. The results of 3 independent experiments, each of which consisted of 3 biological replicates for each condition, are shown. Each experiment contains pooled cells from 2 wild-type mice and 2 Fancc−/− mice. (F-G) CD14+ cells from an FA complementation group A patient and an age-matched healthy donor were isolated from peripheral blood mononuclear cells using magnetic microbeads. Cells were plated at a concentration of 50 000 per mL, pretreated with BIRB 796 (500 nM) for 6 hours, and stimulated with the indicated doses of R848 for 24 hours. Secreted IL-1β (F) and TNF-α (G) were measured in the conditioned media by ELISA. One experiment with 3 biological replicates for each condition is shown. (H-I) T-shNT and T-shFC cells were plated at a concentration of 106 per mL, pretreated with glyburide (Glyb; 50 μM) for 2 hours, and stimulated with R848 (30 μM). Total RNA was isolated at 0, 6, and 24 hours after R848 treatment, and IL-1β (H) and TNF-α (I) mRNA were measured using real-time qRT-PCR and normalized to levels of 18S rRNA. One experiment with 3 technical replicates for each condition is shown. P values were calculated using a paired Student t test.
TLR-induced overproduction of IL-1β by FANCA- and FANCC-deficient cells. (A-C) THP-1 cells expressing shRNA directed against FANCA (T-shFA), FANCC (T-shFC), or a nontargeted shRNA (T-shNT) were plated at a concentration of 106 per mL, pretreated with glyburide (50 μM) for 2 hours, and stimulated with R848 (30 μM) for 24 hours. Secreted IL-1β (A) and TNF-α (B) were measured in the conditioned media by ELISA. One representative experiment of 3 is shown. Each consisted of 3 technical replicates for each condition. (C) Western blotting of indicated proteins from whole cell extracts of the same cells was performed, and molecular weight markers are indicated. (D-E) Wild-type and Fancc−/− BMDMs were plated at a concentration of 50 000 per mL, pretreated with glyburide (50 μM) for 2 hours, and stimulated with R848 (3 μM) for 24 hours. Secreted IL-1β (D) and TNF-α (E) were measured in the culture media by ELISA. The results of 3 independent experiments, each of which consisted of 3 biological replicates for each condition, are shown. Each experiment contains pooled cells from 2 wild-type mice and 2 Fancc−/− mice. (F-G) CD14+ cells from an FA complementation group A patient and an age-matched healthy donor were isolated from peripheral blood mononuclear cells using magnetic microbeads. Cells were plated at a concentration of 50 000 per mL, pretreated with BIRB 796 (500 nM) for 6 hours, and stimulated with the indicated doses of R848 for 24 hours. Secreted IL-1β (F) and TNF-α (G) were measured in the conditioned media by ELISA. One experiment with 3 biological replicates for each condition is shown. (H-I) T-shNT and T-shFC cells were plated at a concentration of 106 per mL, pretreated with glyburide (Glyb; 50 μM) for 2 hours, and stimulated with R848 (30 μM). Total RNA was isolated at 0, 6, and 24 hours after R848 treatment, and IL-1β (H) and TNF-α (I) mRNA were measured using real-time qRT-PCR and normalized to levels of 18S rRNA. One experiment with 3 technical replicates for each condition is shown. P values were calculated using a paired Student t test.
TLR-induced IL-1β overproduction was also observed in primary BMDMs from Fancc−/− mice and in peripheral blood mononuclear phagocytes from an FANCA-deficient patient. Specifically, BMDM from Fancc−/− mice produced significantly more IL-1β in response to R848 than did cells from wild-type mice, and glyburide suppressed both IL-1β (Figure 1D) and TNF-α production (Figure 1E) from both the wild-type and Fancc-deficient cells. Primary peripheral blood CD14+ cells from an FA complementation group A patient (FANCA-deficient) were likewise affected. R848-treated CD14+ patient cells produced 3 to 5 times more IL-1β (Figure 1F) and TNF-α (Figure 1G) than did healthy donor cells. IL-1β secretion from these cells was suppressed six- to sevenfold by the addition of glyburide (supplemental Figure 2).
To rule out the possibility that glyburide directly suppresses IL-1β gene transcription rather than NLRP3 inflammasome function, we measured TLR-induced IL-1β messenger RNA (mRNA) production in T-shNT and T-shFC cells. Although we observed an approximately fourfold increase in IL-1β mRNA levels in FANCC-deficient cells after 24 hours, glyburide did not significantly suppress IL-1β mRNA production (Figure 1H) and had no effect on TNF-α mRNA production (Figure 1I).
IL-1β overproduction by TLR-activated FANCC-deficient macrophages is p38 dependent
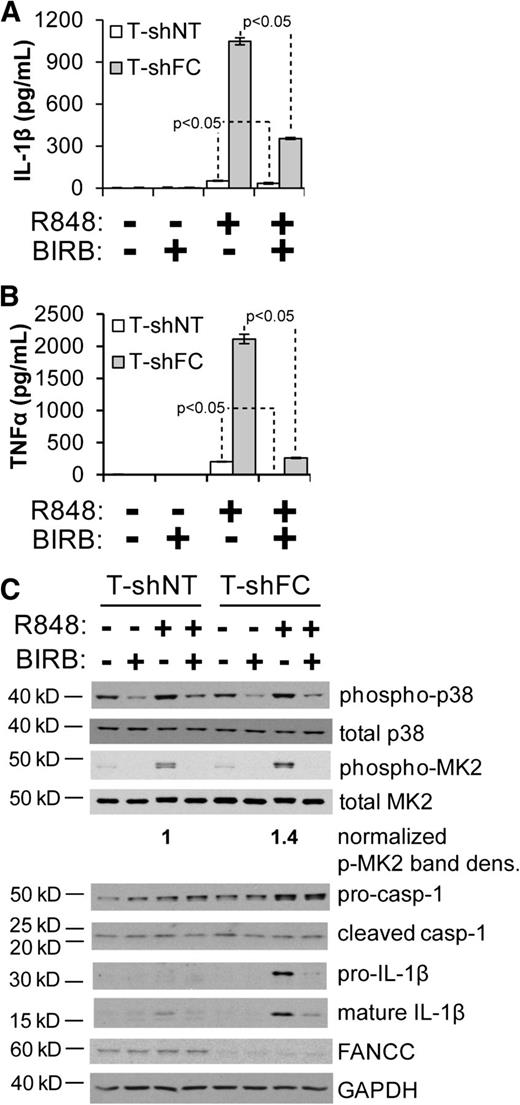
As we previously demonstrated, TLR-induced overproduction of TNF-α by FANCC-deficient macrophages is suppressed by the p38 MAPK inhibitor BIRB 796 and by suppressing expression of the p38 substrate MK2.17 Others have shown that IL-1β expression can be mediated by p38 MAPK.28 Consequently, we sought to determine the degree to which IL-1β overproduction in FA cells is p38 dependent. R848-induced secretion of IL-1β by T-shNT and T-shFC cells was markedly suppressed by BIRB 796 (Figure 2A), suggesting a role for p38 in overproduction of IL-1β. Consistent with our previous report,17 phosphorylation of the p38 substrate MK2 on Thr334 in response to R848 was elevated by 1.4-fold in T-shFC cells and was suppressed by BIRB 796 (Figure 2B). Processing of pro-caspase-1 to the cleaved caspase-1 p20 subunit was equivalent in both cell types and unaffected by BIRB 796 (Figure 2B). Also, R848-induced expression of pro-IL-1β was higher in T-shFC cells than in T-shNT cells (Figure 2B). Although the increased level of mature IL-1β in T-shFC cells is a reflection of the level of pro-IL-1β, the processing of IL-1β was not affected by FANCC deficiency or p38 inhibition (Figure 2B). That 2 markers of inflammasome activation, processing of caspase-1 and of IL-1β, are similar in both cell types and unaffected by BIRB 796 indicates that mature IL-1β production resulting from inflammasome activation occurs independently of FANCC function and p38 activation.
IL-1β overproduction from FANCC-deficient macrophages is p38 dependent. T-shNT and T-shFC cells were plated at a concentration of 106 per mL, pretreated with BIRB 796 (50 nM) for 6 hours, and stimulated with R848 (30 μM) for 24 hours. Secreted IL-1β (A) and TNF-α (B) were measured in the conditioned media by ELISA. One experiment with 3 technical replicates for each condition is shown. (C) Western blotting of indicated proteins from whole cell extracts of the same cells was performed, and molecular weight markers are indicated (casp-1 indicates caspase-1). Band densitometry was performed using ImageJ software. Phosphorylated MK2 band density was normalized to total MK2 band density. P values were calculated using a paired Student t test.
IL-1β overproduction from FANCC-deficient macrophages is p38 dependent. T-shNT and T-shFC cells were plated at a concentration of 106 per mL, pretreated with BIRB 796 (50 nM) for 6 hours, and stimulated with R848 (30 μM) for 24 hours. Secreted IL-1β (A) and TNF-α (B) were measured in the conditioned media by ELISA. One experiment with 3 technical replicates for each condition is shown. (C) Western blotting of indicated proteins from whole cell extracts of the same cells was performed, and molecular weight markers are indicated (casp-1 indicates caspase-1). Band densitometry was performed using ImageJ software. Phosphorylated MK2 band density was normalized to total MK2 band density. P values were calculated using a paired Student t test.
To confirm that production of IL-1β is dependent on p38 pathway activation in primary cells, we measured the effects of BIRB 796 on TLR-activated peripheral blood CD14+ cells from an FANCA-deficient patient. R848-treated FA cells produced 3 to 5 times more IL-1β and TNF-α than did healthy donor cells, and both IL-1β (Figure 1F) and TNF-α (Figure 1G) production by normal and FA cells was suppressed by BIRB 796.
TLR-induced production of IL-1β is not controlled by the p38 substrate MK2
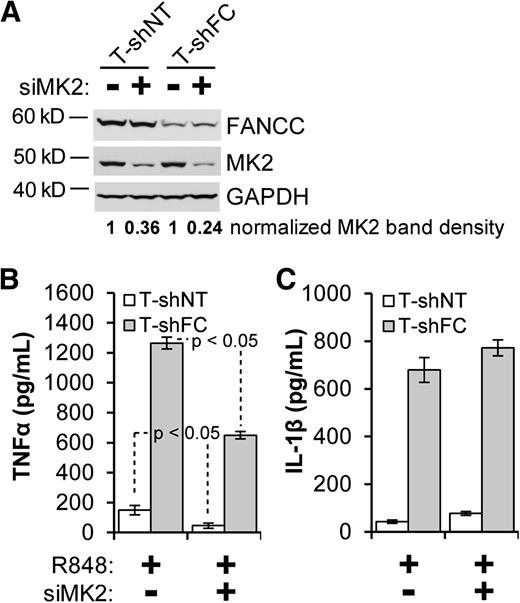
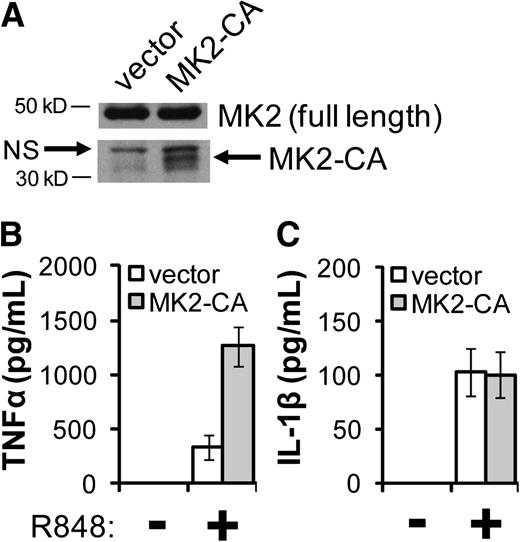
The 3′ untranslated region of the IL-1β mRNA, like that of the TNF-α mRNA, contains AU-rich elements,34 known to be essential for permitting MK2 to enhance transcript stability and translation.35,36 Having established a role for p38-dependent MK2 activation in the TNF-α overproduction phenotype of FA macrophages,17 we tested the hypothesis that MK2 also controls R848-induced IL-1β overproduction in these cells. In T-shNT and T-shFC cells, siRNA targeting MK2 suppressed MK2 expression by 64% and 76%, respectively (Figure 3A), and suppressed R848-induced TNF-α production by ∼50% (Figure 3B). However, knockdown of MK2 did not suppress IL-1β production (Figure 3C). Using a gain-of-function approach, we confirmed that MK2 plays a minor role in the IL-1β phenotype. Specifically, we expressed in THP-1 cells MK2-CA, containing only the catalytic domain (Figure 4A).30 As expected, THP-1 cells transduced with an MK2-CA expression vector produced ∼3 times more TNF-α in response to R848 than did stimulated THP-1 cells transduced with the empty vector (Figure 4B). However, expression of MK2-CA had no effect on R848-induced IL-1β expression in THP-1 cells (Figure 4C).
siRNA knockdown of MK2 suppresses TNF-α production but has only a modest effect on IL-1β production. T-shNT and T-shFC cells were double-transfected with a pool of siRNAs directed against MAPKAPK-2 (siMK2) or a pool of nontargeted siRNAs using Nucleofector II and cultured for 72 hours. (A) Whole cell extracts were then prepared and subjected to western blot analysis of MK2. Band densitometry was performed using ImageJ software. (B-C) After the 72-hour culture period, cells were plated at a concentration of 106 per mL and then stimulated with R848 (30 μM) for 24 hours. Secreted IL-1β (B) and TNF-α (C) were measured in the conditioned media by ELISA. Two experiments were carried out. One representative experiment with 3 biological replicates for each condition is shown. P values were calculated using a paired Student t test.
siRNA knockdown of MK2 suppresses TNF-α production but has only a modest effect on IL-1β production. T-shNT and T-shFC cells were double-transfected with a pool of siRNAs directed against MAPKAPK-2 (siMK2) or a pool of nontargeted siRNAs using Nucleofector II and cultured for 72 hours. (A) Whole cell extracts were then prepared and subjected to western blot analysis of MK2. Band densitometry was performed using ImageJ software. (B-C) After the 72-hour culture period, cells were plated at a concentration of 106 per mL and then stimulated with R848 (30 μM) for 24 hours. Secreted IL-1β (B) and TNF-α (C) were measured in the conditioned media by ELISA. Two experiments were carried out. One representative experiment with 3 biological replicates for each condition is shown. P values were calculated using a paired Student t test.
Ectopic expression of an activating mutant form of MK2 enhances TNF-α production but has no effect on IL-1β production. THP-1 cells were retrovirally transduced with either an expression vector containing complementary DNA coding for the catalytic domain of MK2 (MK2-CA; aa 49-338) or with the empty vector. Transduced cells were then selected using G418. (A) Whole cell extracts were prepared and subjected to western blot analysis to confirm expression of MK2 and MK2-CA. NS indicates a nonspecific band. (B-C) Cells were plated at a concentration of 106 per mL before stimulation with R848 (30 μM) for 24 hours. Secreted TNF-α (B) and IL-1β (C) were measured in the conditioned media by ELISA. Two experiments were carried out, and each consisted of 3 biological replicates for each condition. One representative experiment is shown.
Ectopic expression of an activating mutant form of MK2 enhances TNF-α production but has no effect on IL-1β production. THP-1 cells were retrovirally transduced with either an expression vector containing complementary DNA coding for the catalytic domain of MK2 (MK2-CA; aa 49-338) or with the empty vector. Transduced cells were then selected using G418. (A) Whole cell extracts were prepared and subjected to western blot analysis to confirm expression of MK2 and MK2-CA. NS indicates a nonspecific band. (B-C) Cells were plated at a concentration of 106 per mL before stimulation with R848 (30 μM) for 24 hours. Secreted TNF-α (B) and IL-1β (C) were measured in the conditioned media by ELISA. Two experiments were carried out, and each consisted of 3 biological replicates for each condition. One representative experiment is shown.
TLR-induced IL-1β gene transcription is controlled by a p38-dependent pathway
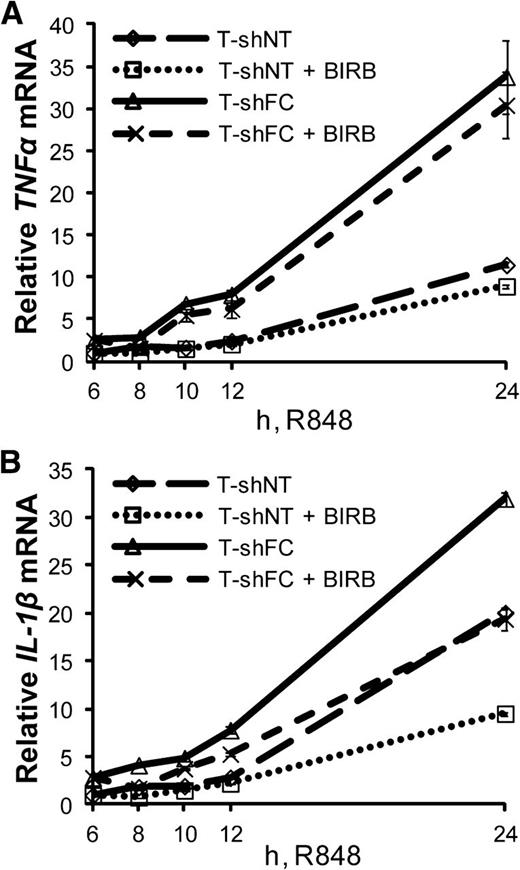
Seeking to identify a transcriptional control point of p38-dependent IL-1β gene expression, we asked whether inhibition of p38 suppresses expression of IL-1β mRNA in TLR-activated FANCC-deficient cells. We measured IL-1β mRNA expression from R848-treated T-shNT and T-shFC cells, with and without exposure to BIRB 796. As expected,17 BIRB 796 had little effect on R848-induced expression of TNF-α mRNA (Figure 5A). However, the inhibitor did suppress IL-1β mRNA expression by ∼40% after 24 hours (Figure 5B). p38 MAPK inhibition did not suppress the half-life of IL-1β transcripts (supplemental Figure 3A), suggesting that p38 controls IL-1β expression in TLR-activated cells at the point of transcription, rather than by enhancing mRNA stability or translation.
TLR-induced IL-1β mRNA expression is controlled by a p38-dependent pathway. T-shNT and T-shFC cells were treated with R848 (30 μM), and 6 hours later, BIRB (50 nM) was added to the culture medium. Total RNA was isolated at 6, 8, 10, 12, and 24 hours after R848 treatment. TNF-α (A) and IL-1β (B) mRNA were measured using real-time qRT-PCR and normalized to levels of 18S rRNA. Shown are the changes in expression of TNF-α (A) and IL-1β (B) mRNA with respect to the expression levels 6 hours after R848 was added, normalized to T-shNT in the absence of BIRB. Two experiments were carried out using 3 technical replicates for each condition and yielded similar results. One representative experiment is shown.
TLR-induced IL-1β mRNA expression is controlled by a p38-dependent pathway. T-shNT and T-shFC cells were treated with R848 (30 μM), and 6 hours later, BIRB (50 nM) was added to the culture medium. Total RNA was isolated at 6, 8, 10, 12, and 24 hours after R848 treatment. TNF-α (A) and IL-1β (B) mRNA were measured using real-time qRT-PCR and normalized to levels of 18S rRNA. Shown are the changes in expression of TNF-α (A) and IL-1β (B) mRNA with respect to the expression levels 6 hours after R848 was added, normalized to T-shNT in the absence of BIRB. Two experiments were carried out using 3 technical replicates for each condition and yielded similar results. One representative experiment is shown.
TLR-induced production of IL-1β and TNF-α is not interdependent
Because TLR-stimulated FANCA- and FANCC-deficient mononuclear phagocytes overproduce both IL-1β and TNF-α, we asked whether production of either cytokine might be dependent on secretion and paracrine signaling by the other. By measuring IL-1β production from R848-treated T-shNT and T-shFC cells incubated with an anti-TNF-α neutralizing antibody or an isotype control antibody, we determined that R848-induced IL-1β production was not significantly affected by neutralization of secreted TNF-α (Figure 6A). Likewise, addition of an anti-IL-1β neutralizing antibody had no effect on TNF-α production from R848-treated T-shNT and T-shFC cells (Figure 6B). Neither recombinant TNF-α nor IL-1β alone was sufficient to induce production of the other cytokine (supplemental Figure 4A-B). In addition, although exogenous recombinant TNF-α enhanced R848-induced IL-1β production from T-shNT and T-shFC cells (supplemental Figure 4A), treatment with recombinant IL-1β had no effect on R848-induced TNF-α production (supplemental Figure 4B).
TLR-induced production of IL-1β and TNF-α is not interdependent. T-shNT and T-shFC cells were plated at a concentration of 106 per mL. Isotype control antibodies or neutralizing antibodies against TNF-α (A) or IL-1β (B) were added to the culture medium at a concentration of 1 µg/mL, and cells were stimulated with R848 (30 μM) for 24 hours. Secreted IL-1β (A) or TNF-α (B) was measured in the conditioned media by ELISA. P values were calculated using a paired Student t test. Two experiments used 3 biological replicates for each condition, and both showed similar results. One experiment is shown.
TLR-induced production of IL-1β and TNF-α is not interdependent. T-shNT and T-shFC cells were plated at a concentration of 106 per mL. Isotype control antibodies or neutralizing antibodies against TNF-α (A) or IL-1β (B) were added to the culture medium at a concentration of 1 µg/mL, and cells were stimulated with R848 (30 μM) for 24 hours. Secreted IL-1β (A) or TNF-α (B) was measured in the conditioned media by ELISA. P values were calculated using a paired Student t test. Two experiments used 3 biological replicates for each condition, and both showed similar results. One experiment is shown.
IL-1β suppresses self-replication of Fancc-deficient LT-HSCs in vitro
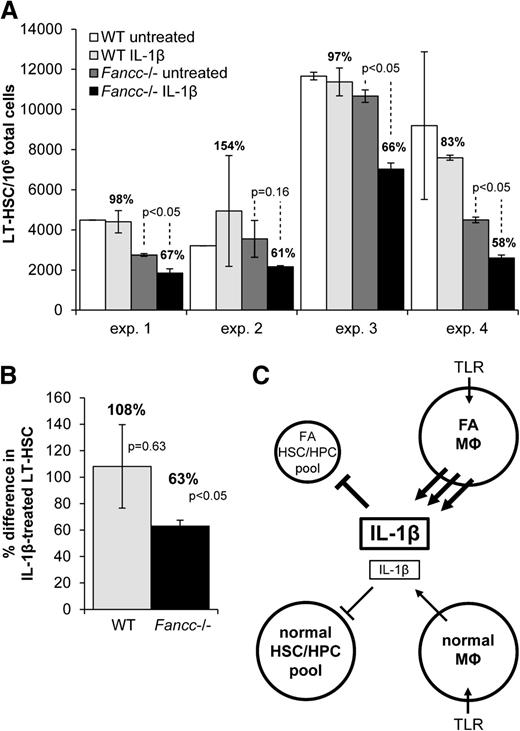
To test the notion that Fancc-deficient LT-HSCs might be uniquely vulnerable to IL-1β, we cultured KLS (c-Kit+Lineage–Sca-1+) cells and quantified the fraction that was CD34–Flt3–29 on days 0 and 7. KLS cells were obtained from wild-type and Fancc−/− mice and were cultured for 7 days in the presence of multilineage growth factors (IL-6, IL-11, SCF, and FLT3L), with or without IL-1β. IL-1β did not affect the expansion of wild-type LT-HSCs but significantly suppressed expansion of Fancc−/− LT-HSCs (Figure 7). When day 7 KLS cells were plated in methylcellulose cultures with SCF, Epo, and IL-3, colony formation of Fancc−/− KLS cells was equivalent to clonal growth of wild-type KLS cells, indicating that the remaining KLS cells were fully capable of undergoing differentiation. The observations suggest that the major suppressive effect of IL-1β involved suppression of self-replication of LT-HSCs.
Fancc-deficient hematopoietic progenitors are hypersensitive to IL-1β. (A) Wild-type or Fancc−/− KLS cells were isolated and, based on percent of the KLS cells also bearing the CD34–Flt3– signature (LT-HSCs), were plated at 1000 LT-HSCs per well in 500 µL RPMI/10% FBS containing IL-6, IL-11, SCF, and FLT3L. Cells were treated with murine IL-1β (30 pg/mL) or medium alone for 7 days. Total cells and LT-HSCs were then counted using flow cytometry. Shown is the mean number of LT-HSCs per 106 total cells in each set of duplicate wells after 7 days. The percent difference in the LT-HSC population in IL-1β–treated cells compared with untreated cells is indicated above each bar that represents the number of LT-HSCs observed after IL-1β treatment. The results of 4 independent experiments are shown. (B) Data from these 4 experiments were pooled and expressed as the percent difference in the LT-HSC population in IL-1β-treated cells compared with untreated cells. (C) A model of the proposed IL-1β–mediated autoinhibitory loop is shown. The FA mononuclear phagocyte (MΦ) pool overproduces IL-1β in response to TLR stimulation, and the FA hematopoietic stem and progenitor cell (HSC/HPC) pool fails to sustain self-replicative activity and therefore contracts as a result of intrinsic IL-1β hypersensitivity. TLR-stimulated normal mononuclear phagocytes produce normal levels of IL-1β, and the normal HSC/HPCs are more tolerant of IL-1β.
Fancc-deficient hematopoietic progenitors are hypersensitive to IL-1β. (A) Wild-type or Fancc−/− KLS cells were isolated and, based on percent of the KLS cells also bearing the CD34–Flt3– signature (LT-HSCs), were plated at 1000 LT-HSCs per well in 500 µL RPMI/10% FBS containing IL-6, IL-11, SCF, and FLT3L. Cells were treated with murine IL-1β (30 pg/mL) or medium alone for 7 days. Total cells and LT-HSCs were then counted using flow cytometry. Shown is the mean number of LT-HSCs per 106 total cells in each set of duplicate wells after 7 days. The percent difference in the LT-HSC population in IL-1β–treated cells compared with untreated cells is indicated above each bar that represents the number of LT-HSCs observed after IL-1β treatment. The results of 4 independent experiments are shown. (B) Data from these 4 experiments were pooled and expressed as the percent difference in the LT-HSC population in IL-1β-treated cells compared with untreated cells. (C) A model of the proposed IL-1β–mediated autoinhibitory loop is shown. The FA mononuclear phagocyte (MΦ) pool overproduces IL-1β in response to TLR stimulation, and the FA hematopoietic stem and progenitor cell (HSC/HPC) pool fails to sustain self-replicative activity and therefore contracts as a result of intrinsic IL-1β hypersensitivity. TLR-stimulated normal mononuclear phagocytes produce normal levels of IL-1β, and the normal HSC/HPCs are more tolerant of IL-1β.
Discussion
The molecular pathogenesis of BM failure in FA remains enigmatic. Because cross-linking agent hypersensitivity is the sine qua non of this disease, it has been widely presumed that the loss of stem cells over time simply reflects genotoxic cell death as a result of exposure to either environmental or endogenous cross-linking agents.9,10 However, FA HSCs are also hypersensitive to endogenous cytokines.11-15 Moreover, FANCA- and FANCC-deficient macrophages overproduce 1 of these proinflammatory cytokines, TNF-α,16,17 a factor to which FA marrow progenitors are uniquely hypersensitive.11,22,37,38 How these FA proteins modulate TLR pathway activation is unknown, but it is clear that p38 MAPK is an essential mediator of the TLR pathway hyperactivation state.17 Because prolonged exposure of stem cells to TNF-α ex vivo results in the selection of TNF-α–resistant neoplastic clones,22 this cytokine may play a role not only in marrow failure but also in clonal evolution.21,22
Given that TLR activation in macrophages leads to the secretion of several other factors, including IL-1β,23-25 a major mediator of inflammation known to promote carcinogenesis,26,27,39 we reasoned that IL-1β might also be overproduced by FANCA- and FANCC-deficient macrophages and may also negatively influence survival of Fancc-deficient multipotent BM progenitor cells. Here we show TLR7/8 induced IL-1β overproduction in FANCA- and FANCC-deficient cell lines (Figure 1A), primary Fancc−/− BMDM (Figure 1D), and CD14+ cells from an FA complementation group A patient (Figure 1F).
IL-1β production in monocytes is known to be controlled by the inflammasome, a multimeric complex that can be activated by a wide range of stress signals, including the TLR7/8 ligand R848.24,31,32 Upon inflammasome activation, caspase-1 is cleaved to its active form, which in turn cleaves the pro-IL-1β polypeptide, allowing for secretion of mature IL-1β.31 The involvement of the canonical inflammasome in the IL-1β overproduction phenotype was confirmed by the results of our studies using glyburide, a known inhibitor of the NLRP3 inflammasome.33 That is, glyburide suppressed IL-1β overproduction from both T-shFA and T-shFC cells (Figure 1A), Fancc−/− BMDM (Figure 1D), and FANCA-deficient patient CD14+ cells (supplemental Figure 2). However, there were no quantitative differences in cleaved caspase-1 between FANCC-deficient and control cells, indicating that the control point for the FA proteins was likely upstream of posttranslational IL-1β processing (Figure 2C). Glyburide also unexpectedly40 suppressed TLR-induced TNF-α production from wild-type and Fancc−/− BMDM (Figure 1D). The mechanism for this result is unknown, but because this effect was only noted in murine BMDM and not in human cells, it may reflect a species-specific enhancement of TNF-α production by autocrine IL-1 receptor activation or an off-target effect of glyburide. Although it has been shown that glyburide can reduce TNF-α expression in monocytes after LPS treatment of whole blood, this effect was not observed when isolated monocytes were treated.40
We have previously shown that TNF-α production is inhibited by the p38 MAPK inhibitor BIRB 796.17 IL-1β expression is also known to be controlled by a p38 MAPK-dependent pathway in macrophages,28 and we confirmed that inhibition of p38 suppressed TLR-induced IL-1β overproduction by T-shFC cells and by FA patient CD14+ cells (Figures 1F and 2A). The p38 inhibition did not, however, suppress inflammasome activity (Figure 2C). Taken together, these results suggest that FANCC, FANCA, and p38 MAPK do not modulate TLR-induced IL-1β production at the level of the inflammasome but do influence IL-1β production at a more proximal control point.
In the presence of TLR agonists, activation of the p38 substrate MK2 is required to induce production of the cytokines interferon γ, macrophage-inflammatory protein 1α, and TNF-α in selected cell types.17,35,41,42 MK2 influences TNF-α production posttranscriptionally by enhancing stability and translation of TNF-α mRNA.35,36 Specifically, activated MK2 phosphorylates and inactivates the 3′ AU-rich element (AURE)-binding protein tristetraprolin, a translational suppressor,35 thus promoting the translation of AURE-containing mRNAs. Given that the IL-1β mRNA also contains a 3′ AURE,34 and that MK2 was hyperphosphorylated in TLR-stimulated T-shFC cells (Figure 2C and Anur et al17 ), we asked whether MK2 hyperactivation in FANCC-deficient cells induces the overproduction of IL-1β in a similar fashion. As expected,17 we found that MK2-targeted siRNA suppressed TNF-α production, but we found no suppression of IL-1β production (Figure 3B-C). Likewise, ectopic expression of a constitutively active MK2 mutant in THP-1 cells enhanced TLR-stimulated TNF-α production but had no effect on IL-1β production (Figure 4B-C). Therefore, both gain- and loss-of-function studies confirmed that IL-1β production is controlled by a p38-dependent, but largely MK2-independent pathway. The point of IL-1β control is clearly transcriptional because p38 inhibition diminished TLR-induced IL-1β mRNA abundance (Figure 5B) without suppressing the half-life of IL-1β transcripts (supplemental Figure 3A). That inhibition of p38 had little effect on TNF-α mRNA (Figure 5A and supplemental Figure 3B) indicates that although control of p38 activation is a point of control for the FA proteins studied, the influence of p38 on expression of TNF-α and IL-1β is mechanistically distinct.
The transcription factor(s) that enhance IL-1β gene expression in response to p38 have not yet been identified. It has been suggested that the p38 substrate C/EBPβ may act as a transcriptional activator for IL-1β in macrophages,28,43,44 but we have not observed increased C/EBPβ phosphorylation in response to TLR activation in THP-1 cells (data not shown). β-catenin has also been implicated in the transcriptional control of IL-1β mRNA,45 and p38 has been shown to contribute to increased β-catenin activity in thymocytes through direct phosphorylation and inactivation of GSK3β, the inhibitor of β-catenin.46 However, in experiments in which we inactivated GSK3β pharmacologically using BIO (0.2-2 μM), we did not observe an increase in IL-1β expression (data not shown).
IL-1β mRNA overexpression has been described in unstimulated FA patient peripheral blood mononuclear cells, T lymphocytes, and FA lymphoblast cell lines,45 but the mechanisms involved are unclear. Likewise, although increased levels of IL-1β exist in the serum of many FA patients,45 the cellular source of the protein has not been defined. Here we show that mononuclear phagocytes, a major in vivo source of IL-1β,39 overproduce both TNF-α17 and IL-1β in patients with FA because the TLR-dependent p38 MAPK signaling pathway is aberrantly controlled. We suspect that IL-1β contributes to FA pathophysiology (Figure 7C) because exposure of multipotental progenitor cells from Fancc-deficient mice to IL-1β ex vivo resulted in exaggerated suppression of LT-HSC (c-Kit+Lineage–Sca-1+CD34–Flt3–) expansion (Figure 7A-B). The involvement of multiple inflammatory cytokines in stem cell suppression may also create selective pressure for the emergence of cytokine-resistant preneoplastic cells,47 as has been shown to be the case with TNF-α.22
The mechanism by which FANCA and FANCC modulate TLR-agonist–induced p38 MAPK activation remains unclear, but it is apparent that one of its substrates, MK2, is consistently hyperphosphorylated in TLR-activated FA macrophages. We have carefully sought evidence that FANCC and FANCA interact with regulatory elements of the TLR pathway but have found that neither FANCA nor FANCC interacts with A20, TAK1, TRAF6, ASK1, RIP1, HSP90, MKK3, MKK6, p38, or MK2 in co-immunoprecipitation experiments (not shown). We have previously shown that FANCC interacts with HSP70,11,37 and recent evidence suggests that HSP70 modulates p38 activity in fibroblasts.48 However, we have not observed a p38/HSP70 interaction in THP-1 cells using co-immunoprecipitation methods (data not shown). We have, however, demonstrated TLR hypersensitivity in FA monocytes resulting in IL-1β and TNF-α overproduction.
That each cytokine has the potential, independently, of suppressing FA stem cell function suggests that targeting a single factor alone will not be sufficient to enhance hematopoietic activity. Indeed, in a preliminary clinical trial, treatment of 6 FA patients with the TNF-α inhibitor etanercept did not ameliorate pancytopenia in subjects with FA.49 In considering preclinical strategies designed to enhance BM function in FA, we must now take into account that overproduction of both of these inhibitory cytokines, and likely others, is controlled by a state of p38 MAPK hyperactivity. Therefore, we suggest that p38 MAPK represents a more tractable therapeutic target than does either of these cytokines alone. Potential toxicities need to be considered as well. For example, cytoplasmic p38/MK2 signaling is known to be activated in response to cytotoxic chemotherapeutic agents and UV radiation and plays a role in checkpoint maintenance.50-52 Although the pathway is reportedly most essential in cells with defective p53 function,53 suppression of a checkpoint response in FA stem cells could conceivably enhance stem cell death or permit the undesirable survival of cells with higher levels of genomic damage. Therefore, the evaluation of such compounds as potential therapeutic agents must include careful assessment of potentially adverse genomic consequences in the FA stem cell pool.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Peter Kurre for referring patients for participation in this study.
This work was supported by the National Institutes of Health National Heart, Lung, and Blood Institute (P01 HL048546) (G.C.B.) (T32 HL 7781-19) (M.R.G. and G.C.B.), the Department of Veterans Affairs (Merit Review) (G.C.B.), the Fanconi Anemia Research Fund (G.C.B.), and the National Institutes of Health National Cancer Institute (1 R01 CA138237) (G.C.B.).
Authorship
Contribution: M.R.G. and G.C.B. designed the project, developed the assays, performed the experiments, analyzed the primary data, and wrote the manuscript; L.E.H. developed the murine BM macrophage experiments, maintained Fancc-deficient mice, and proofread and revised the manuscript; W.K. maintained all cell lines and conducted gene transduction experiments; and R.K.R. and J.E.Y. performed KLS cell isolation, flow cytometry, survival assays, and colony assays and provided scientific advice and proofread and revised the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Grover C. Bagby Jr, Knight Cancer Institute at Oregon Health & Science University, CR145, HRC 14D40, 3181 SW Sam Jackson Park Rd, Portland, OR 97239; e-mail: grover@ohsu.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal