In this issue of Blood, McNerney and colleagues identify CUX1 as a tumor suppressor gene (TSG) on the long arm of chromosome 7, showing frequent inactivation in acute myeloid leukemia (AML).1
Complete loss of chromosome 7 (monosomy 7) and partial deletion of the long arm of chromosome 7 [del(7q)] are recurring karyotypic abnormalities in malignant myeloid disorders, including the myelodysplastic syndromes (MDS) and AML, and are associated with a poor prognosis.2,3 The del(7q) in MDS and AML is considered to mark the location for a gene or genes the loss of which may affect important processes such as growth control and normal hematopoiesis. The identification of significant genes associated with chromosome deletions, including the del(7q), in human leukemia has proven challenging, however.
The basis for research on deletions such as the del(7q) in MDS and AML is well known. The first step is to characterize the deletions and to identify the commonly deleted region (CDR), the region of deletion shared by all patients, as this localizes the gene(s) for further study. Over the years several CDRs mapping to 7q have been identified in MDS and AML, including CDRs at 7q22, 7q32-33, and 7q35-36.2-4 The next step typically involves the sequencing of all the candidate genes that map within the CDR in a group of affected patients. The gene sequencing is critical to our understanding of the disease pathogenesis; if the Knudsen 2-hit model applies, there would be loss of 1 allele of a gene and a mutation of the remaining copy of the same gene. The lack of recurrent mutations identified in the genes mapping to the various CDRs identified on 7q in MDS and AML patients with −7/del(7q) suggests that haploinsufficiency, a dosage effect resulting from the loss of a single allele of a gene,5 may be the molecular mechanism relevant in this group of malignancies. There has been growing recognition of haploinsufficiency as a cancer model over the past decade and the importance of this model in the context of myeloid disorders is supported by recent studies concerning MDS patients with the 5q− syndrome.6
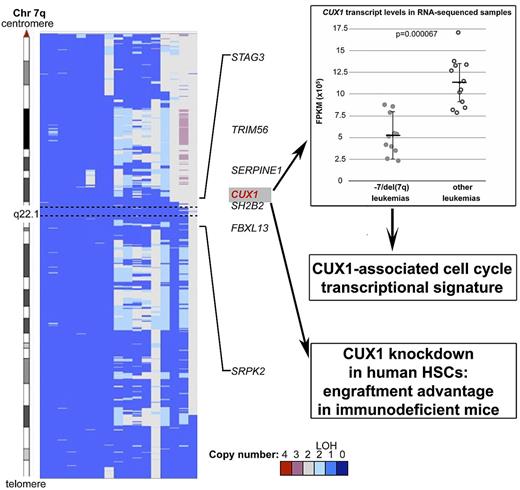
Interest in the CUX1 (CUTL1) gene, encoding a transcription factor, as a possible candidate gene in malignant myeloid disorders with abnormalities of chromosome 7, stretches back to the mid-1990s when it was first mapped to the CDR at 7q22 by investigators.2,7 Most recently CUX1, normally highly expressed in multipotent hematopoietic progenitors, was shown to be expressed at reduced levels in CD34+ cells from patients with MDS with −7/del(7q).4 McNerney and colleagues have used SNP array analysis to refine the mapping of the CDR at 7q22, identifying a 2.17Mb CDR at 7q22.1 containing the CUX1 gene in patients with de novo and therapy-related malignant myeloid disorders with −7/del(7q) (see figure).1 Transcriptional profiling using a sensitive RNA sequencing method revealed that CUX1 was haploinsufficient in this patient group and was, moreover, the most significantly differentially expressed transcript within the CDR (see figure).1 Intriguingly, in 1 patient the CUX1 gene was shown to be disrupted by a translocation, resulting in a loss-of-function RNA fusion transcript.1
Copy number analysis of 7q derived from SNP arrays of leukemia samples with −7/del(7q) shows that CUX1 maps within the 2.17 Mb CDR at 7q22.1. RNA-sequencing data showed that CUX1 is expressed at haploinsufficient levels in −7/del(7q) leukemias and this is associated with a cell-cycle transcriptional gene signature. Haploinsufficiency of CUX1 gave human hematopoietic cells a significant engraftment advantage upon transplantation into immunodeficient mice. Adapted from Figure 1 and Figure 3A in the article by McNerney et al that begins on page 975.1
Copy number analysis of 7q derived from SNP arrays of leukemia samples with −7/del(7q) shows that CUX1 maps within the 2.17 Mb CDR at 7q22.1. RNA-sequencing data showed that CUX1 is expressed at haploinsufficient levels in −7/del(7q) leukemias and this is associated with a cell-cycle transcriptional gene signature. Haploinsufficiency of CUX1 gave human hematopoietic cells a significant engraftment advantage upon transplantation into immunodeficient mice. Adapted from Figure 1 and Figure 3A in the article by McNerney et al that begins on page 975.1
The CUX1 transcription factor regulates many genes including several involved in DNA replication and chromosome segregation.8 Cell-based assays have established a role for CUX1 in the control of cell-cycle progression, cell motility, and invasion.8 Within the RNA-sequencing data obtained from the −7/del(7q) leukemia patients studied by McNerney et al there was enrichment of gene transcripts known to play a role in the mitotic cell cycle, many of which were direct targets of CUX1.1 Thus CUX1 might act as a TSG in myeloid cells through the regulation of genes involved in the control of the cell cycle.
CUX1 is highly conserved between humans and Drosophila and to investigate the hypothesis that CUX1 acts as a TSG in vivo, McNerney et al performed animal modeling experiments using Drosophila. A targeted RNAi approach in developing Drosophila hemocytes demonstrated that haploinsufficiency of the ortholog cut resulted in increased hemocyte proliferation and melanotic tumor formation in developing larvae. Similarly, partial knockdown of CUX1 in human hematopoietic progenitor cells resulted in increased hematopoietic engraftment upon transplantation into immunodeficient mice.1 These data suggest that cut/CUX1 acts as a TSG important in the regulation of normal hematopoietic cell growth in Drosophila and humans.
The study by McNerney et al provides good evidence that CUX1 is a haploinsufficient TSG involved in malignant myeloid disorders with −7/del(7q).1 However, whether CUX1 is a causal gene in relation to producing a clonal human hematopoietic disorder, and whether haploinsufficiency of an additional gene (or genes) mapping to chromosome 7 play a role in disease pathogenesis, remain important questions. Deletions of chromosome 7q are typically large and it is highly likely that the loss of several genes along chromosome 7q will have various phenotypic effects in patients thus affected. Clearly, mouse knockout models might prove very informative in relation to this question. Mutation of other genes on 7q may also play an important role in disease pathogenesis. A good example is the EZH2 gene, mapping to 7q36, mutation of which is associated with 7q uniparental disomy or 7q36.1 microdeletion, but not generally with monosomy 7 and del(7q), in malignant myeloid disorders.9 Whether CUX1 is inactivated as a result of point mutations in malignant myeloid disorders remains to be fully determined; to date, only 1 missense mutation has been described in a secondary AML patient.10
Finally, in contrast to malignant myeloid disorders with abnormalities of chromosome 7, frequent over-expression of CUX1 has been reported in solid tumors of various types, including breast tumors and cancer cell lines, and in this setting it has been proposed that elevated CUX1 expression plays an important role in tumor progression.8 Clearly, aberrant expression of CUX1, either increased or reduced compared with normal levels in a given cell type, may play an important role in human tumorigenesis: CUX1 dosage matters!
Conflict-of-interest disclosure: The author declares no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal