In this issue of Blood, Mildner et al report elucidation of a transcriptome-wide miRNA map for in vivo mononuclear phagocyte populations. Using this approach, they identify that the miRNA miR-142 was critical for dendritic cell (DC) development, particularly for homeostasic regulation of the CD4+ DC subset.1
Dendritic cells (DCs) are a rare type of white blood cell that play a critical role in bridging the innate and adaptive immune system. They are essential for presentation of both endogenous (self-) antigens and foreign (pathogen-derived) antigens and as such constitute a frontline defense against micro-organisms. To effectively provide immune protection, different DC subsets have evolved. The genetic program that coordinates the development of these subsets is controlled largely by cytokines and transcription factors.2 Interferon regulatory factors (Irf), namely Irf4 and Irf8, are essential for the development of CD4+ and CD8+ DC lineages, respectively, in mice, and PU.1 is a pioneering transcription factor for myeloid cell development (see figure). While the fundamental differentiation program for DCs is laid down by specific transcription factors, increasingly it is recognized that fine-tuning of this program is likely to depend on an added layer of control mediated through microRNA (miRNA) expression.3,4
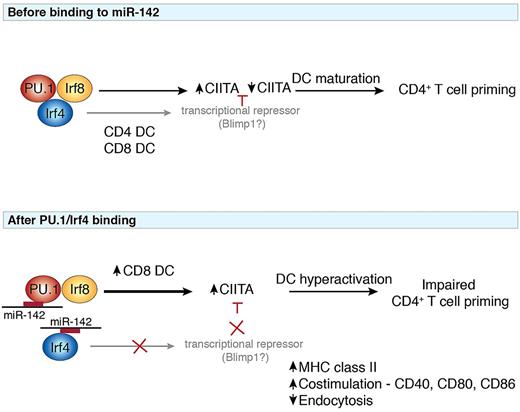
Potential model of the interactions between miR-142 and key transcription factors implicated in the pathway. At steady-state (top panel), the interplay between PU.1, Irf8, and Irf4 orchestrates the differentiation of CD4 and CD8 DCs. Heterodimeric binding of PU.1 and Irf8 induces expression of the MHC class II transactivator CIITA, which may be repressed by transcription factors such as Blimp1 to allow efficient induction of DC antigen presentation required for T-cell priming. Modulation of the PU.1/Irf4 complex by miR-142 (bottom panel) disrupts the generation of CD4 DCs and favors the generation of CD8 DCs. Because Irf4 is important for induction of other transcription factors including Blimp1, failure of CIITA repression leads to elevated DC activation and maturation and impaired CD4+ T-cell priming ability. miR-142 may thus regulate the balance of different transcription factors to guide the outcome of an immune response.
Potential model of the interactions between miR-142 and key transcription factors implicated in the pathway. At steady-state (top panel), the interplay between PU.1, Irf8, and Irf4 orchestrates the differentiation of CD4 and CD8 DCs. Heterodimeric binding of PU.1 and Irf8 induces expression of the MHC class II transactivator CIITA, which may be repressed by transcription factors such as Blimp1 to allow efficient induction of DC antigen presentation required for T-cell priming. Modulation of the PU.1/Irf4 complex by miR-142 (bottom panel) disrupts the generation of CD4 DCs and favors the generation of CD8 DCs. Because Irf4 is important for induction of other transcription factors including Blimp1, failure of CIITA repression leads to elevated DC activation and maturation and impaired CD4+ T-cell priming ability. miR-142 may thus regulate the balance of different transcription factors to guide the outcome of an immune response.
MicroRNAs are a class of small noncoding RNAs that repress translation of target genes through their complementary binding to the 3′ untranslated regions of mRNAs by repressing translation initiation, induction of transcript decapping and deadenylation, and degradation of the mRNA. Canonical miRNAs are initially transcribed as long primary transcripts that are then processed into mature 20-24 nucleotide (nt)–long miRNAs. In mammals, posttranscriptional modulation of gene expression depends on an imperfect matching between the miRNA and target. However, perfect complementarity between the 5′-proximal seed sequence of the miRNA with the mRNA target is thought to be particularly important in metazoans. This feature introduces a level of promiscuity in which each miRNA can bind multiple targets (hundreds to thousands) and reciprocally, each mRNA can be targeted by many different miRNAs. Some understanding of miRNA expression in DCs has been gained through investigation mainly of in vitro GM-CSF–derived DCs. This has resulted in identification of several miRNAs, mainly in human cultured DCs, that target cytokine production (miR-10a, miR-21, miR-142-3p), DC activation (miR-155 and miR146), and antigen presentation (miR-511, miR-99b, miR-211). Understanding miRNA expression and requirements in different in vivo DC subsets, and which components of the differentiation and maturation pathways they target, has been a significant challenge. Deletion of the gene encoding Dicer, a keyenzyme necessary for the biogenesis of miRNAs, specifically in CD11c+ cells, resulted in only a minimal impact on DC differentiation.4 These mice displayed a perturbed Langerhans cell compartment, but conventional DCs were relatively normal. The lack of phenotype observed in the conventional DC compartment may have been due to the fact that miRNAs are very stable. Combined with the quite short half-life of DCs, this is likely to result in insufficient time for the miRNAs to be efficiently depleted in mature cells. Thus, to fully evaluate the involvement of miRNAs in DC approaches to deplete miRNAs from earlier stages of differentiation will be important.
Mildner et al have isolated in vivo mononuclear cells including myeloid progenitors, monocytes, and splenic DCs to identify that each population displayed a distinct miRNA profile.1 This suggested that a signature miRNA expression for each subset might be important in establishing or maintaining the identity of each population. Focusing on miRNAs that affected DC development, Mildner et al went on to demonstrate that miR-142, originally identified to inhibit IL-6,5 was expressed at low levels in all DC subsets but differentially affected CD4+ DCs. Furthermore, this finding could be reproduced in vitro, providing an opportunity to probe the molecular regulation of the miR-142 using chromatin immune-precipitation followed by sequencing (ChIP-seq) and revealed constitutive binding of PU.1, Runx1, and IRF4. To extend this study, the authors analyzed a newly developed miR-142–deficient mouse that revealed a approximately 2-fold reduction in plasmacytoid and CD8α+ DCs and a somewhat stronger reduction in the CD4+ DC compartment in vivo. Earlier reports strongly implicated miR-142-3p as playing an important role in IL-6 signaling in DCs5 but had not investigated in detail additional roles of this miRNA.
Although the loss of CD4+ DCs was not complete, repopulation experiments revealed that CD4+ DCs, but not other subsets, failed to develop in a competitive reconstitution setting. The selective loss of CD4+ DCs in the absence of miR-142 at least partly phenocopied the loss of CD4+ DCs observed in Irf4−/− mice. Irf4 is required for the development of CD4+ DCs and is an interacting partner of PU.1.6 Loss of miR-142 in DCs was also accompanied by elevated expression of Irf8 in CD4-equivalent DCs and in vitro favored the development of CD8-equivalent DCs (see figure). Irf4 and Irf8 have been identified to direct CD4+ and CD8α+ DC differentiation, respectively, and intriguingly share similar DNA binding sites. In immature DCs, PU.1 binds with Irf8 as a heterodimer to induce CIITA expression that is subsequently repressed by expression of Blimp1 (PRDM1) on DC maturation. Loss of miR-142 was accompanied by elevated expression of co-stimulatory molecules such as CD40, CD80, CD86, and MHC class II normally associated with maturation of DCs, similar to the phenotype observed in mice deficient for the negative regulatory transcription factor Zbtb46.7,8 In the absence of miR-142, this hyperactivated status may occur because of the enhanced Irf8 combined with dampened Irf4 expression, which could push DCs toward a more activated phenotype similar to that induced by TLR signaling. This in turn results in an impaired DC capacity to acquire and present antigens to prime CD4+ T cells (see figure). Irf8 also directs the expression of IL-129 and IL-18,10 thereby promoting Th1 responses in contrast to the Th2 bias induced by Irf4.
A more detailed understanding of how miRNAs influence transcriptional regulation will allow further refinement of the miRNA regulatory circuits that influence DC differentiation and function. In this study by Mildner et al, miR-142 clearly regulates the differentiation and homeostasis of CD4+ DCs potentially positioning it in regulating the balance between interferon regulatory factors to tune the expression of each of these transcription factors in the complex.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal