Key Points
Ex vivo isolated myeloid populations of the mononuclear phagocyte network display specific microRNA expression signatures.
miR-142–deficient mice display a reduction of splenic CD4+ dendritic cells resulting in impaired priming of CD4 T-cell responses.
Abstract
The mononuclear phagocyte system comprises cells as diverse as monocytes, macrophages, and dendritic cells (DCs), which collectively play key roles in innate immune responses and the triggering of adaptive immunity. Recent studies have highlighted the role of growth and transcription factors in defining developmental pathways and lineage relations within this cellular compartment. However, contributions of miRNAs to the development of mononuclear phagocytes remain largely unknown. In the present study, we report a comprehensive map of miRNA expression profiles for distinct myeloid populations, including BM-resident progenitors, monocytes, and mature splenic DCs. Each of the analyzed cell populations displayed a distinctive miRNA profile, suggesting a role for miRNAs in defining myeloid cell identities. Focusing on DC development, we found miR-142 to be highly expressed in classic FLT3-L–dependent CD4+ DCs, whereas reduced expression was observed in closely related CD8α+ or CD4−CD8α− DCs. Moreover, mice deficient for miR-142 displayed an impairment of CD4+ DC homeostasis both in vitro and in vivo. Furthermore, loss of miR-142–dependent CD4+ DCs was accompanied by a severe and specific defect in the priming of CD4+ T cells. The results of our study establish a novel role for miRNAs in myeloid cell specification and define miR-142 as a pivotal genetic component in the maintenance of CD4+ DCs.
Introduction
The mononuclear phagocyte system is a body-wide network of nongranulocytic myeloid cells that collectively perform critical roles in tissue remodeling, homeostasis, and stimulatory and regulatory aspects of innate and adaptive immunity. Mononuclear phagocytes are currently divided into 3 cell types: highly plastic monocytes with precursor and effector potential and the more terminally differentiated macrophages and dendritic cells (DCs), which themselves comprise multiple subpopulations.
Development of the myeloid cell lineage involves the generation of intermediate myeloid progenitors (MPs) and macrophage/DC progenitors (MDPs), which lost the potential to give rise to granulocytes.1 MDPs represent BM-resident clonotypic founder cells of mononuclear phagocytes, which differentiate locally into monocytes2 or committed DC progenitors (CDPs), thus generating the monocyte/macrophage and classic DC (cDC) lineage, respectively. CDPs further develop into plasmacytoid DCs (pDCs) or pre-DCs, which exit the BM to the blood circulation.3 Pre-DCs seed lymphoid and nonlymphoid tissues to differentiate into FLT3-L–dependent cDCs, which share an unrivaled potential to prime naive T lymphocytes.4 However, the existence of multiple DC subpopulations highlights further specialization of this cellular compartment with at least 3 prominent cDC populations detected in murine spleens: CD8α+ DCs, CD4+ DCs, and CD4−CD8α− double-negative (DN) cDCs. Whereas splenic CD8α+ DCs have been studied intensively in recent years and are now known to be the primary DC population with in vivo cross-presenting activity and superior capacity to produce IL-12 under infectious conditions,5-7 specific in vivo functions of CD4+ DCs remain less well defined.
The development of myeloid cells is tightly controlled by temporal and sequential expression of various transcription factors. Expression of the ETS family transcription factor PU.1 commits hematopoietic stem cells to the myeloid cell fate8 ; the basic-region leucine zipper transcription factor C/EBPα is critical for the transition from MPs to granulocytes9 ; and a deficiency of Kruppel-like factor 4 (Klf-4) affects monocyte development.10 In addition, the generation of DC subsets is controlled by transcription factors. Mice deficient for Id2,11 IFN-γ responsive factor 8 (IRF8),12 or the basic leucine zipper transcription factor BatF313 are characterized by a lack of CD8α+ cDCs, whereas deficiencies of the transcription factor IRF4 and Notch-2 affect the development of CD4+ DCs.14,15 However, the underlying mechanisms as to why the respective transcription factors are required for DC generation remain to be elucidated.
Studies addressing mononuclear phagocyte specification have so far focused on the role of transcription and growth factors; the possible critical roles of miRNAs in this process were not addressed systematically. miRNAs are a class of short, noncoding RNAs that modulate the proteome through binding to complementary mRNAs by repressing translation initiation or inducing mRNA degradation.16,17 Posttranscriptional regulation of gene expression by the 20- to 24-nt long miRNAs depends on an imperfect match of 5′-proximal “seed” sequences (positions 2-8) with their target mRNA. Therefore, each miRNA has the potential to suppress multiple, even thousands, of targets and one mRNA can be targeted by many different miRNAs.
miRNAs are assumed to fine-tune cellular mRNA expression levels,18 predisposing them for the control of cell development and cell fates. Indeed, miRNAs have been shown to play critical roles in the development of the adaptive immunity.19,20 However, despite increasing knowledge on the role of miRNAs in controlling myeloid cell functions21-23 and some evidence for their contribution in in vitro monocyte differentiation,24,25 the in vivo role of specific miRNAs in the development and homeostasis of myeloid cells remains to be investigated.
In the present study, we characterized miRNA expression in mononuclear phagocytes, including the 3 mentioned BM myeloid precursor subsets, monocytes, and pDCs and splenic cDCs. We identified miRNA clusters specifically expressed by each subtype, indicating the existence of distinct miRNA-based regulatory circuits in the respective populations and cell-type–specific roles of miRNAs. Focusing on DC development, we found miR-142 to be highly expressed in cDCs. Analysis of the splenic DC compartment of newly generated miR-142–deficient mice revealed a severe, cell-intrinsic homeostatic defect of CD4+ DCs in vivo that could be recapitulated in in vitro cultures. We provide a comprehensive fingerprint analysis of the miRNome of mononuclear phagocytes under physiologic conditions and identify miR-142 as a critical regulator of CD4+ DC homeostasis and maintenance.
Methods
Mice
C57BL/6 Ly5.2 mice were purchased from Harlan Laboratories. C57BL/6 Ly5.1 mice and TCR-transgenic mice harboring ovalbumin (OVA)–specific CD4+ T cells were bred in the Weizmann animal facility. Heterozygous embryonic C57BL/6 stem cells carrying a LacZ gene trap in the miR-142 locus were purchased from Texas A&M Institute of Genomic Medicine. Heterozygous ES cell lines were injected into host blastocysts to produce chimeras. Transmission of the targeted allele through the male germline was confirmed by PCR, LacZ staining, and quantitative RT-PCR analysis. For transplantation experiments, recipient mice were lethally irradiated (10.5 Gy) using a cesium radiation source and maintained under antibiotics (Ciproxin; Bayer) for 10 days. A total of 5 × 106 cells for transplantation was injected IV into the tail vein. All mice used in this study were maintained under specific pathogen-free conditions and handled according to protocols approved by the Weizmann Institute Animal Care Committee as per international guidelines.
Cell sorting
C57BL/6 mice 6 weeks of age were purchased from Harlan Laboratories. For BM precursor isolation, ACK (0.15M NH4Cl, 0.1M KHCO3, and 1mM EDTA in PBS)–lysed BM cells from femurs and tibias were pooled from 15 mice and enriched by MACS with biotinylated CD135 (A2F10), followed by anti-biotin MACS beads (Miltenyi Biotec). The enriched fraction was further stained with streptavidin-PerCP, CD117 (2B8), CD115 (AFS98), and lineage Ab cocktail: CD11b (M1/70), CD3 (145-2C11), CD4 (GK1.5), CD8α (53-6.7), Gr1 (RB6-8C5), Sca-1 (D7), B220 (RA3-6B2), Ter-119, CD11c (N418), and NK1.1 (PK136). Splenic DCs were preenriched from 8 mice by CD11c MACS beads (Miltenyi Biotec) and further stained for CD8α, CD4, CD11c, and CD86 (PO3). BM pDCs were isolated from 5 mice, Ficoll enriched, and stained for CD11c, CD317 (927), and Siglec H (eBio440c). BM monocytes were isolated from Ficoll-enriched BM cells from 5 mice. Staining markers were CD11b, Gr1, and CD115. All Abs were purchased from BioLegend or eBiosciences if not indicated otherwise. A FACSAria flow cytometer (BD Biosciences) was used for sorting and duplets were excluded by their forward scatter height versus foward scatter width appearance.
RNA isolation and microarray analysis
Total RNA from sorted cells was extracted using the miRNeasy Mini Kit (QIAGEN) including DNase digest (QIAGEN). RNA purity was assessed with a BioAnalyzer 2100 (Agilent Technologies). Expression levels of miRNAs were assayed by Agilent miRNA microarrays (Release 12.0 and 15.0), according to the manufacturer's protocols. Then, 100 ng of total RNA per sample (duplicates for each cell population from independent sorts) was labeled and hybridized according to the manufacturer's instructions. For K-Means clustering with Pearson correlation, only miRNAs with a ≥ 2-fold differential expression in at least 1 population were used. As a target prediction algorithm, TargetScan 5.126 was applied. For mRNA microarray analysis, total RNA was extracted and subjected to gene-expression profiling using the Mouse Genome Gene 1.0 ST Affymetrix Exon Microarray according to the manufacturer's instructions. For RNA sequencing (RNA-Seq) and ChIP followed by massive parallel sequencing (ChIP-Seq), total RNA was extracted with QIAzol reagent following the miRNeasy kit's procedure (QIAGEN), and sample quality was tested on a 2100 Bioanalyzer (Agilent). RNA-A+-Seq libraries were prepared using the ′dUTP second-strand (strand-specific) protocol. For detailed information, see the Methods section in Garber et al.33 Microarray data may be found at the Gene Expression Omnibus (GEO) under accession numbers GSE42325 (mRNA chip) and GSE42434 (miRNA chip).
Flow cytometric analysis
Surface staining for the DC maturation markers CD40 (3/23), I-Ab (AF6-120.1), and CD86 (GL-1) was conducted on ACK-lysed splenic cell suspensions. For detection of b-galactosidase activity in miR142-heterozygous DCs, splenic DCs were first stained for CD11c, I-Ab, CD4, and CD8α. After labeling, the cells were loaded with 2mM fluorescein di-β-D-galactopyranoside (F-1179; Molecular Probes) at 37°C.27 After 1 minute, the cell suspension was diluted 10-fold in cold FACS buffer and incubated on ice for 45 minutes. The apoptotic cell assay was performed by annexin V and propidium iodide staining according to the manufacturer's protocol (eBiosciences). The cells were analyzed either on an LSR Fortessa or LSRII flow cytometer (BD Biosciences) using FACSDiva Version 6.2 software (BD Biosciences). FACS data were further analyzed using FlowJo Version 9.3.2 software (TreeStar).
T-cell proliferation assays
CD4+ and CD8+ OVA-specific T cells were isolated from spleens and lymph nodes of the respective TCR-transgenic OT-II and OT-I mice and enriched by MACS with CD4 or CD8 beads (Miltenyi Biotec). Cells were labeled with CFSE (Invitrogen) and coinjected into the tail veins of recipient mice (2 × 106 cells/mouse). Twenty-four hours later, 20 μg of soluble OVA (Sigma-Aldrich) per mouse was injected. Analysis of T-cell proliferation within the spleens of recipients was performed 96 hours after the T-cell transfer.
BM cell cultures
For the generation of in vitro BM-derived DCs, 5 × 106 BM cells were cultured for 8 days in full RPMI medium (Gibco-BRL) containing 10% FCS (Biochrom), 1% Pen/Strep, 1% MEM-EAGLE nonessential amino acids, 1% l-glutamine solution, 1% sodium pyruvate solution (all from Biologic Industries) and supplemented with 200 ng/mL of FLT3-L (PeproTech). Every third day, the cells received 1/3 volume fresh media with FLT3-L. For the identification of in vitro derived CD4+ and CD8α+ DC equivalents, the cell cultures were stained for B220, CD11c, CD11b, CD24 (M1/69), and CD172a (P84; BD Pharmingen).
Quantitative RT-PCR
To quantify miRNA expression, 50-250 ng of total RNA was reverse transcribed with the miScript reverse transcription kit (QIAGEN) according to the manufacturer's instructions. The miScript SYBR Green kit (QIAGEN) was used to detect amplification in a LightCycler 480 (Roche) machine. The following primers were used in combination with the universal primer (QIAGEN): U6, 5′-GATGACACGCAAATTCGTGAA-3′; miR-155-5p, 5′-TTAATGCTAATTGTGATAGGG-3′; miR-223-3p, 5′-TGTCAGTTTGTCAAATACCC-3′; miR-146a-5p, 5′-TGAGAACTGAATTCCATGGGT-3′; miR-196b-5p, 5′-TAGGTAGTTTCCTGTTGTTG-3′; miR-532-5p, 5′-CATGCCTTGAGTGTAGGACC-3′; miR-22-3p, 5′-AAGCTGCCAGTTGAAGAACTG-3′; miR-142-3p, 5′-TGTAGTGTTTCCTACTTTATGGA-3′; miR-142-5p, 5′-CATAAAGTAGAAAGCACTACT-3′.
Results
miRNome analysis of the mononuclear phagocyte system
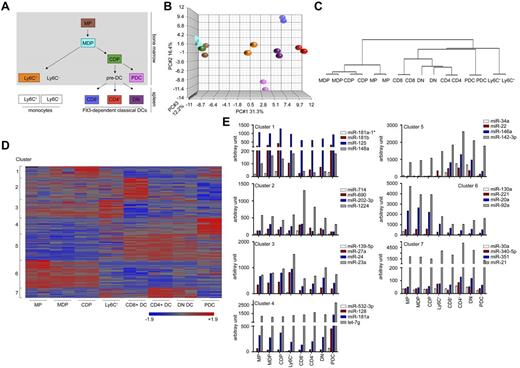
To identify miRNAs that are expressed in the murine mononuclear phagocyte system and might potentially regulate cell differentiation, we isolated 8 different members of this particular myeloid network for miRNA profiling. This included recently characterized BM-resident myeloid precursor populations (ie, MPs, CDPs, and MDPs),3 Ly6C+ BM monocytes, classic splenic DC subsets (CD4+ DCs, CD8α+ DCs, and DN DCs), and pDCs (Figure 1A). RNA of sorted cells was subjected to miRNA microarray analysis (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Principal component analysis of microarray duplicates showed a high reproducibility of the miRNA arrays for all cell populations (Figure 1B). Quantitative RT-PCR analysis for a subset of miRNAs further confirmed the sensitivity and quality of the miRNA array data (supplemental Figure 2).
Mononuclear phagocyte populations are characterized by specific miRNA-expression profiles. (A) Schematic of the development and relationship between the populations of the mononuclear phagocyte network. Framed and colored populations were sorted and investigated in this study. (B) Principal component analysis of the miRNA microarray results obtained from the 8 phagocyte populations. Each symbol represents a microarray dataset and for each population, color-coded as in panel A, duplicates were performed. (C) Hierarchal clustering of miRNA-profiling data revealed a clear separation of the various cells and reflects the developmental relationships consistent with the established tree. (D) K-means clustering of miRNAs that showed an at least 2-fold differential expression in 1 of the 8 cell populations tested. A total of 136 miRNAs could be divided into 7 defined clusters. Intensities of red and blue refer to increased or decreased miRNA expression, respectively. The full list of miRNAs and expression values can be found in supplemental Table 1. (E) Mean arbitrary expression signal intensities for 4 representative miRNAs of cluster 1-7. Normalized and standardized expression levels obtained from the 2 individual miRNA chips as depicted in supplemental Table 1 were averaged and converted to anti-log arbitrary expression values. Note the high expression of miR-142 in the DC compartment.
Mononuclear phagocyte populations are characterized by specific miRNA-expression profiles. (A) Schematic of the development and relationship between the populations of the mononuclear phagocyte network. Framed and colored populations were sorted and investigated in this study. (B) Principal component analysis of the miRNA microarray results obtained from the 8 phagocyte populations. Each symbol represents a microarray dataset and for each population, color-coded as in panel A, duplicates were performed. (C) Hierarchal clustering of miRNA-profiling data revealed a clear separation of the various cells and reflects the developmental relationships consistent with the established tree. (D) K-means clustering of miRNAs that showed an at least 2-fold differential expression in 1 of the 8 cell populations tested. A total of 136 miRNAs could be divided into 7 defined clusters. Intensities of red and blue refer to increased or decreased miRNA expression, respectively. The full list of miRNAs and expression values can be found in supplemental Table 1. (E) Mean arbitrary expression signal intensities for 4 representative miRNAs of cluster 1-7. Normalized and standardized expression levels obtained from the 2 individual miRNA chips as depicted in supplemental Table 1 were averaged and converted to anti-log arbitrary expression values. Note the high expression of miR-142 in the DC compartment.
On average, we detected in each cell population approximately 160 expressed miRNAs, which was consistent with the estimated number,28 and confirmed experimentally the miRNA numbers expressed in hematopoietic cells.29,30 Unsupervised clustering of the miRNA-profiling data revealed distinctive miRNA expression signatures for the individual cell types. Dendrogram stratification of these profiles indicated relationships consistent with the established developmental tree31 (Figure 1C). Therefore, the miRNA profiles of MDPs and CDPs were more similar to each other than to MPs. Moreover, within the cDC compartment, we detected a closer relationship of CD4+ DCs with DN cDCs than with CD8α+ DCs, which is consistent with published mRNA profiles of these cell types.32
A total of 136 miRNAs were differentially expressed (> 2 fold) between the tested cell populations. To group the miRNAs into similar regulatory circuits, we performed K-means clustering (k = 7; Figure 1D-E; a full list of miRNAs is provided in supplemental Table 1). A cluster of miRNAs specifically expressed in the precursor populations (cluster 6) comprised, among other miRNAs, members of the miR-17∼92 family and miR-222, both of which were proposed to play a role in in vitro monocytopoiesis.24,25 Established innate immune cell–associated miRNAs such as miR-155 and miR-146a could be detected in cluster 5 defined by miRNAs highly expressed in the 3 tested cDC populations. Additional well-defined clusters could be identified for CD8α+ cDCs (cluster 2) and pDCs (cluster 4).
Our global analysis enables a new definition of mononuclear phagocyte ontogeny based on miRNA expression profiles, suggesting a role for miRNAs in defining cell identities.
miR-142 is highly expressed in CD4+ DCs and its absence affects DC development
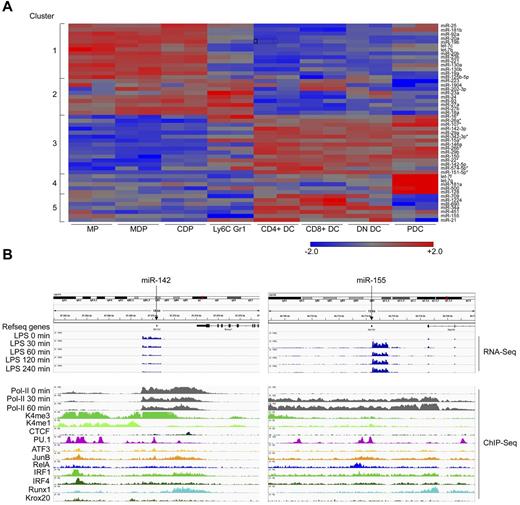
To select miRNAs potentially involved in mononuclear phagocyte differentiation, we focused on miRNAs differentially expressed by the myeloid cell populations. Clustering of the 50 most prominent miRNAs revealed a similar, although less defined, separation pattern compared with the clustering based on all detected miRNAs (Figure 2A). Focusing on DC development, we concentrated on miRNAs that are derived from independent transcription units and differentially expressed between DC subsets and the precursors, a criterion fulfilled by miR-142. Specifically, both forms of this miRNA, miR-142-3p and miR-142-5p, were expressed in all cDC subsets, though most prominently in CD4+ DCs. miR-142-3p could be detected at much higher levels than miR-142-5p (Figure 3A and supplemental Table 1). ChIP-Seq of the miR-142 locus in GM-CSF culture–derived BM DCs33 revealed constitutive binding of the myeloid-specific pioneering transcription factor PU.1, as well as Runx1 and IRF4 (Figure 2B left panel), all of which are associated with myeloid cell development.8,14,34,35 In contrast, no constitutive binding of the TLR-regulated RelA component was found, which is characteristic of, for example, the activation-related miRNA-155 (Figure 2B right panel). Rather, analysis of miR-142 loci of LPS challenged BM-DCs revealed that, unlike miRNA-155, miR-142 expression was repressed on stimulation with LPS (Figure 2B). These data suggest that miR-142 may act as a negative regulator of TLR-triggered responses. miR-142 was reported to regulate IL-6 production by BM-culture–derived DCs,36 supporting the notion that miR-142 might be important for DC function.
miR-142 is a candidate miRNA governing DC subset specification. (A) K-means clustering of the 50 highest expressed miRNAs in the 8 mononuclear phagocyte subsets tested. DC-specific miRNAs (cluster 3) that are located inside protein-coding transcriptional units are indicated by asterisks (*). (B) Genomic localization and transcriptional control of miR-142 (left) and miR-155 (right). ChIP-Seq data of mRNA obtained from murine BM-derived DCs under steady-state conditions and at various time points (30, 60, 120, and 240 minutes) after 100 ng/mL of lipopolysaccharide exposure (shown only for transcript reads and polymerase activity). The binding of transcription factors is shown under physiologic conditions. A full description of the RNA-Seq and ChIP-Seq data can be found in Garber et al.33
miR-142 is a candidate miRNA governing DC subset specification. (A) K-means clustering of the 50 highest expressed miRNAs in the 8 mononuclear phagocyte subsets tested. DC-specific miRNAs (cluster 3) that are located inside protein-coding transcriptional units are indicated by asterisks (*). (B) Genomic localization and transcriptional control of miR-142 (left) and miR-155 (right). ChIP-Seq data of mRNA obtained from murine BM-derived DCs under steady-state conditions and at various time points (30, 60, 120, and 240 minutes) after 100 ng/mL of lipopolysaccharide exposure (shown only for transcript reads and polymerase activity). The binding of transcription factors is shown under physiologic conditions. A full description of the RNA-Seq and ChIP-Seq data can be found in Garber et al.33
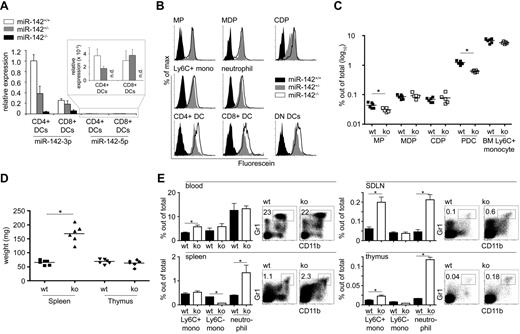
Loss of miR-142 affects the composition of the myeloid compartment in vivo. (A) Quantitative real-time PCR of splenic CD4+ and CD8α+ DCs isolated from miR-142+/+, miR-142+/−, and miR-142−/− mice. The detection of miR-142-3p and miR-142-5p was normalized to endogenous U6 levels. All expression levels were calculated to the miR-142-3p level in miR-142+/+ CD4+ DCs. Note the reduced expression of miR-142-3p in WT CD8α+ DCs compared with CD4+ DCs. (B) β-Galactosidase activity in ex vivo myeloid cell populations isolated from miR-142+/+ (black filled), heterozygous mutant miR-142 LacZ knock-in mouse (gray filled), or homozygous mutant miR-142 LacZ knock-in mouse (white filled) as determined by the FACS-FDG assay. (C) Quantification of mononuclear phagocyte populations in the BM of miR-142–deficient mice and control animals. Only MPs (0.044% ± 0.009% and 0.031% ± 0.005% of total BM cells in WT and miR-142−/− mice, respectively) and pDCs (1.2% ± 0.18% and 0.6% ± 0.05% of total BM cells in WT and miR-142−/− mice, respectively) showed significant reduced cell numbers in miR-142−/− mice. (D) Splenomegaly in 6-week-old miR-142−/− mice (65.2 ± 8.5 vs 168 ± 30.2 mg in WT and miR-142−/− mice, respectively). Thymi showed no obvious weight changes (68.5 ± 8.5 vs 63.3 ± 11.2 mg in WT and miR-142−/− mice, respectively). (E) Flow cytometric analysis of primary (thymi) and secondary lymphoid organs (skin draining lymph nodes, mesenteric lymph nodes, and spleens) for the infiltration of myeloid cells in miR-142−/− and miR-142+/+ mice. Neutrophils were identified as CD115−CD11b Gr1+; Ly6C+ monocytes as CD115+CD11b+Gr1+; and Ly6C− monocytes as CD115+CD11b+Gr1−. Three animals in an age of 6 weeks were used in each group. *P < .05 was considered significant using a Student 2-tailed t test.
Loss of miR-142 affects the composition of the myeloid compartment in vivo. (A) Quantitative real-time PCR of splenic CD4+ and CD8α+ DCs isolated from miR-142+/+, miR-142+/−, and miR-142−/− mice. The detection of miR-142-3p and miR-142-5p was normalized to endogenous U6 levels. All expression levels were calculated to the miR-142-3p level in miR-142+/+ CD4+ DCs. Note the reduced expression of miR-142-3p in WT CD8α+ DCs compared with CD4+ DCs. (B) β-Galactosidase activity in ex vivo myeloid cell populations isolated from miR-142+/+ (black filled), heterozygous mutant miR-142 LacZ knock-in mouse (gray filled), or homozygous mutant miR-142 LacZ knock-in mouse (white filled) as determined by the FACS-FDG assay. (C) Quantification of mononuclear phagocyte populations in the BM of miR-142–deficient mice and control animals. Only MPs (0.044% ± 0.009% and 0.031% ± 0.005% of total BM cells in WT and miR-142−/− mice, respectively) and pDCs (1.2% ± 0.18% and 0.6% ± 0.05% of total BM cells in WT and miR-142−/− mice, respectively) showed significant reduced cell numbers in miR-142−/− mice. (D) Splenomegaly in 6-week-old miR-142−/− mice (65.2 ± 8.5 vs 168 ± 30.2 mg in WT and miR-142−/− mice, respectively). Thymi showed no obvious weight changes (68.5 ± 8.5 vs 63.3 ± 11.2 mg in WT and miR-142−/− mice, respectively). (E) Flow cytometric analysis of primary (thymi) and secondary lymphoid organs (skin draining lymph nodes, mesenteric lymph nodes, and spleens) for the infiltration of myeloid cells in miR-142−/− and miR-142+/+ mice. Neutrophils were identified as CD115−CD11b Gr1+; Ly6C+ monocytes as CD115+CD11b+Gr1+; and Ly6C− monocytes as CD115+CD11b+Gr1−. Three animals in an age of 6 weeks were used in each group. *P < .05 was considered significant using a Student 2-tailed t test.
We next studied the in vivo role of miR-142 in DCs by analyzing newly developed miR-142–deficient C57BL/6 mice generated from embryonic stem cells harboring a LacZ gene trap insertion in their miR-142 locus. We focus herein on the myeloid phenotype of the miR-142−/− mice; a full description of the strain will be published elsewhere (unpublished observations).
Targeted insertion of the β-galactosidase gene into the miR-142 locus abrogated miR-142 expression, as revealed by analysis of splenic DCs isolated from wild-type (WT) and mutant animals (Figure 3A). Moreover, flow cytometric analysis of splenic DCs and other myeloid cell populations isolated from miR-142+/− and miR-142−/− mice using the nonquantitative FDG assay revealed β-galactosidase activity in all tested myeloid subsets (Figure 3B), indicating a functional knock-in of the LacZ gene and confirming endogenous miR-142 promoter activity in these cell populations. Flow cytometric analysis of the BM of miR-142–deficient mice revealed normal numbers of macrophage and DC precursors, including MDPs, CDPs, and monocytes, compared with littermate controls (Figure 3C). Only early MPs were found to be slightly (approximately 30%) decreased in the absence of miR-142. Conversely, the loss of miR-142 expression resulted in a 2-fold reduction of BM pDCs, which also showed high levels of miR-142 expression (Figure 1E). The miR-142 deficiency also resulted in macroscopic signs of splenomegaly starting from the age of 6 weeks (Figure 3D), which was accompanied by a slight infiltration of secondary and primary lymphoid organs by myeloid cells (Figure 3E). Neutrophils in particular dominated the infiltration of spleens, skin-draining lymph nodes, and thymi in miR-142−/− mice.
FACS analysis of spleens revealed an approximately 60% reduction of CD11chiMHCII+ cells in miR-142−/− mice accompanied by a strikingly distorted DC composition (Figure 4A). Although frequencies of DN DCs were not affected by the mir-142 deficiency (Figure 4B), their absolute numbers were increased mainly because of the splenomegaly (Figure 4C). In contrast, CD4+ DCs and CD8α+ DCs numbers in miR-142−/− spleens were reduced by 2.5- and 1.7-fold, respectively, relative to WT littermates and the absolute cell numbers were even reduced independently of the enlarged spleens (Figure 4B-C). Analysis of the DC maturation state revealed that all 3 DC subtypes isolated from miR-142−/− mice displayed elevated expression of the costimulatory molecules CD40, CD80, and CD86, whereas MHCII expression was slightly reduced (Table 1). Analysis of the apoptotic rate indicated that only miR-142−/− CD4+ DCs, but neither CD8α+ nor DN DCs, underwent more apoptosis than their littermate counterparts, although these results did not reach statistical significance (P = .06; Figure 4D).
Loss of miR-142 affects the composition of lymphoid tissue–resident DCs in vivo. (A) Flow cytometric analysis of splenic DC composition in miR-142–deficient mice and control animals. (B) Quantification of FLT3L-dependent cDCs in miR-142−/− mice and littermate controls per milligram of spleen tissue. (C) Data from panel B were calculated for total splenic weight to account for splenomegaly. Total cell numbers were: DN DCs: 3.1 × 105 ± 5.4 × 104 in WT versus 6.3 × 105 ± 2 × 105 in miR142−/−, P = .003; CD8α+ DCs: 1.2 × 105 ± 2.4 × 104 in WT versus 6.9 × 104 ± 1.9 × 104 in miR142−/−, P = .003; and CD4+ DCs: 7 × 105 ± 2.4 × 105 in WT versus 2.8 × 105 ± 6.4 × 104 in miR142−/−, P = .002. Each dot represents an independent animal. Five to 6 animals at an age of 6 weeks were used in each group. *P < .05 was considered significant using a Student 2-tailed t test. (D) Analysis of apoptotic miR-142–deficient and miR-142–competent cDCs determined by annexin V and propidium iodide staining. Each dot represents an independent animal. (E) Flow cytometric analysis of mesenteric lymph node- and small intestine–resident DCs isolated from miR-142+/+ and miR-142−/− animals. Each dot represents an independent animal. *P < .05 was considered significant using a Student 2-tailed t test.
Loss of miR-142 affects the composition of lymphoid tissue–resident DCs in vivo. (A) Flow cytometric analysis of splenic DC composition in miR-142–deficient mice and control animals. (B) Quantification of FLT3L-dependent cDCs in miR-142−/− mice and littermate controls per milligram of spleen tissue. (C) Data from panel B were calculated for total splenic weight to account for splenomegaly. Total cell numbers were: DN DCs: 3.1 × 105 ± 5.4 × 104 in WT versus 6.3 × 105 ± 2 × 105 in miR142−/−, P = .003; CD8α+ DCs: 1.2 × 105 ± 2.4 × 104 in WT versus 6.9 × 104 ± 1.9 × 104 in miR142−/−, P = .003; and CD4+ DCs: 7 × 105 ± 2.4 × 105 in WT versus 2.8 × 105 ± 6.4 × 104 in miR142−/−, P = .002. Each dot represents an independent animal. Five to 6 animals at an age of 6 weeks were used in each group. *P < .05 was considered significant using a Student 2-tailed t test. (D) Analysis of apoptotic miR-142–deficient and miR-142–competent cDCs determined by annexin V and propidium iodide staining. Each dot represents an independent animal. (E) Flow cytometric analysis of mesenteric lymph node- and small intestine–resident DCs isolated from miR-142+/+ and miR-142−/− animals. Each dot represents an independent animal. *P < .05 was considered significant using a Student 2-tailed t test.
Increased expression of costimulatory molecules on miR-142 deficient DCs
| Surface marker . | Splenic DC subset . | miR-142+/+ . | miR-142−/− . | t test . |
|---|---|---|---|---|
| CD40 | CD4+ DC | 942.3 ± 86.1 | 1082.2 ± 53.7 | .012 |
| CD8a+ DC | 1513.5 ± 160.1 | 1790.2 ± 66.0 | .006 | |
| DN DC | 827.8 ± 53.8 | 801.0 ± 96.2 | .572 | |
| CD80 | CD4+ DC | 1160.7 ± 81.5 | 1865.2 ± 162.9 | .00001 |
| CD8a+ DC | 961.5 ± 119.1 | 784.0 ± 84.8 | .021 | |
| DN DC | 758.7 ± 83.9 | 982.4 ± 77.0 | .001 | |
| CD86 | CD4+ DC | 233.8 ± 65.3 | 345.8 ± 30.9 | .007 |
| CD8a+ DC | 365.7 ± 27.4 | 517.0 ± 15.7 | 2 × 10−6 | |
| DN DC | 155.7 ± 67.0 | 197.0 ± 18.1 | .217 | |
| MHCII | CD4+ DC | 10 811.7 ± 1865.0 | 7037.8 ± 708.7 | .002 |
| CD8a+ DC | 7372 ± 1667.5 | 6206.4 ± 620.8 | .176 | |
| DN DC | 8878.5 ± 1380.1 | 5150.6 ± 536.9 | .0003 |
| Surface marker . | Splenic DC subset . | miR-142+/+ . | miR-142−/− . | t test . |
|---|---|---|---|---|
| CD40 | CD4+ DC | 942.3 ± 86.1 | 1082.2 ± 53.7 | .012 |
| CD8a+ DC | 1513.5 ± 160.1 | 1790.2 ± 66.0 | .006 | |
| DN DC | 827.8 ± 53.8 | 801.0 ± 96.2 | .572 | |
| CD80 | CD4+ DC | 1160.7 ± 81.5 | 1865.2 ± 162.9 | .00001 |
| CD8a+ DC | 961.5 ± 119.1 | 784.0 ± 84.8 | .021 | |
| DN DC | 758.7 ± 83.9 | 982.4 ± 77.0 | .001 | |
| CD86 | CD4+ DC | 233.8 ± 65.3 | 345.8 ± 30.9 | .007 |
| CD8a+ DC | 365.7 ± 27.4 | 517.0 ± 15.7 | 2 × 10−6 | |
| DN DC | 155.7 ± 67.0 | 197.0 ± 18.1 | .217 | |
| MHCII | CD4+ DC | 10 811.7 ± 1865.0 | 7037.8 ± 708.7 | .002 |
| CD8a+ DC | 7372 ± 1667.5 | 6206.4 ± 620.8 | .176 | |
| DN DC | 8878.5 ± 1380.1 | 5150.6 ± 536.9 | .0003 |
Mean fluorescence intensity (MFI) analysis of the co-stimulatory molecules CD40, CD80, and CD86 on splenic classical DCs isolated from miR-142−/− mice and littermate controls. At least 5 animals per group were used for the analysis. P < .05 considered to be significant using the Student 2-tailed t test.
To determine whether miR-142 deficiency also affects the development of other CD8α− CD11b+ lymphoid and nonlymphoid DCs, we investigated CD4+ DCs in mesenteric lymph nodes and the corresponding CD103+ CD11b+ DCs in the small intestine.15 Interestingly and comparable to the spleen, we found a decrease in the frequency of miR-142–deficient CD4+ DCs in the mesenteric lymph nodes, whereas intestinal CD103+ CD11b+ DCs were represented in higher numbers (Figure 4E).
These findings suggest that miR-142 expression is critical for the maintenance of lymphoid tissue DC quiescence and that its lack interferes with DC homeostasis.
Cell-intrinsic and cell-specific miR-142 requirement for the generation of CD4+ DCs
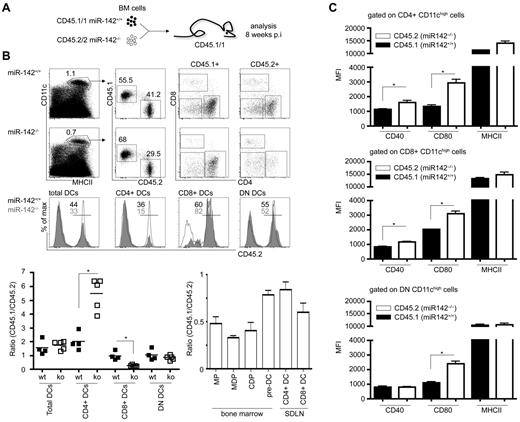
The absence of miR-142 results in a significant reduction of splenic CD4+ and CD8α+ DCs. However, DC homeostasis and prevalence can be influenced by other hematopoietic cells such as T-regulatory cells37 and by environmental factors. Therefore, we conducted competitive repopulation experiments to determine whether the DC impairment in miR-142−/− mice reflects a cell-intrinsic phenomenon. BM cells of either homozygote miR-142 mutant mice (CD45.2+) or littermates were mixed at a 1:1 ratio with congenic WT BM cells (CD45.1+) and transferred into lethally irradiated CD45.1+ recipient mice. Eight weeks after transplantation, chimeric miR-142+/+/WT > WT and miR-142−/−/WT > WT spleens were analyzed for reconstitution of their DC compartment. In the control group, miR-142+/+ cells efficiently reconstituted all 3 DC populations (Figure 5A). DN and CD8α+ DCs in miR-142−/−/WT > WT mice developed equally well from both genotypes, with miR-142−/−/CD8α+ DCs even showing a slight reconstitution advantage over their WT competitor (Figure 5A). In stark contrast, miR-142−/− BM failed to reconstitute CD4+ DCs, which in miR-142−/−/WT > WT chimeras were almost exclusively derived from WT cells (Figure 5A). However, no developmental defect of miR-142−/− skin-draining lymph node DCs and DC precursors, such as MPs, MDPs, CDPs, and pre-DCs, was observed in these animals (Figure 5B). Next, we investigated the expression of costimulatory molecules on DCs in miR-142−/−/WT > WT BM chimeras. Comparable to miR-142–deficient mice (Table 1), miR-142−/− BM-derived DCs showed in the mixed chimeras an up-regulation of CD40, CD80, and even MHCII compared with their WT counterparts (Figure 5C). These data suggest that the reduction of CD8α+ DCs in miR-142−/− is largely indirect, but is associated with cell-intrinsic signs of hyperactivation. In contrast, miR-142 is specifically and cell intrinsically required for the homeostasis of lymphoid organ–resident CD4+ DCs.
The developmental defect of CD4+ DCs in the absence of miR-142 is cell intrinsic. (A) Schematic experimental outline of mixed BM reconstitution experiment. (B) Flow cytometric analysis of miR-142+/+ (CD45.2)/WT (CD45.1) > WT (CD45.1) and miR-142−/− (CD45.2)/WT (CD45.1) > WT (CD45.1) chimeric animals 8 weeks after transplantation for contribution of distinct miR-142 genotypes to the 3 cDC populations (left panel) and BM precursors, respectively. For miR-142+/+/WT > WT mice: CD45.1/2 ratios: for CD8α+ DCs, 0.97 ± 0.31; for CD4+ DCs, 2.01 ± 0.64; for DN DCs, 1.03 ± 0.39; for miR-142−/−/WT > WT mice: CD45.1/2 ratios: for CD8α+ DCs, 0.26 ± 0.07; for CD4+ DCs, 5.48 ± 1.15; and for DN DCs, 0.84 ± 0.17. CD45.1/CD45.2 ratios were calculated for each investigated cell population. Values > 1 indicate out-competition of the mutant by WT (CD45.1) cells, whereas values < 1 show an advantage of miR-142−/− (CD45.2) cells. Representative results from 1 of 2 independent experiments are shown (means ± SD) with at least 4 animals in each group. SDLN indicates skin draining lymph nodes. (C) Mean fluorescence intensity (MFI) of CD40, CD80, and MHCII expression on miR-142–competent (CD45.1+) and miR-142–deficient (CD45.2+) CD4+, CD8α+, and DN DCs isolated from mixed chimeras. Representative results from 1 of 2 independent experiments are shown (means ± SD) with 3 animals in each group.
The developmental defect of CD4+ DCs in the absence of miR-142 is cell intrinsic. (A) Schematic experimental outline of mixed BM reconstitution experiment. (B) Flow cytometric analysis of miR-142+/+ (CD45.2)/WT (CD45.1) > WT (CD45.1) and miR-142−/− (CD45.2)/WT (CD45.1) > WT (CD45.1) chimeric animals 8 weeks after transplantation for contribution of distinct miR-142 genotypes to the 3 cDC populations (left panel) and BM precursors, respectively. For miR-142+/+/WT > WT mice: CD45.1/2 ratios: for CD8α+ DCs, 0.97 ± 0.31; for CD4+ DCs, 2.01 ± 0.64; for DN DCs, 1.03 ± 0.39; for miR-142−/−/WT > WT mice: CD45.1/2 ratios: for CD8α+ DCs, 0.26 ± 0.07; for CD4+ DCs, 5.48 ± 1.15; and for DN DCs, 0.84 ± 0.17. CD45.1/CD45.2 ratios were calculated for each investigated cell population. Values > 1 indicate out-competition of the mutant by WT (CD45.1) cells, whereas values < 1 show an advantage of miR-142−/− (CD45.2) cells. Representative results from 1 of 2 independent experiments are shown (means ± SD) with at least 4 animals in each group. SDLN indicates skin draining lymph nodes. (C) Mean fluorescence intensity (MFI) of CD40, CD80, and MHCII expression on miR-142–competent (CD45.1+) and miR-142–deficient (CD45.2+) CD4+, CD8α+, and DN DCs isolated from mixed chimeras. Representative results from 1 of 2 independent experiments are shown (means ± SD) with 3 animals in each group.
Loss of miR-142–dependent CD4+ DCs is associated with impaired CD4+ T-cell priming
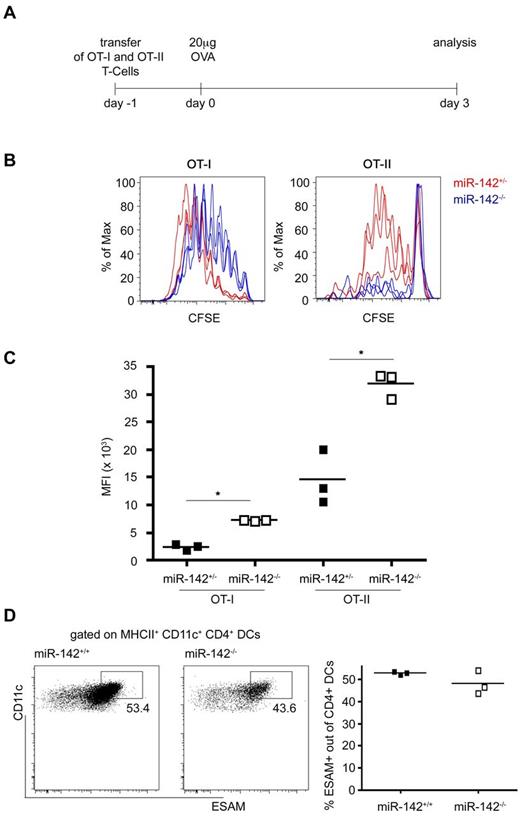
All cDC subtypes share the capacity to uptake antigen and present antigen-derived peptides for naive T-cell stimulation. However, CD8α+ DCs were demonstrated to be specialized in priming CD8+ T cells, whereas CD4+ DCs are superior in presenting MHC class II–restricted antigens to CD4+ T cells.38,39 To probe for functional consequences of the CD4+ DC loss, we tested mice harboring a miR-142−/−–deficient immune system for their ability to respond to antigen challenge. Specifically, miR-142−/− > WT] and miR-142+/− > WT BM chimeras were engrafted with CD8+ or CD4+ T cells harboring transgene-encoded TCRs reactive to OVA. To monitor and quantify T-cell responses, grafts were labeled with CFSE before transfer. Flow cytometric analysis of recipient spleens 3 days after IV OVA challenge (Figure 6A) revealed a partially impaired proliferation of CD8+ T cells in WT and miR-142−/− mice (Figure 6B-C). In contrast, grafted CD4+ T cells proliferated only in the challenged WT recipients, but retained their CFSE label in miR-142–deficient mice, indicating the absence of CD4+ T-cell division.
CD4+ T-cell priming defect in miR-142−/− mice. (A) Schematic of the experimental protocol. (B) Flow cytometric analysis of T-cell grafts retrieved from immunized recipient mice indicating proliferated OT-I CD8+ T cells (left) and OT-II CD4+ T cells (right) cells in miR-142+/− (red) and miR-142−/− mice (blue). (C) Quantification of CFSE mean fluorescence intensity (MFI) of proliferated OT-I and OT-II cells. OT-II CFSE MFI WT 14 473 ± 4961 versus CFSE MFI knockout (ko) 31 812 ± 2316, P = .005. Each dot represents an animal. *P < .05 was considered significant using a Student 2-tailed t test. (D) Flow cytometric analysis of splenic CD4+ DCs isolated from miR-142−/− mice and littermate controls for the cell-surface molecule ESAM. Cells were gated on CD11c+ MHCIIhi and CD4+. Each dot represents an animal.
CD4+ T-cell priming defect in miR-142−/− mice. (A) Schematic of the experimental protocol. (B) Flow cytometric analysis of T-cell grafts retrieved from immunized recipient mice indicating proliferated OT-I CD8+ T cells (left) and OT-II CD4+ T cells (right) cells in miR-142+/− (red) and miR-142−/− mice (blue). (C) Quantification of CFSE mean fluorescence intensity (MFI) of proliferated OT-I and OT-II cells. OT-II CFSE MFI WT 14 473 ± 4961 versus CFSE MFI knockout (ko) 31 812 ± 2316, P = .005. Each dot represents an animal. *P < .05 was considered significant using a Student 2-tailed t test. (D) Flow cytometric analysis of splenic CD4+ DCs isolated from miR-142−/− mice and littermate controls for the cell-surface molecule ESAM. Cells were gated on CD11c+ MHCIIhi and CD4+. Each dot represents an animal.
To investigate the underlying mechanism, we analyzed the CD4+ DC compartment for a subpopulation of ESAMhi cells. The Notch2 receptor controls the differentiation of a unique splenic CX3CR1lo ESAMhi CD4+ DC subset that is required for efficient priming of CD4+ T cells.15 However, despite the reduction of CD4+ DCs in miR-142−/− animals, no differences in the frequency of ESAMhi CD4+ DCs were observed (Figure 6D).
These data corroborate the earlier notion of the supremacy of CD4+ DCs to stimulate naive CD4+ T cells and provide functional evidence for the DC defect in miR-142–deficient animals.
miR-142–deficient BM cells fail to develop into CD4+ DCs in vitro
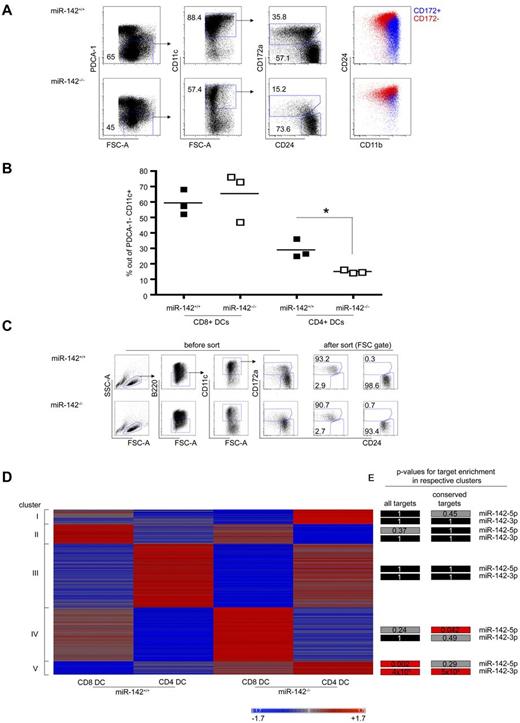
We also evaluated the potential of miR-142−/− DC precursors to develop into CD4+ DCs in vitro. BM cells from miR-142−/− mice and their miR-142+/+ littermates were isolated and cultured for 7 days in the presence of FLT3-L. In the absence of CD4 and CD8α antigen expression by the in vitro–generated cells, equivalents of splenic CD4+ and CD8α+ DCs in these cultures were identified as CD11c+CD172+CD24lowCD11b+ and CD11c+CD172−CD24+CD11blow cells, respectively.40 CD8α+ DC equivalents were generated in equal efficiency from both WT and miR-142−/− BM in these cultures (59.1% ± 8.1% in WT vs 65.3% ± 15.9% in miR-142−/−, P = .58). In contrast, miR-142–deficient BM cells were significantly impaired in their potential to give rise to the CD4+ DC equivalent in vitro (29.1% ± 6.0% in WT vs 14.9% ± 1.1% in miR-142−/−, P = .016, Figure 7A-B). These results corroborate our in vivo data showing that miR-142 is intrinsically required for CD4+ DC homeostasis and establish that the defect observed in miR-142−/− mice is independent of tissue context and DC migration.
Impaired development of miR-142–deficient CD4+ DCs in vitro and enrichment of miR-142 target expression in miR-142–deficient DCs. (A) Flow cytometric analysis of BM cells of miR-142−/− mice and WT littermates cultured for 6 days in presence of 200 ng/mL of FLT3-L quantifying percentages of CD4+ and CD8α+ DC equivalents. CD4+ DC equivalents were identified as PDCA-1−CD11c+CD24intCD172a+CD11bhigh cells and CD8α+ DC equivalents were identified as PDCA-1−CD11c+CD24highCD172a−CD11bint cells. Each symbol represents BM cells derived from independent mice. One representative experiment of 2 is shown. (B) Graphic summary of data. *P < .05 was considered significant using a Student 2-tailed t test. (C) Sorting of in vitro FLT3-L–cultured CD4+ and CD8α+ DC equivalents generated from miR-142−/− mice and WT littermates. Notice the strong reduction of CD172+ DCs in miR-142−/− culture. (D) Heat map depicting expression of genes showing at least a 2-fold expression difference in 1 of the 4 cell populations tested. Clustering was performed using the Pearson correlation as the distance metric. Intensities of red and blue indicate increased or decreased mRNA levels, respectively. (E) Statistical analysis showing the P values for the enrichment of miR-142-3p and miR-142-5p targets within the 5 detected clusters. Predicted and conserved targets taken from TargetScan, P values were calculated using the hypergeometric test. Black color indicates a P value of 1, gray a P value of < 1, and red a statistically significant enrichment with P < .05.
Impaired development of miR-142–deficient CD4+ DCs in vitro and enrichment of miR-142 target expression in miR-142–deficient DCs. (A) Flow cytometric analysis of BM cells of miR-142−/− mice and WT littermates cultured for 6 days in presence of 200 ng/mL of FLT3-L quantifying percentages of CD4+ and CD8α+ DC equivalents. CD4+ DC equivalents were identified as PDCA-1−CD11c+CD24intCD172a+CD11bhigh cells and CD8α+ DC equivalents were identified as PDCA-1−CD11c+CD24highCD172a−CD11bint cells. Each symbol represents BM cells derived from independent mice. One representative experiment of 2 is shown. (B) Graphic summary of data. *P < .05 was considered significant using a Student 2-tailed t test. (C) Sorting of in vitro FLT3-L–cultured CD4+ and CD8α+ DC equivalents generated from miR-142−/− mice and WT littermates. Notice the strong reduction of CD172+ DCs in miR-142−/− culture. (D) Heat map depicting expression of genes showing at least a 2-fold expression difference in 1 of the 4 cell populations tested. Clustering was performed using the Pearson correlation as the distance metric. Intensities of red and blue indicate increased or decreased mRNA levels, respectively. (E) Statistical analysis showing the P values for the enrichment of miR-142-3p and miR-142-5p targets within the 5 detected clusters. Predicted and conserved targets taken from TargetScan, P values were calculated using the hypergeometric test. Black color indicates a P value of 1, gray a P value of < 1, and red a statistically significant enrichment with P < .05.
Molecular impact of the miR-142 deficiency on DC gene expression
To gain insight into the molecular mechanism underlying the impaired homeostasis of CD4+ DCs in miR-142–deficient mice, we performed an Affymetrix Gene microarray analysis. Given the almost complete absence of CD4+ DCs in miR-142−/− mice, we resorted to CD4+ and CD8α+ DC equivalents from in vitro FLT3-L cultures of WT and miR-142−/− BM (Figure 7C). Consistent with a previous report,40 WT CD4+ and CD8α+ DC equivalents showed distinct mRNA profiles, including differential expression of IRF4 and IRF8. Comparison of the expression profiles of WT and miR-142−/− CD4+ DC equivalents using ingenuity pathway analysis revealed an up-regulation of the transcription factors HoxA9 (10.99-fold), IRF8 (2.44-fold), and Meis1 (1.94-fold) in miR-142−/− CD4+ DCs (supplemental Figure 3A) accompanied by prominent expression alterations for the partially HoxA9-dependent “hematologic system development network” and the IRF8-associated “inflammatory response network” (supplemental Figure 3A). The up-regulation of IRF8, a gene important for the differentiation of CD8α+ DCs,12 may suggest a functional role for miR-142 in the specification of CD4+ versus CD8α+ DCs through regulation of the IRF8 pathway. However, neither these networks nor the analysis of the mean expression levels of all detectable and predicted miR-142-3p and miR-142-5p targets (approximately 4100 genes) in WT and miR-142−/− samples yielded significant enrichment of targets for either of the 2 miRNAs (supplemental Figure 3B). We therefore next performed a bioinformatics analysis by comparing the expression of genes that were at least 2-fold differentially expressed in 1 cell population of the 4 tested. Pearson correlation analysis of these genes revealed a total of 5 distinctive clusters (Figure 7D), including specific ones for CD4+ DCs (cluster III) and CD8α+ DCs (cluster IV and, less specific, cluster II), independently of the genotype. Surprisingly, we also detected genes co-up-regulated in WT CD8α+ DCs and miR-142−/− CD4+ DCs (cluster I), indicating a transcriptional relationship between these populations. Finally, we identified genes displaying specific increased expression only in the miR-142−/−, but not in the WT DCs (cluster V, containing 131 genes). Analysis of these clusters for enrichment of predicted miR-142-3p and miR-142-5p targets by TargetScan revealed significant enrichment in cluster V all predicted miR-142-3p targets among genes up-regulated in both CD4+ and CD8α+ miR-142−/− DCs over WT cells (cluster V; P = 3.6*10−5), as well as conserved miR-142-3p targets (P = .0005, Figure 7E). In addition, the predicted miR-142-5p targets were also enriched in this cluster (P = .002). Overall, 36% of the genes belonging to cluster V were predicted targets of miR-142. Despite the up-regulation of these specific miR-142 targets, no impaired gene signatures with relevance for DC development or biology were detected (supplemental Figure 3C-D). Our data suggest that the impaired development of CD4+ DCs of miR-142–deficient mice both in vitro and in vivo results from a complex dysregulation of multiple targets repressed by either miR-142-3p or miR-142-5p.
Discussion
In the present study, we report a comprehensive miRNome analysis of mononuclear phagocytes and their BM-resident precursor cells, revealing discrete miRNA-expression profiles of all populations analyzed. Focusing on DC differentiation, we identified a specific miRNA, miR-142, as a critical regulator of CD4+ DC homeostasis.
Although miRNAs are known to participate in the control of function and maturation of myeloid cells,41,42 their contribution to myeloid cell differentiation, and in particular the in vivo generation of phagocyte populations, remains poorly defined. Therefore, although miR-146a and miR-223 were shown to have a functional impact on DCs and neutrophils, respectively,21,22 their absence does not seem to affect the development of these cells. In addition, the analysis of CD11c-Cre:Dicerfl/fl mice generated to address the general role of miRNAs in cDCs yielded limited insights,43 most likely because of a complicated interplay among the unknown average miRNA half-life, the time needed for the CD11c-promoter–controlled Cre recombinase-mediated loss of Dicer, and the limited cDC lifespan.
Our comprehensive miRNA-expression profiling revealed that recently characterized BM-resident myeloid precursor populations, monocytes, classic splenic DC subsets, and pDCs can each be defined by unique miRNA-expression patterns. For example, we found high expression of the miR-17∼92 cluster in myeloid precursor cells (MPs, MDPs, and CDPs), suggesting its role in early myeloid development in vivo, possibly upstream of the Runx1 transcription factor.24 Similarly, miR-196b and miR-221/222 were highly expressed in replicating progenitors, which may be linked to their reported function in leukemia pathogenesis.44,45 However, in contrast to a previous study, we could not detect higher levels of these miRNAs in the cDC compartment compared with pDCs,43 highlighting differences between ex vivo isolates and in vitro–cultured cells. BM Ly6C+ monocytes showed a less specific miRNA expression pattern compared with MPs and DCs, but interestingly characterized by a complete absence of miR-155 expression and prominent expression of the miR-23a∼miR-24 cluster.
cDCs were found to be enriched for miRNAs well known to be involved in the regulation of immune responses, including miR-146a, which regulates TLR-signaling pathways by targeting Irak1 and Traf6,46 and miR-155, the expression of which is triggered by inflammatory stimulation and which can act as a pro- and anti-inflammatory regulator.23,41,47
In the present study, we focused on the miR-142 gene, which is an independent transcription unit on mouse chromosome 11. The 2 miRNAs derived from the same pre-miRNA, miR-142-3p and miR-142-5p, display differential expression among DC subsets and reduced expression in their BM-resident precursors. Given the abundance of miR-142 in splenic CD4+ DCs revealed by miRNA profiling, we analyzed the impact of miR-142 deficiency on the distribution of splenic DC subsets. Flow cytometric analysis of miR-142–deficient mice, as well as BM chimeras generated with miR-142–deficient BM, established that this miRNA is intrinsically required for the homeostasis of classic splenic CD4+ DCs. The observed cell-intrinsic phenotype of miR-142–deficient mice in the development of CD4+ DCs is reminiscent of the phenotype observed in IRF4−/− mice.14 Further analysis of secondary lymphoid organs revealed that miR-142 also seems to influence the development of CD4+ mesenteric lymph node DCs—again resembling the IRF4−/− phenotype—whereas in contrast to IRF4−/− mice (William Agace, University of Lund, personal verbal communication, 2012), CD4+ DC equivalent small intestinal CD11b+ CD103+ DCs appeared in increased numbers. These results demonstrate that miR-142 deficiency does not affect a general genetic differentiation program that blocks the development of the classic CD4+ DC lineage in all organs and therefore does not seem to be connected to IRF4. Instead, our data suggest that the miR-142 deficiency results in a functional defect that manifests itself in an organ-specific manner. Splenic miR-142–deficient DCs showed an abnormal high expression of costimulatory molecules, indicating increased maturation and activation that might affect DC homeostasis. A precedent for such a scenario was recently provided with the analysis of mice deficient for the negative regulatory transcription factor Zbtb46. Therefore, Zbtb46-deficient cDCs showed increased levels of activation and a distorted splenic DC composition with decreased numbers of CD4+ DCs,48 which is similar to miR-142−/− mice. However as for the miR-142−/− mice, the mechanism leading to decreased numbers of CD4+ DCs in Zbtb46−/− mice remains unclear.
We consistently detected a small fraction of residual CD4+ DCs in the absence of miR-142, which suggests unimpaired DC development but a defective DC homeostasis/maintenance. Supporting this notion, we observed a trend toward more DC death, particular in miR-142–deficient CD4+ DCs. This finding is consistent with the mixed BM chimera experiment in which we were able to demonstrate an out-competition of the miR-142−/− CD4+ DCs by miR-142+/+ CD4+ DCs.
DCs represent the primary APC population in the immune system and are therefore indispensable for the initiation of the adaptive immune response. The reduced numbers and functional dysregulation of CD4+ DCs in miR-142–deficient mice resulted in specific impairment of CD4+ T-cell responses, corroborating the supremacy of CD4+ DCs as APCs for MHCII-restricted antigens.38
What might be the mechanism behind the observed phenotypes? As mentioned earlier, orchestrated expression of the IRF4 and IRF8 facilitates the development of CD4+ and CD8α+ cDCs, respectively,12,14 and dysregulated expression of these transcription factors might affect the distribution of the subsets. Affymetrix Gene microarray analysis of FLT3-L–driven in vitro BM cultures revealed a developmental shift of miR-142−/− DC precursors toward a CD8+ DC fate. Indeed, analysis of the mixed BM chimeras indicated that miR-142–deficient CD8α+ DCs displayed a slight competitive advantage over their WT counterparts in vivo. Expression of the transcription factor IRF8, known to be critical for CD8α+ DC development,12 was down-regulated in the WT CD4+ DC equivalents, which is consistent with published expression data of splenic CD4+ and CD8α+ DCs.49 However, IRF8 expression was elevated in the miR-142–deficient CD4+ DC equivalents and overexpression of IRF8 can facilitate inflammatory gene expression.50 This suggests that part of miR-142 function in CD4+ DCs may be to repress IRF8, the expression of which might be incompatible with the maintenance and maturation of CD4+ DC identity. However, according to available target prediction algorithms, IRF8 is not a direct miR-142 target.
The miR-142 mRNA targets required for the development and maintenance of CD4+ DCs remain unclear. miRNAs can target multiple mRNAs. Therefore, we assume that the observed phenotype of miR-142–deficient mice is the result of an orchestrated interplay of multiple targets repressed by either miR-142-3p or miR-142-5p. Future research will be required for uncovering the precise mechanism by which miR-142 influences the development of CD4+ DCs.
The results of the present study demonstrate that individual mononuclear phagocyte populations and their precursors can be defined by specific miRNA signatures. Furthermore, our analysis identified miR-142 as a specific regulator for CD4+ DC homeostasis and identified an miRNA—in addition to transcription factors and cytokines—that is necessary for the maintenance of an innate immune cell type.
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the staff members of the Weizmann sorting facility, in particular Ayala Sharp and Eitan Ariel, for excellent technical support; the members of the biologic service unit, David Pfilzer and Gilgi Friedlander; the other Jung laboratory members for stimulating discussions; and Rita Krauthgamer for technical assistance.
A.M. is a fellow of the Minerva Foundation. This study was supported by the Leir Charitable Foundation, the Wolfson Family Charitable Trust, the Israeli Science Foundation, and the Deutsche Forschungsgemeinschaft Research Unit 1336.
Authorship
Contribution: A.M. performed the experiments; E.C. and E.H. generated the miR-142−/− mice; O.M., Z.B.I., and E.F. helped with the bioinformatics analysis; S.Y., K.-W.K., T.A., and D.V. provided expert technical help; G.B. established the mutant mouse strain;. I.A. provided data; and A.M. and S.J. designed the experiments and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Steffen Jung, PhD, Department of Immunology, The Weizmann Institute of Science, PO Box 26, Rehovot 76100, Israel; e-mail: s.jung@weizmann.ac.il.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal