Key Points
Paroxysmal nocturnal hemoglobinuria identifies a role for GPI-linked proteins in the homeostasis of human NK cell subsets.
GPI-deficient NK cells exhibit impaired chemotactic responses.
Abstract
In paroxysmal nocturnal hemoglobinuria (PNH), hematopoietic cells lacking glycosylphosphatidylinositol (GPI)-linked proteins on their surface (GPIneg) exist alongside normal (GPI+) cells. Analysis of natural killer (NK) cell subsets in 47 PNH patients revealed that the ratio of CD56bright:CD56dim NK cells differed in the GPI+ and GPIneg populations, with GPInegCD56bright NK cells significantly more abundant in peripheral blood than their normal GPI+ counterparts. Indeed, GPI+CD56bright NK cells were not detected in the peripheral blood of some patients, suggesting their trafficking to a niche unavailable to the GPInegCD56bright NK cell population. Defective cellular trafficking in this disease was supported by findings showing differential chemokine receptor expression between GPI+ and GPIneg NK cells and impaired stromal cell–derived factor 1 (SDF-1)–induced chemotaxis of GPIneg NK cells. Our results indicate a role for GPI-linked proteins in NK cell subset homeostasis and suggest that differential chemokine responses might contribute to the balance of GPI+ and GPIneg populations in this disease.
Introduction
The glycosylphosphatidylinositol (GPI) anchor secures numerous proteins to the cell surface.1 In paroxysmal nocturnal hemoglobinuria (PNH), there is expansion of hematopoietic stem cells (HSCs) harboring a somatic mutation in the X-linked PIGA gene, disrupting a key step in GPI anchor biosynthesis.2,3 These mutant HSCs generate substantial populations of mature blood cells lacking GPI-linked proteins; red blood cells and platelets lacking the GPI-linked complement protectors CD55 and CD59 are highly susceptible to complement-mediated lysis, resulting in many of the clinical features of PNH.2,3 Accordingly, inhibiting the formation of the complement membrane attack complex using a humanized anti-C5 antibody (eculizumab) is an effective therapy.2,3
Human natural killer (NK) cells are classified into 2 main subsets.4 The weakly cytotoxic CD56bright cells constitute 10% of peripheral blood NK cells and are the likely precursors of the highly cytotoxic CD56dim cells, which constitute the remaining 90% of peripheral NK cells in the blood. However, CD56bright NK cells express lymph node homing molecules, and the ratio of CD56dim:CD56bright cells is reversed in secondary lymphoid tissue.5,6 We analyzed NK cells from 47 PNH patients and identified a role for GPI-linked molecules in NK cell chemotactic responses and the homeostasis of NK cell subsets.
Study design
Peripheral blood samples were collected, after informed consent in accordance with the Declaration of Helsinki, from 47 PNH patients (32 receiving eculizumab; supplemental Figure 1, available on the Blood Web site) attending the PNH National Service clinic in Leeds, United Kingdom. The study was approved by the Leeds Teaching Hospitals National Health Service Trust ethical review board. Cells were stained with antibodies defining NK cells (CD56+CD3neg) and GPI-linked molecules (CD48, CD59, CD160; see supplemental Methods). Discrimination of GPI+ and GPIneg NK cells was also performed using fluorescent aerolysin, a modified bacterial toxin that binds to intact GPI anchors.7 For migration assays, NK cells (purified by indirect selection) were applied to 5-μM membrane transwells (Corning) and allowed to migrate in response to 100 nM stromal cell–derived factor 1 (SDF-1; R&D Systems). The GPI+:GPIneg NK cell ratio was analyzed in the input and migrated populations by flow cytometry. Further details are provided in supplemental Methods.
Results and discussion
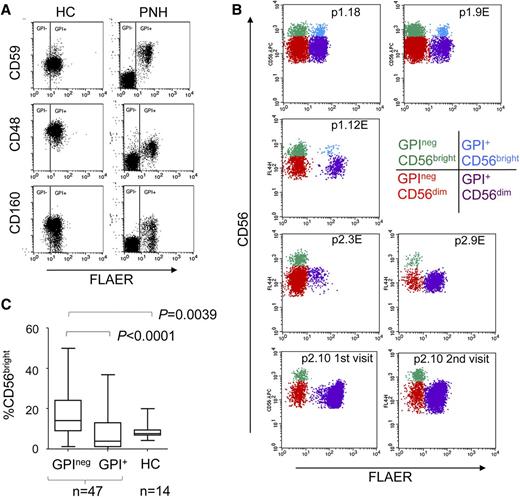
The GPIneg NK cells in PNH patients lacked cell surface expression of the GPI-linked molecules CD48, CD59, and CD160 (Figure 1A). GPI+ and GPIneg populations of CD56dim and CD56bright NK cell subsets were detectable in the majority of patients (Figure 1B; supplemental Figure 1). However, in 10 PNH patients, the GPI+CD56bright NK cells made up less than 0.02% of the total GPI+ cells (Figure 1B; supplemental Figure 1). This was not associated with eculizumab treatment, and 4 patients analyzed on multiple occasions revealed this to be a stable phenotype (Figure 1B; supplemental Figures 1-2). CD56bright NK cells differentiate into CD56dim NK cells,9-11 and we reasoned that GPI+CD56bright NK cells must be present in these individuals to generate the GPI+CD56dim population. Furthermore, GPI+CD56bright NK cells have the normal, not the GPI-deficient, phenotype. We speculated that the GPI+CD56bright NK cells were present in all PNH patients but that GPI+CD56bright and GPInegCD56bright NK cells were differentially distributed between the peripheral blood and other tissues. Indeed, CD56bright NK cells have lymph node homing properties, and inflammation results in their loss from the periphery.12,13 We suggest that the GPInegCD56bright NK cells are outcompeted by the normal GPI+CD56bright NK cells for entrance to (or occupancy of) a niche such as the secondary lymphoid tissue. Such competition would reduce the occupancy of this niche by GPInegCD56bright NK cells, resulting in their enrichment in peripheral blood. Accordingly, analysis of the entire PNH cohort revealed that GPInegCD56bright NK cells were significantly more abundant in peripheral blood compared with their GPI+ counterparts and with the CD56bright NK cells from healthy donors (Figure 1C).
Altered NK cell subset homeostasis in PNH. (A) Cell surface expression of the GPI-linked proteins CD48, CD59, and CD160 on the NK cells (gated on CD56+CD3neg cells) of a healthy control (HC) and a PNH patient. Staining with fluorescent aerolysin (FLAER) demarcates GPI+ from GPIneg cells. (B) CD56bright and CD56dim NK cell subsets present in the peripheral blood of PNH patients (plots gated on CD56+CD3neg cells). The top 2 rows show 3 patients with all 4 subsets of NK cells (denoted p1.number), and the bottom 2 rows show 3 patients in which the GPI+CD56bright NK cells made up less than 0.02% of the total GPI+ NK cells (denoted p2.number). The 4 subsets of NK cells are indicated in different colors, according to the key. NK cells from p2.10 were analyzed on 3 occasions; the first 2 visits are shown here, and the third visit, together with sequential analysis of 3 other patients, is shown in supplemental Figure 2. Across the cohort of 47 patients, the size of the GPIneg NK cell population varied from ∼1% to ∼99% of the total NK cells (supplemental Figure 1), in agreement with previous findings.8 An “E” after the patient number indicates the patient was receiving eculizumab at the time of analysis. (C) Frequency of CD56bright NK cells as a percentage of total GPIneg or total GPI+ NK cells compared with healthy controls. P values (calculated using the Mann-Whitney nonparametric test) are shown for statistically different data.
Altered NK cell subset homeostasis in PNH. (A) Cell surface expression of the GPI-linked proteins CD48, CD59, and CD160 on the NK cells (gated on CD56+CD3neg cells) of a healthy control (HC) and a PNH patient. Staining with fluorescent aerolysin (FLAER) demarcates GPI+ from GPIneg cells. (B) CD56bright and CD56dim NK cell subsets present in the peripheral blood of PNH patients (plots gated on CD56+CD3neg cells). The top 2 rows show 3 patients with all 4 subsets of NK cells (denoted p1.number), and the bottom 2 rows show 3 patients in which the GPI+CD56bright NK cells made up less than 0.02% of the total GPI+ NK cells (denoted p2.number). The 4 subsets of NK cells are indicated in different colors, according to the key. NK cells from p2.10 were analyzed on 3 occasions; the first 2 visits are shown here, and the third visit, together with sequential analysis of 3 other patients, is shown in supplemental Figure 2. Across the cohort of 47 patients, the size of the GPIneg NK cell population varied from ∼1% to ∼99% of the total NK cells (supplemental Figure 1), in agreement with previous findings.8 An “E” after the patient number indicates the patient was receiving eculizumab at the time of analysis. (C) Frequency of CD56bright NK cells as a percentage of total GPIneg or total GPI+ NK cells compared with healthy controls. P values (calculated using the Mann-Whitney nonparametric test) are shown for statistically different data.
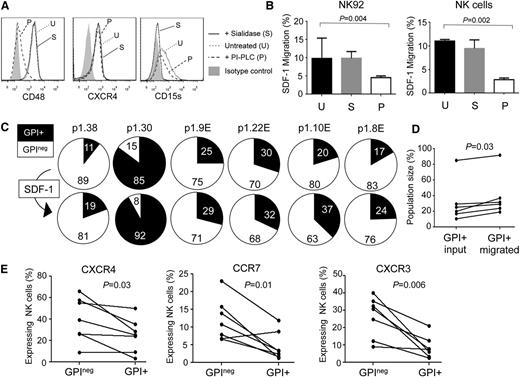
The importance of chemokines in leukocyte trafficking suggests that impaired chemotactic responses might underlie altered NK cell subset distribution in PNH. The expression of chemokine receptors by NK cells is complex,14,15 and it is difficult to perform chemotaxis assays on the small numbers of CD56bright cells available from these patients. We therefore analyzed the responses of GPI+ and GPIneg NK cell populations to SDF-1/CXCL12, the CXCR4 ligand. CXCR4 is expressed by the majority of peripheral blood NK cells, aiding these studies.14,15 Treatment of NK cells with phosphatidylinositol-specific phospholipase C (PI-PLC) removed GPI anchor–linked proteins but left CXCR4 expression intact. These PI-PLC treated NK cells exhibited significantly reduced migration in response to SDF-1, whereas sialidase treatment had no effect (Figure 2A-B). This suggested that GPI-deficient NK cells have impaired chemotactic responses. We repeated the SDF-1 migration assays using NK cells from 6 PNH patients in which all 4 GPI+/GPIneg and CD56bright/CD56dim subsets were present in peripheral blood. We analyzed the ratio of GPI+:GPIneg NK cells in the starting population and in the migrating cells and found that the SDF-1–induced migration significantly enriched the GPI+ NK cells (Figure 2C-D).
Defective chemotaxis in GPInegNK cells. (A) Cell surface expression of CD48, CXCR4, and CD15s on NK92 cells, with and without enzyme treatment. PI-PLC hydrolyses GPI anchors (removing CD48 but preserving CXCR4 and CD15s), whereas sialidase treatment leaves CD48 and CXCR4 expression intact but removes sialic acid and destroys the CD15s antigen. (B) Migration of enzyme-treated NK92 cells (left) or primary NK cells (right) to SDF-1. Cells were treated with either sialidase (S) or PI-PLC (P) or were left untreated (U). The data were analyzed using the Mann-Whitney nonparametric test. SDF-1 induced migration by a factor of 4 over media lacking the chemokine (data not shown). (C) Migration of NK cells from 6 PNH patients in response to SDF-1. Pie charts show the percentage of GPI+ (black slice) and GPIneg (white slice) NK cells applied to the assay (top; input population) and those that migrated in response to the chemokine (bottom; migrated population). Patient p1.38 was an addition to the 47 patients in the cohort shown. (D) Analysis of migration of NK cells from the 6 PNH patients according to the size of the GPI+ population in the input and migrated fractions. The P value was calculated using the Wilcoxon matched-pairs signed rank test. (E) CXCR4, CCR7, and CXCR3 expression on the GPI+ and GPIneg NK cells of 7 PNH patients. The lines link the NK cells from a particular patient. The data show the percentage of NK cells expressing these chemokine receptors (determined by flow cytometry) as a fraction of the total GPI+ or GPIneg NK cell population. Analysis of the expression of these molecules on the CD56dim and CD56bright NK cell subsets is shown in supplemental Figure 3. P values were calculated using Wilcoxon matched-pairs signed rank test.
Defective chemotaxis in GPInegNK cells. (A) Cell surface expression of CD48, CXCR4, and CD15s on NK92 cells, with and without enzyme treatment. PI-PLC hydrolyses GPI anchors (removing CD48 but preserving CXCR4 and CD15s), whereas sialidase treatment leaves CD48 and CXCR4 expression intact but removes sialic acid and destroys the CD15s antigen. (B) Migration of enzyme-treated NK92 cells (left) or primary NK cells (right) to SDF-1. Cells were treated with either sialidase (S) or PI-PLC (P) or were left untreated (U). The data were analyzed using the Mann-Whitney nonparametric test. SDF-1 induced migration by a factor of 4 over media lacking the chemokine (data not shown). (C) Migration of NK cells from 6 PNH patients in response to SDF-1. Pie charts show the percentage of GPI+ (black slice) and GPIneg (white slice) NK cells applied to the assay (top; input population) and those that migrated in response to the chemokine (bottom; migrated population). Patient p1.38 was an addition to the 47 patients in the cohort shown. (D) Analysis of migration of NK cells from the 6 PNH patients according to the size of the GPI+ population in the input and migrated fractions. The P value was calculated using the Wilcoxon matched-pairs signed rank test. (E) CXCR4, CCR7, and CXCR3 expression on the GPI+ and GPIneg NK cells of 7 PNH patients. The lines link the NK cells from a particular patient. The data show the percentage of NK cells expressing these chemokine receptors (determined by flow cytometry) as a fraction of the total GPI+ or GPIneg NK cell population. Analysis of the expression of these molecules on the CD56dim and CD56bright NK cell subsets is shown in supplemental Figure 3. P values were calculated using Wilcoxon matched-pairs signed rank test.
These results reveal that optimal chemotactic responses require GPI-linked molecules: GPI-linked proteins are abundant in lipid rafts, PIGA mutations disrupt raft formation, and CXCR4 requires incorporation into these structures for optimal activity.16-18 In GPIneg NK cells, SDF-1 responses may be impaired because of the redistribution of CXCR4 to membrane microdomains less able to support receptor signaling or into a local membrane environment that alters CXCR4 conformation.17 Impaired SDF-1 responses were not caused by defective CXCR4 expression, as GPIneg NK cells expressed significantly higher levels of cell surface CXCR4 compared with their GPI+ counterparts (Figure 2E; supplemental Figure 3). Increased chemokine receptor expression has previously been reported in GPI-deficient cells,19 and we also found elevated expression of CXCR3 and CCR7 on GPIneg NK cells (Figure 2E; supplemental Figure 3). Indeed, CCR7 is preferentially expressed by CD56bright NK cells, and this receptor exhibited greater expression on the GPInegCD56bright NK cells (supplemental Figure 3). Elevated receptor expression may reflect in vivo selection, whereby GPIneg NK cells respond to chemokines only if they express high levels of the receptor, favoring their survival. Alternatively, defective chemokine receptor activity may prevent activation-induced internalization in GPIneg NK cells, increasing cell surface expression.20
Why the GPI+CD56bright NK cells were undetectable in the blood of just a fifth of PNH patients is unclear. However, NK subset homeostasis is regulated by both environmental (eg, infection/inflammation9-13 ) and genetic factors,21 and separating their respective contributions within the confines of the rare PNH phenotype represents a considerable challenge. Defective SDF-1/CXCR4 activity cannot itself account for the altered subset homeostasis we observed in the patients, but it does highlight defective chemotaxis in PNH. Several models have been proposed to account for the abundance of GPIneg cells in PNH, focusing on the maintenance and expansion of GPIneg HSC.3,22,23 Our results indicate that the balance of GPI+ and GPIneg cells within a particular hematopoietic lineage is governed not only by the abundance of GPIneg HSC but also by in vivo processes operating on mature cells. GPI-linked molecules are known to play a role in cellular trafficking and homeostasis of other hematopoietic cell types.24,25 Furthermore, GPInegCD34+ cells exhibit enhanced SDF-1 chemotaxis but reduced adhesion to SDF-1.23 Our results appear contradictory to these findings, but such differences may be explained by the presence of cell-type–specific factors that regulate chemokine receptor activity.20 Despite their differences, our data, and those of Ratajczak et al,23 reveal that differential chemotactic responses of GPI+ and GPIneg cells result in unequal competition for a particular niche, contributing to the balance and persistence of GPI+ and GPIneg cells in PNH.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful to other members of our laboratory for technical advice and assistance and to Alan Melcher and Pam Jones for encouragement.
Authorship
Contribution: R.J.K., A.H., and P.H. manage the PNH clinic and recruited the patient cohort; Y.M.E.-S. performed the experimental work; Y.M.E.-S., G.M.D., and G.P.C. analyzed the data; G.P.C. and Y.M.E.-S. designed the study; and G.P.C. wrote the article with input from all authors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Graham Cook; Leeds Institute of Cancer and Pathology, University of Leeds, Wellcome Brenner Building, St James's University Hospital, Leeds LS9 7TF, UK; e-mail: g.p.cook@leeds.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal