Abstract
t(8;21) is one of the most frequent chromosomal translocations occurring in acute myeloid leukemia (AML) and is considered the leukemia-initiating event. The biologic and clinical significance of microRNA dysregulation associated with AML1/ETO expressed in t(8;21) AML is unknown. Here, we show that AML1/ETO triggers the heterochromatic silencing of microRNA-193a (miR-193a) by binding at AML1-binding sites and recruiting chromatin-remodeling enzymes. Suppression of miR-193a expands the oncogenic activity of the fusion protein AML-ETO, because miR-193a represses the expression of multiple target genes, such as AML1/ETO, DNMT3a, HDAC3, KIT, CCND1, and MDM2 directly, and increases PTEN indirectly. Enhanced miR-193a levels induce G1 arrest, apoptosis, and restore leukemic cell differentiation. Our study identifies miR-193a and PTEN as targets for AML1/ETO and provides evidence that links the epigenetic silencing of tumor suppressor genes miR-193a and PTEN to differentiation block of myeloid precursors. Our results indicated a feedback circuitry involving miR-193a and AML1/ETO/DNMTs/HDACs, cooperating with the PTEN/PI3K signaling pathway and contributing to leukemogenesis in vitro and in vivo, which can be successfully targeted by pharmacologic disruption of the AML1/ETO/DNMTs/HDACs complex or enhancement of miR-193a in t(8;21)–leukemias.
Key Points
AML1/ETO triggers the heterochromatic silencing of miR-193a and PTEN by binding at AML1-binding sites and recruiting epigenetic enzymes.
A feedback circuitry involving miR-193a and AML1/ETO/DNMTs/HDACs, cooperating with the PTEN/PI3K pathway and contributing to leukemogenesis.
Introduction
t(8;21) is one of the most common recurrent chromosomal translocation in acute myeloid leukemia (AML). It involves the AML1 (also known as RUNX1) gene, which is a DNA-binding transcription factor required for hematopoiesis, and the ETO (also known as RUNX1T1) gene, which is a corepressor molecular expressed in a variety of tissues.1 Many studies have documented the complex and multifaceted functions of the AML1/ETO fusion protein created by t(8;21), including inhibiting differentiation and subsequent apoptosis, and activating signals for cell proliferation.2,3 However, the fusion gene alone is not sufficient for inducing leukemia and additional genetic abnormalities are probably involved.4
MicroRNAs (miRNAs) have been recently found to play an important role in the mechanisms that regulate the lineage differentiation fate of hematopoietic cells by modulating the expression of oncogenes or tumor suppressors.5,6 Deregulation of miRNA expression has been shown to be involved in multistep carcinogenesis and has rapidly emerged as a novel therapeutic target.7,8 In AML, the miR-223 activity is linked to the differentiation fate of myeloid precursors, and the expression of AML1/ETO triggers heterochromatic silencing of genomic regions generating miR-223.5 Another miRNA, miR-29b contributes to myeloid leukemogenesis through up-regulation of DNMTs and KIT.9 The expression level of KIT is higher in AML1/ETO positive t(8;21)–AML than in other leukemias and predicts poor outcome, thereby supporting a critical role of KIT overexpression in t(8;21) leukemogenesis.4 Despite these observations, the mechanistic link between AML1/ETO fusion proteins and overexpression of KIT receptors has not yet been determined.
Recent studies have shown that PTEN/PI3K signaling is frequently activated in blast cells of AML patients and strongly contributes to the proliferation, survival and drug resistance of these cells.10 The activity of PTEN, a negative regulator of oncogenic PTEN/PI3K signaling, is lost through mutations, deletions or promoter methylation silencing at high frequency in many human cancers including leukemia.11 However, in AML, the mutations in the PTEN gene are very infrequent, which suggest that the down-regulation of PTEN expression is probably because of PTEN epigenetic deregulation.12 If this is proven to be true in future studies, the epigenetic silencing of PTEN promoter would represent a novel therapeutic target.
In this study, we provide evidence that epigenetic silencing of miR-193a is the mechanistic link between AML1/ETO fusion proteins and overexpression of KIT receptor, down-regulation of PTEN, and activation of the PTEN/PI3K signal pathway. Our findings also identify the AML1/ETO corepressor complex, miR-193a, and PTEN as novel and crucial players in the molecular circuitry controlling human myelopoiesis.
Methods
Clinical samples
The study was approved by the local internal review boards and ethics committees. Written informed consent was obtained from all subjects in accordance with the Declaration of Helsinki. Normal mononuclear cells were isolated from the bone marrow (BM) or peripheral blood (PB) of consenting healthy donors. Leukemia blasts were obtained from the BM of 123 leukemia patients. Cases were classified according to the FAB classification and showed an initial percentage of greater than 60% circulating blasts.13
Cell lines and cell cultures
HL60, U937, U937-A/E-HA, Kasumi-1, SKNO-1, SKNO-1-siA/E-RNA, and KG1 cell lines were maintained in RPMI 1640 medium supplemented with 50 μg/mL streptomycin, 50 IU penicillin, and 10% FCS. SKNO-1 infection with the pRRLcPPT.hPGK, the lentiviral vector encoding the previously described siAGF1 oligonucleotides (sense, 5′-CCUCGAAAUCGUACUGAGAAG-3′, antisense, 5′-UCUCAGUACGAUUUCGAGGUU-3′) against the AML1/ETO mRNA fusion site (siA/E-RNAs) allowed the silencing of AML1/ETO in SKNO-1 cells.5,14-16 The U937-A/E-HA clone was obtained by electroporation into U937 wild-type (WT) cells of an HA-tagged AML1/ETO cDNA subcloned into a vector carrying the Zn2+-inducible mouse MT-1 promoter. Human embryonic kidney (HEK) 293T cells were cultured in DMEM medium supplemented with 50 μg/mL streptomycin, 50 IU penicillin, and 10% FCS. The lentiviral vector Lenti-193a (System Biosciences) was used for the ectopic induction of miR-193a into the SKNO-1 to generate the Lenti-193a cells. Treatment with 5-Azacytidine (Sigma-Aldrich) and SAHA (Sigma-Aldrich) was performed at a concentration of 2.5μM and 3.0μM, respectively for 40 hours.
RNA extraction and analysis
Total RNA was extracted from cells using the TriPure isolation system (Roche). The relative quantity of miR-193a was measured on 200 ng of total RNA by qRT-PCR using the mir-Vana qRT-PCR detection kit (Ambion, Applied Biosystems) in the ABI PRISM 7500 sequence detection system (Applied Biosystems), and it was determined by the comparative CT method using snRNA U6 levels for normalization as recommended in the manufacturer's instructions.
Immunoblot assays
Immunoblot assays were performed on total cell lysates (50 μg) using the monoclonal antibodies anti-AML1/RHD (Calbiochem), anti-ETO (Santa Cruz Biotechnology), anti-KIT (Santa Cruz Biotechnology), anti–p-PDK1 (Cell Signaling Technology), anti-p-AKT (Cell Signaling Technology), anti–p-Gsk3β (Cell Signaling Technology), the anti–Cyclin D1 (Cell Signaling Technology), anti-MDM2 (Cell Signaling Technology) or anti-PTEN (Cell Signaling Technology). The anti–β-actin (Santa Cruz Biotechnology) was used to normalize the amount of the samples analyzed. The immunoreactivity was determined by the ECL method (Amersham Biosciences).
Transactivation assays
DNA fragments were PCR amplified from human genomic DNA. Primer sequences are shown in supplemental Table 5 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). All fragments were inserted in the pGL3-LUC reporter vector (Promega). The mutants were generated by the overlap-extension PCR method. HEK293T cells (2 × 105) were plated in 24-well plates and transiently cotransfected by the SuperFect (QIAGEN) with 10, 50, or 100 ng of the pcDNA3 vectors with or without the AML1/ETO cDNAs, and 400 ng of the LUC reporter constructs described above. A cotransfected pRL-TK renilla luciferase reporter vector (Promega) was used as an internal control. Cells were harvested 48 hours after transfection and assayed using the dual luciferase assay (Promega) according to the manufacturer's instructions.
ChIP assay
Formaldehyde (1% final concentration) was added to cultured cells (2 × 106) and incubated for 10 minutes at 37°C to crosslink the proteins to DNA. After sonication, the chromatin was immunoprecipitated overnight with 5 μg of the following antibodies recognizing AML1 (Santa Cruz Biotechnology), ETO (Santa Cruz Biotechnology), DNMT1 (Abcam), DNMT3a (Abcam), DNMT3b (Abcam), and HDAC1 (Abcam). Chromatin immunoprecipitation (ChIP) using the normal mouse IgG (Abcam) antibody was performed on naked and sonicated DNA extracted from the same cell samples and assayed with the EZ-ChIP chromatin immunoprecipitation kit (Millipore) according to the manufacturer's instructions. Genomic pre-miR-193a and PTEN upstream regions close to the putative AML1-binding site were amplified. Primers sequences are shown in supplemental Table 4. GAPDH served as a control for nonspecific precipitated sequences.
Bisulfite modification and genomic sequencing
The methylation status of the CpG dinucleotides within regions (nt − 557 to − 256) relative to the pre-miR-193a gene, and (nt − 1180 to − 755) relative to the transcriptional start site (TSS) of the PTEN gene were analyzed. Bisulfite sequencing assay was performed on 1 μg of bisulfite-treated genomic DNA from the indicated cell lines. After bisulfite conversion performed with EpiTect bisulfite kit (QIAGEN) as previously described,17 the fragments of interest were amplified. Primers sequences are shown in supplemental Table 4. PCR products were gel purified and cloned into the pGEM-T vector systems (Promega). Individual bacterial colonies were used for PCR using vector-specific primers, and the products were sequenced for the analyses of DNA methylation.
Reverse screening the miRNAs regulating ETO, DNMT3A, HDAC3, KIT, CCND1, and MDM2 by luciferase assay
To screen the miRNAs that directly regulate the expression of ETO, DNMT3A, HDAC3, KIT, CCND1, and MDM2, the 3′-UTRs of these genes were cloned into a modified pGL-3 control vector, placing the 3′-UTRs with all potential miRNA binding sites downstream of the coding sequence of luciferase. The transfection mixtures contained 100 ng of firefly luciferase reporter and 400 ng of plasmids expressing miR-193a primary transcripts, derived from a constructed miRNA expression library. pRL-TK (Promega) was transfected as an internal control. The firefly luciferase activities were assayed using the dual luciferase assay (Promega) 48 hours after transfection.
Cell differentiation, cell cycle, apoptosis, and colony forming unit analysis
Cell differentiation was evaluated by light microscopic morphologic examination of Wright-Giemsa stained cytospins. The cells were also stained with anti-CD11b (BD Bioscience, PharMingen). A minimum of 50 000 events were collected for each sample by FACScan flow cytometry (Becton Dickinson) using CellQuest Pro software (Becton Dickinson) for data acquisition and analysis. For cell cycle analysis, cells transfected with vector or lenti-193a, were harvested and fixed overnight in ice-cold ethanol (70% vol/vol). The cells were stained for 3 hours at 4°C with 50 μg/mL PI (Calbiochem) in PBS containing 0.5 μg/mL RNase A (Sigma-Aldrich). For apoptosis assay, the cells were stained with annexin V and 7AAD (BD Bioscience) and analyzed by FACSCalibur flow cytometer (Becton Dickinson). Colony forming unit assays were performed using the methylcellulose H4230 (StemCell Technologies) culture system according to the manufacturer's instructions. Localized clusters of > 50 cells showing morphologic hematopoietic characteristics were counted as colonies after incubation for 10 days at 37°C and 5% CO2 in a humidified atmosphere.
Murine models
All mice used in this study were bred and maintained in a pathogen-free environment. The xenograft model in nude mice was established by subcutaneous inoculation of 2 × 107 SKNO-1 cells into the right flank. Tumor size was measured daily until it reached 50 mm3 after which 5 μg of synthetic miR-193a or scramble (supplemental Table 6) diluted in lipofectamine (Invitrogen) solution (100 μL total volume) was injected directly into the tumors on days 1, 3, 7, and 10. Tumors were measured on the day of the injections and 4 days after the last injection when the animals were killed. Necropsies were performed and tumors were weighed. Tumor volumes were calculated using the equation V (in mm3) = A × B2/2, where A is the largest diameter and B is the perpendicular diameter.
Statistical analysis
SPSS 13.0 software was used to process the data. The Wilcoxon signed-rank test was selected to determine the differences of miR-193a, PTEN, and CCND1 expression among clinical samples. Eighty-nine patients were grouped into quartiles according to miR-193a expression levels and divided into high (miR-193a-H, n = 22) and low (miR-193a-L, n = 67) based on the trend observed in clinical outcome after performing a Cox regression analysis event-free survival (EFS) and overall survival (OS) with miR-193a quartile grouping as the independent variable. Multivariate survival analysis was carried out using a Cox regression model. Student t test was used to determine the statistical significance of experimental results and a P value of .05 or less was considered significant.
Results
miR-193a expression is down-regulated in AML1/ETO-positive leukemia cells
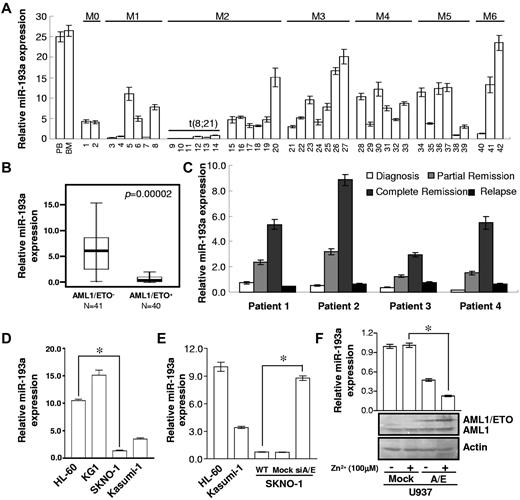
We previously demonstrated that KIT, a tyrosine kinase receptor, is a target gene of miR-193a.18 The expression levels of miR-193a were quantified from human hematopoietic cells isolated from healthy donor (n = 3) and from diagnostic samples of AML patients (n = 42; supplemental Table 1). We found that miR-193a is expressed at higher levels in total nucleated cells from PB and BM from healthy donors than in cells isolated from AML patients. The lowest level of miR-193a expression was observed in 6 t(8;21) AML-M2 subtype (by FAB classification13 ; Figure 1A).
miR-193a levels in leukemia and cell lines. Relative qRT-PCR quantification of miR-193a level in mononucleated cells (MNC) isolated from 42 leukemia patients (A) and 81 AML FAB M2 subtype patients (B). Cases were grouped into 2 categories: those with (AML1/ETO+) and without (AML1/ETO−) t(8;21). AML1/ETO− cases had higher miR-193a expression levels (P = .0005). (C) Serial analyses of miR-193a levels in samples isolated from 4 individual leukemia patients with disease progression. (D) Relative quantification of miR-193a levels in the indicated leukemia cell lines (*P = .0007). (E) Relative quantification of miR-193a levels in HL-60, Kasumi-1, SKNO-1 (WT, mock, and siA/E). The results represent the average of 3 independent evaluations ± SD (*P = .001). (F) Top panel: Relative quantification of miR-193a levels in U937 (Mock and A/E-HA) cells. The results represent the average of 3 independent evaluations ± SD (*P = .006). Bottom panels: Immunoblot analysis for AML1 and AML1/ETO with an anti-AML1 antibody. β-actin was used for protein loading control. Zn2+ treatment (100μM for 16 hours) was used to increase the expression of AML1/ETO in U937 cells.
miR-193a levels in leukemia and cell lines. Relative qRT-PCR quantification of miR-193a level in mononucleated cells (MNC) isolated from 42 leukemia patients (A) and 81 AML FAB M2 subtype patients (B). Cases were grouped into 2 categories: those with (AML1/ETO+) and without (AML1/ETO−) t(8;21). AML1/ETO− cases had higher miR-193a expression levels (P = .0005). (C) Serial analyses of miR-193a levels in samples isolated from 4 individual leukemia patients with disease progression. (D) Relative quantification of miR-193a levels in the indicated leukemia cell lines (*P = .0007). (E) Relative quantification of miR-193a levels in HL-60, Kasumi-1, SKNO-1 (WT, mock, and siA/E). The results represent the average of 3 independent evaluations ± SD (*P = .001). (F) Top panel: Relative quantification of miR-193a levels in U937 (Mock and A/E-HA) cells. The results represent the average of 3 independent evaluations ± SD (*P = .006). Bottom panels: Immunoblot analysis for AML1 and AML1/ETO with an anti-AML1 antibody. β-actin was used for protein loading control. Zn2+ treatment (100μM for 16 hours) was used to increase the expression of AML1/ETO in U937 cells.
To confirm this observation, 81 additional AML-M2 subtype cases (supplemental Table 2) were sorted into 2 categories, positive for AML1/ETO (AML1/ETO+) and negative (AML1/ETO−). The expression level of miR-193a was decreased by 7.5-fold in AML1/ETO+ cases (Figure 1B). In 4 AML1/ETO+ patients, the miR-193a expression levels were high in cells isolated after induction chemotherapy and at achievement of complete remission, and low at relapse (Figure 1C, supplemental Table 3). In analogy to this, lower expression levels of miR-193a were found in AML1/ETO+ Kasumi-1 and SKNO-1 cell lines, compared with AML1/ETO− HL-60 and KG1 cell lines (Figure 1D).
To evaluate its clinical significance, miR-193a expression was assessed in 89 patients with t(8;21) AML-M2. These cases were accordingly divided into the miR-193a-H group (n = 22, with higher level of miR-193a by quantitative RT-PCR) and the miR-193a-L group (n = 67, with lower but still detectable level of miR-193a by quantitative RT-PCR (supplemental Table 4, supplemental Figure 1B). Clinically, miR-193a overexpression was related to long event-free and overall survival time (P = .046 and P = .039, respectively; supplemental Figure 1C-E). Taken together, these data suggest that miR-193a overexpression indicates good disease outcome in t(8;21) AML-M2.
To test the putative role of AML1/ETO in the regulation of miR-193a expression, AML1/ETO was knocked-out in SKNO-1 cells (SKNO-1-siA/E) using lentiviral vector. It was observed that the expression of endogenous levels of miR-193a increased by 9-fold in SKNO-1 siA/E cells compared with SKNO-1 or mock cells, and reached to similar measurable levels as observed in HL-60 cell lines (Figure 1E). Similar results were observed in Kasumi-1 cells when the AML1/ETO was inhibited by synthesis siRNA (supplemental Figure 2A). In U937 cells ectopically expressing an HA-tagged AML1/ETO (U937-A/E-HA) in a zinc-inducible manner,5,15 the miR-193a measurable levels were reduced up to 25% relative to U937 mock cells (Figure 1F). These results suggested that a functionally negative correlation exists between miR-193a and AML1/ETO levels.
AML1/ETO protein localizes at an AML1-binding site and triggers the epigenetic silencing of miR-193a genomic region
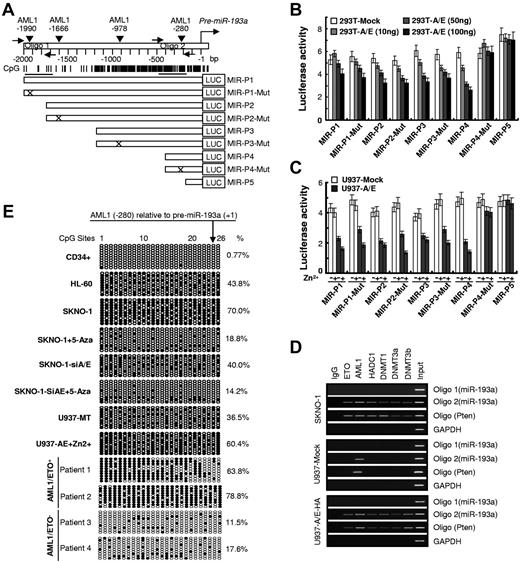
AML1/ETO acts as a transcriptional repressor of AML1 target genes. A bioinformatics search (http://www.cbrc.jp/research/db/TFSEARCH.html) at the 5′ end of the predicted “core promoter” sequence revealed the presence of 4 putative AML1 binding sites surrounded by “CpG islands” on the pre-miR-193a upstream region (nt − 2000 to +1 relative to pre-miR-193a; Figure 2A). When luciferase reporter constructs containing WT (MIR-P1, MIR-P2, MIR-P3, MIR-P4, and MIR-P5) and mutated (MIR-P1-Mut, MIR-P2-Mut, MIR-P3-Mut, and MIR-P4-Mut) sequences of the pre-miR-193a regulatory region (Figure 2A bottom panel) were cotransfected with increasing amounts of AML1/ETO-containing or empty vectors into 293T cells, the expression of AML1/ETO caused a dose-dependent decrease in the luciferase reporter activity of the MIR-P1, MIR-P2, MIR-P3, and MIR-P4 and their mutants MIR-P1-Mut, MIR-P2-Mut, MIR-P3-Mut, but not of the MIR-P4-mut and MIR-P5 that lack any functional AML1 binding site (Figure 2B). When these reporter vectors were transfected into U937 (Mock and −A/E-HA) cells under Zn2+ treatment, similar results were obtained (Figure 2C). These results suggested that the proximal AML1 binding site on the pre-miR-193a putative regulatory region contributes to the AML1/ETO-dependent silencing of miR-193a.
AML1/ETO/DNMTs/HDACs complex acts on the AML1 dna-binding site present on the upstream sequence of pre-miR-193a gene and alters its epigenetic status. (A) Schematic diagrams of the AML1 binding sites and of the CpG islands along the miR-193a genes. Numbers are the nucleotides relative to pre-miR-193a (+1). Vertical arrows indicate AML1 binding sites, horizontal arrows indicate the location of the primers used in the ChIP assay. Vertical lines indicate CpG dinucleotides, horizontal bars below the CpG sites show the regions analyzed by bisulfite sequencing. (B) Human 293T cells were transiently cotransfected for 48 hours with luciferase reporter containing the WT sequence of the miR-193a regulatory regions or its counterpart mutants, and increasing amounts (10, 50, and 100 ng) of pcDNA3.0 with AML1/ETO cDNA or without (293T-mock). (C) U937 (mock and −A/E-HA) cells were transiently transfected with luciferase reporter and treated with Zn2+ (100μM for 16 hours). (D) Chromatin was immunoprecipitated using the indicated antibodies or IgG. PCR was performed using oligo 1 primers designed for the amplification of a distal region on miR-193a gene containing the predicted AML1 binding site to evaluate the specificity of protein binding. Oligo 2 primers were designed to amplify DNA sequences surrounding the proximal AML1-binding site. PTEN primers were designed to amplify DNA sequences surrounding the AML1-binding site (− 2033) and the CpGs in the PTEN gene regulatory region. Input shows the amplification from sonicated chromatin. Amplification of GAPDH was a control for nonspecific precipitated sequences. (E) Genomic bisulfite sequencing was performed to detect the methylation status of the DNA sequences surrounding the AML1-binding site (− 280) in the pre-miR-193a gene upstream region from CD34+ hematopoietic progenitors isolated from PB of healthy donors (CD34+) and the indicated leukemia cell lines and leukemic blasts. Each row of circles represents the sequence of an individual clone. Black circles and empty circles represent methylated and unmethylated CpG dinucleotides, respectively. Cells were either untreated or treated with 2.5μM 5-Aza for 40 hours. For each sample, the percentages of global methylation level of these regions on the miR-193a gene are indicated.
AML1/ETO/DNMTs/HDACs complex acts on the AML1 dna-binding site present on the upstream sequence of pre-miR-193a gene and alters its epigenetic status. (A) Schematic diagrams of the AML1 binding sites and of the CpG islands along the miR-193a genes. Numbers are the nucleotides relative to pre-miR-193a (+1). Vertical arrows indicate AML1 binding sites, horizontal arrows indicate the location of the primers used in the ChIP assay. Vertical lines indicate CpG dinucleotides, horizontal bars below the CpG sites show the regions analyzed by bisulfite sequencing. (B) Human 293T cells were transiently cotransfected for 48 hours with luciferase reporter containing the WT sequence of the miR-193a regulatory regions or its counterpart mutants, and increasing amounts (10, 50, and 100 ng) of pcDNA3.0 with AML1/ETO cDNA or without (293T-mock). (C) U937 (mock and −A/E-HA) cells were transiently transfected with luciferase reporter and treated with Zn2+ (100μM for 16 hours). (D) Chromatin was immunoprecipitated using the indicated antibodies or IgG. PCR was performed using oligo 1 primers designed for the amplification of a distal region on miR-193a gene containing the predicted AML1 binding site to evaluate the specificity of protein binding. Oligo 2 primers were designed to amplify DNA sequences surrounding the proximal AML1-binding site. PTEN primers were designed to amplify DNA sequences surrounding the AML1-binding site (− 2033) and the CpGs in the PTEN gene regulatory region. Input shows the amplification from sonicated chromatin. Amplification of GAPDH was a control for nonspecific precipitated sequences. (E) Genomic bisulfite sequencing was performed to detect the methylation status of the DNA sequences surrounding the AML1-binding site (− 280) in the pre-miR-193a gene upstream region from CD34+ hematopoietic progenitors isolated from PB of healthy donors (CD34+) and the indicated leukemia cell lines and leukemic blasts. Each row of circles represents the sequence of an individual clone. Black circles and empty circles represent methylated and unmethylated CpG dinucleotides, respectively. Cells were either untreated or treated with 2.5μM 5-Aza for 40 hours. For each sample, the percentages of global methylation level of these regions on the miR-193a gene are indicated.
ChIP assay performed in U937-A/E and SKNO-1 cells using specific antibodies, revealed the presence of AML1/ETO, HDAC1, DNMT1, DNMT3a, and DNMT3b at the pre-miR-193a chromatin regulatory regions surrounding the AML1-binding site (Figure 2D). In this region, the higher frequency of methylated CpG dinucleotides encompassing the endogenous pre-miR-193a gene sequences in U937-A/E-HA (60.4%) and in SKNO-1 cells (70.0%) compared with U937 (36.5%), HL60 cells (43.8%), and normal human bone marrow blasts (0.77%) was demonstrated by genomic bisulfite sequencing, thereby confirming the basal hypermethylated status of these CpGs in AML1/ETO+ cells (Figure 2E). Knockout of AML1/ETO (SKNO-1-siA/E cells) induced a 40% decrease in CpGs methylation levels of the pre-miR-193a gene and reactivated miR-193a expression (Figures 1E, 2E). In AML-M2 patients, the higher frequency of methylated CpG dinucleotides encompassing the endogenous pre-miR-193a gene sequences in AML1/ETO+ blasts compared with AML1/ETO− blasts (Figure 2E). Thus, the chromatin remodeling complex aberrantly formed by AML1/ETO, and the hypermethylation of the “CpG islands” present in the AML1 binding sites on the pre-miR-193a regulatory region appear to be key regulatory mechanisms for transcriptional silencing of miR-193a.
miR-193a regulates AML1/ETO/DNMT3a/HDAC3 complex, KIT, CCND1, and MDM2 expression directly, and increases PTEN expression indirectly
By computational analysis using miRNA target prediction programs, such as Target Scan (http://www.targetscan.org) and miRanda (http://www.microrna.org), we found that ETO, DNMT3a, and HDAC3 are putative targets of miR-193a (Figure 3A). Cells cotransfected with ETO, DNMT3a, and HDAC3 reporters and pre-miR-193a exhibited approximately 40% to 60% reduction of luciferase activity with respect to that cotransfected with the scramble oligonucleotide (Figure 3B). Consistent with these results, we also showed that overexpression of miR-193a did not reduce the luciferase activity from the reporter construct containing mutants of ETO, DNMT3a, and HDAC3 3′-UTRs (Figure 3A-B). Western blot analysis revealed that miR-193a markedly reduced the endogenous levels of ETO, DNMT3a, and HDAC3 proteins compared with scramble transfected cells (Figure 3C). AML1/ETO is the fusion product of the t(8;21), wherein the C-terminal contains almost full-length ETO including its intact 3′-UTR.19 Western blot analysis revealed that the AML1/ETO protein level was markedly reduced by synthetic miR-193a (Figure 3C). All these results demonstrated that miR-193a regulates the expression of AML1/ETO, DNMT3a, and HDAC3 by targeting its putative binding sites and indicated that a feedback minicircuitry consisting of AML1/ETO/DNMTs/HDACs corepressor and miR-193a is operative in AML1/ETO+ blasts.
miR-193a targets ETO, DNMT3a, HDAC3, KIT, CCND1, and MDM2 directly, and PTEN indirectly. (A) The sequences indicate the putative interaction sites between the miR-193a seed region (nucleotides 2-8) and the 3′ UTRs of ETO, DNMT3a, HDAC3, KIT, CCND1, and MDM2 WT or mutated derivatives. (B) pCDNA3.0 control plasmids and miR-193a–expressing plasmids were cotransfected with a modified luciferase pGL-3 control vector containing ETO, DNMT3a, HDAC3, KIT, CCND1, and MDM2 3′-UTRs or their mutants as indicated. Luciferase assays were performed 48 hours after transfection. Each histogram shows normalized mean values of relative luciferase activity from 3 independent experiments. Normalized luciferase activity in the absence of miR-193a expression vector was set to 100%. (C) The effectiveness of synthetic miR-193a on ETO, DNMT3a, HDAC3, AML1/ETO, KIT, CCND1, and MDM2 proteins was analyzed by Western blot. (D) AML1/ETO+ had higher KIT levels (P = .0004). (E) miR-193a increases PTEN expression indirectly. SKNO-1 cells (mock, and si-A/E) were transfected with synthetic miR-193a or scramble oligonucleotides for 48 hours. Relative PTEN levels were evaluated by qRT-PCR. The results represent the average of 3 independent evaluations ± SD.
miR-193a targets ETO, DNMT3a, HDAC3, KIT, CCND1, and MDM2 directly, and PTEN indirectly. (A) The sequences indicate the putative interaction sites between the miR-193a seed region (nucleotides 2-8) and the 3′ UTRs of ETO, DNMT3a, HDAC3, KIT, CCND1, and MDM2 WT or mutated derivatives. (B) pCDNA3.0 control plasmids and miR-193a–expressing plasmids were cotransfected with a modified luciferase pGL-3 control vector containing ETO, DNMT3a, HDAC3, KIT, CCND1, and MDM2 3′-UTRs or their mutants as indicated. Luciferase assays were performed 48 hours after transfection. Each histogram shows normalized mean values of relative luciferase activity from 3 independent experiments. Normalized luciferase activity in the absence of miR-193a expression vector was set to 100%. (C) The effectiveness of synthetic miR-193a on ETO, DNMT3a, HDAC3, AML1/ETO, KIT, CCND1, and MDM2 proteins was analyzed by Western blot. (D) AML1/ETO+ had higher KIT levels (P = .0004). (E) miR-193a increases PTEN expression indirectly. SKNO-1 cells (mock, and si-A/E) were transfected with synthetic miR-193a or scramble oligonucleotides for 48 hours. Relative PTEN levels were evaluated by qRT-PCR. The results represent the average of 3 independent evaluations ± SD.
In AML, there is overexpression of KIT gene in a majority of the cases where these mutations are associated with activation of PTEN/PI3K signal pathways.20 Consistent with this finding, we observed that KIT is overexpressed by 2.7-fold in AML1/ETO+ cells compared with AML1/ETO− cells (Figure 3D, supplemental Table 2). Interestingly, computational analysis using miRNA target prediction programs showed that the 2 genes in PTEN/PI3K pathway, CCND1 and MDM2, are putative targets of miR-193a (Figure 3A). Both luciferase assay and Western blot further confirmed that miR-193a regulates the expression of CCND1 and MDM2 by targeting its putative binding sites (Figure 3B-C).
The expression level of PTEN was increased by 5-fold in SKNO-1 cells transfected with miR-193a, whereas it was slightly increased in the SKNO-1-siA/E cell line (Figure 3E). The 3′-UTR of PTEN did not contain miR-193a complementary binding sites. In view of the feedback minicircuitry between miR-193a and AML1/ETO corepressor complex, we speculate that the AML1/ETO corepressor complex triggers the epigenetic silencing of the PTEN genomic region, and ectopic miR-193a expression increases the PTEN expression indirectly through disruption of the AML1/ETO-dependent corepressor complex.
AML1/ETO localizes at an AML1-binding site on the PTEN gene affecting its transcriptional regulation
We showed a 6.7-fold decrease in expression level of PTEN in AML1/ETO+ compared with AML1/ETO− bone marrow samples from patients with AML-M2 (Figure 4A, supplemental Table 2). The expression levels of PTEN were increased by 7-fold in SKNO-1-siA/E cell lines. In contrast, PTEN was suppressed in U937-A/E cell lines on Zn2+-induced AML1/ETO expression (Figure 4B). The similar results were observed in Kasumi-1 cells when the AML1/ETO was inhibited by synthesis siRNA (supplemental Figure 2B). These results suggested that the AML1/ETO oncoprotein specifically triggers the transcriptional silencing of PTEN.
AML1/ETO/DNMTs/HDACs complex alters the epigenetic status of PTEN genomic regions. (A) Relative qRT-PCR quantification of PTEN level in mononucleated cells (MNC) isolated from 81 AML FAB M2 subtype patients (supplemental Table 2; P = .0007). (B) Relative quantification of PTEN levels in SKNO-1, SKNO-1-siA/E, and U937 c treated or untreated with Zn2+. The results represent the average of 3 independent evaluations ± SD (*P = .003, #P = .007). (C) Schematic diagrams of the AML1 binding sites and of the CpG islands along the PTEN genes. Numbers are the nucleotides relative to PTEN (ATG). Vertical arrows indicate AML1 binding sites, horizontal arrows indicate the location of the primers used in the ChIP assay. Vertical lines indicate CpG dinucleotides, horizontal bars below the CpG sites show the regions analyzed by bisulfite sequencing. (D) HEK293T cells were transiently cotransfected for 48 hours with luciferase reporter containing the sequence of the PTEN regulatory regions or its counterpart mutants (E), and increasing amounts (10, 50, and 100 ng) of pcDNA3.0 with AML1/ETO cDNA or without (293T-mock). The data are expressed as activity relative to that of the empty pGL3-LUC vector alone. pRL-TK was used as an internal control. The results shown are the average of 3 independent evaluations ± SD. (F) Genomic bisulfite sequencing assay was performed to detect the methylation status of the DNA sequences surrounding the AML1-binding site (− 2033) in PTEN promoter from CD34+ hematopoietic progenitors isolated from healthy donors PB (CD34+) and the indicated leukemia cell lines and leukemic blasts. Cells were either untreated or treated with 2.5μM 5-Aza for 40 hours. For each sample, the percentages of global methylation level of these regions on the PTEN genes are indicated. (G) SKNO-1 cells (mock, and si-A/E) were treated or untreated with 2.5μM 5-Aza or/and SAHA 3.0μM for 40 hours. miR-193a and PTEN relative expression levels were evaluated by qRT-PCR. The results represent the average of 3 independent evaluations ± SD.
AML1/ETO/DNMTs/HDACs complex alters the epigenetic status of PTEN genomic regions. (A) Relative qRT-PCR quantification of PTEN level in mononucleated cells (MNC) isolated from 81 AML FAB M2 subtype patients (supplemental Table 2; P = .0007). (B) Relative quantification of PTEN levels in SKNO-1, SKNO-1-siA/E, and U937 c treated or untreated with Zn2+. The results represent the average of 3 independent evaluations ± SD (*P = .003, #P = .007). (C) Schematic diagrams of the AML1 binding sites and of the CpG islands along the PTEN genes. Numbers are the nucleotides relative to PTEN (ATG). Vertical arrows indicate AML1 binding sites, horizontal arrows indicate the location of the primers used in the ChIP assay. Vertical lines indicate CpG dinucleotides, horizontal bars below the CpG sites show the regions analyzed by bisulfite sequencing. (D) HEK293T cells were transiently cotransfected for 48 hours with luciferase reporter containing the sequence of the PTEN regulatory regions or its counterpart mutants (E), and increasing amounts (10, 50, and 100 ng) of pcDNA3.0 with AML1/ETO cDNA or without (293T-mock). The data are expressed as activity relative to that of the empty pGL3-LUC vector alone. pRL-TK was used as an internal control. The results shown are the average of 3 independent evaluations ± SD. (F) Genomic bisulfite sequencing assay was performed to detect the methylation status of the DNA sequences surrounding the AML1-binding site (− 2033) in PTEN promoter from CD34+ hematopoietic progenitors isolated from healthy donors PB (CD34+) and the indicated leukemia cell lines and leukemic blasts. Cells were either untreated or treated with 2.5μM 5-Aza for 40 hours. For each sample, the percentages of global methylation level of these regions on the PTEN genes are indicated. (G) SKNO-1 cells (mock, and si-A/E) were treated or untreated with 2.5μM 5-Aza or/and SAHA 3.0μM for 40 hours. miR-193a and PTEN relative expression levels were evaluated by qRT-PCR. The results represent the average of 3 independent evaluations ± SD.
A bioinformatics search of the PTEN promoter sequence revealed the presence of 8 putative AML1 binding sites (Figure 4C). Luciferase reporter constructs containing different portions of the PTEN promoter and their mutants (Figure 4C bottom panel) were cotransfected with increasing amounts of expression plasmids encoding for AML1/ETO or the empty vector into 293T cells. The expression of AML1/ETO caused a dose-dependent decrease in the activities of PTEN-P1, PTEN-P2, PTEN-P3, PTEN-P4, and PTEN-P5, the lowest activities were observed in PTEN-P6, PTEN-P7, PTEN-P8, and PTEN-P9. Notably, most of the AML1/ETO dose-dependent decrease of luciferase activity was blocked in the PTEN-P5 mutant (Figure 4E). Thus, we concluded that the AML1 binding site (− 2033 nt relative to ATG) in the promoter of PTEN is involved in the AML1/ETO-dependent silencing of PTEN.
ChIP analysis revealed the presence of AML1/ETO, HDAC1, DNMT1, DNMT3a, and DNMT3b at the PTEN promoter region surrounding this AML1-binding sites (Figure 2D). A higher frequency of methylated CpG dinucleotides in U937-A/E-HA (75.6%) and in SKNO-1 cells (89.4%) compared with U937 (47.8%), HL60 (47.2%) cells or normal human bone marrow blasts (1.1%) was demonstrated, thereby confirming the basal hypermethylated status of these CpGs in AML1/ETO-positive cells (Figure 4F). SKNO-1-siA/E cells induced a 46.1% decrease in CpGs methylation level at these sites on PTEN gene and reactivated its expression (Figure 4B-F). In AML-M2 patients, the higher frequency of methylated CpG dinucleotides encompassing the endogenous PTEN gene sequences in AML1/ETO+ blasts compared with AML1/ETO− blasts (Figure 4F).
SKNO-1 cells were treated with the DNMT inhibitor 5-azacytidine (5-Aza; 2.5μM) or/and HDAC inhibitor SAHA (3.0μM) for 40 hours. It was observed that pharmacologic 5-Aza treatment decreased the methylation status of the endogenous pre-miR-193a CpG islands (from 70.0%-18.8% in SKNO-1 cells and 40.0%-14.2% in SKNO-1-siA/E), and PTEN CpG islands (from 89.4%-18.3% in SKNO-1 cells and 46.1%-14.4% in SKNO-1-siA/E; Figures 2E, 4F). In SKNO-1 cells, treatment with 5-Aza increased the miR-193a expression levels by approximately 3.7-fold and PTEN mRNA expression by 2.5-fold. However, the highest expression levels of these genes were induced by SAHA treatment combined with 5-Aza (Figure 4G). Notably, in the SKNO-1-siA/E cell line, the expression levels of miR-193a and PTEN were increased 8-fold and 10.6-fold, respectively, on 5-Aza treatment and 10.8-fold and 14.3-fold, respectively, by 5-Aza combined with SAHA treatment (Figure 4G). All these results suggested that the chromatin remodeling complex aberrantly formed by AML1/ETO and the hypermethylation of the “CpG islands” present in the AML1 binding sites on the pre-miR-193a and PTEN regulatory region appear to be key regulatory mechanisms for transcriptional silencing of miR-193a and PTEN.
Both miR-193a and PTEN epigenetic silencing contribute to the activities of CCND1 and MDM2 directly or via PTEN/PI3K signal pathways
Our data also showed that CCND1 is overexpressed by 5-fold in AML1/ETO+ compared with AML1/ETO− cells (Figure 5A, supplemental Table 2). The expression levels of MDM2 in clinical samples and cell lines were similar to those of the CCND1 (supplemental Figure 3A). The expression levels of CCND1 were suppressed by 3-fold in SKNO-1-siA/E cell lines and increased by 4.5-fold in U937-A/E cell lines on Zn2+-induced AML1/ETO expression (Figure 5B, supplemental Figure 3B). The knocking down of AML1/ETO in SKNO-1-siA/E cell lines not only down-regulated KIT expression, but also that of KIT-dependent downstream signaling effectors, p-PDK1, p-AKT, p-GSK3β, CCND1, and MDM2 (Figure 5C left panel). PI3K inhibition by Ly294002 was able to reduce Akt levels in SKNO-1-siA/E cell lines, and resulted in a stronger inhibition of KIT and PTEN/PI3K pathway effectors than that induced by Ly294002 alone (Figure 5C left panel). Interestingly, in U937-A/E cell lines with Zn2+-induced AML1/ETO expression, the expression levels of KIT and KIT-dependent downstream signaling effectors were found to be up-regulated, whereas there was a down-regulation seen in the expression levels of these signaling proteins in this cell line on exposure to Ly294002 treatment (Figure 5C right panel). All these results support the notion that epigenetic silencing of both miR-193a and PTEN contribute to the activation of CCND1 and MDM2 directly or via P13K/Akt signal pathways in AML1/ETO+ leukemia.
Epigenetic silencing of both miR-193a and PTEN contributes partially to the activation of CCND1 and MDM2 via the P13K/AKT signal pathway. (A) Relative qRT-PCR quantification of CCND1 level in MNC isolated from 81 AML FAB M2 patients. P = .02. (B) Relative quantification of AML1/ETO and CCND1 mRNA levels in the indicated cell lines. Zn2+ (100μM for 16 hours) were used to increase the AML1/ETO levels in U937 cells. The results represent the average of 3 independent evaluations ± SD. (C) Immunoblot analysis demonstrated the down-regulation of KIT and its downstream effectors as indicated in SKNO-1 (WT, mock, and siA/E; left panels) and U937 (mock and A/E; right panels) cell lines treated with PI3K inhibitor Ly294002 or/and with Zn2+.
Epigenetic silencing of both miR-193a and PTEN contributes partially to the activation of CCND1 and MDM2 via the P13K/AKT signal pathway. (A) Relative qRT-PCR quantification of CCND1 level in MNC isolated from 81 AML FAB M2 patients. P = .02. (B) Relative quantification of AML1/ETO and CCND1 mRNA levels in the indicated cell lines. Zn2+ (100μM for 16 hours) were used to increase the AML1/ETO levels in U937 cells. The results represent the average of 3 independent evaluations ± SD. (C) Immunoblot analysis demonstrated the down-regulation of KIT and its downstream effectors as indicated in SKNO-1 (WT, mock, and siA/E; left panels) and U937 (mock and A/E; right panels) cell lines treated with PI3K inhibitor Ly294002 or/and with Zn2+.
Demethylation, RNAi inhibition of AML1/ETO, or ectopic miR-193a expression reprogram myeloid differentiation in SKNO-1 cells
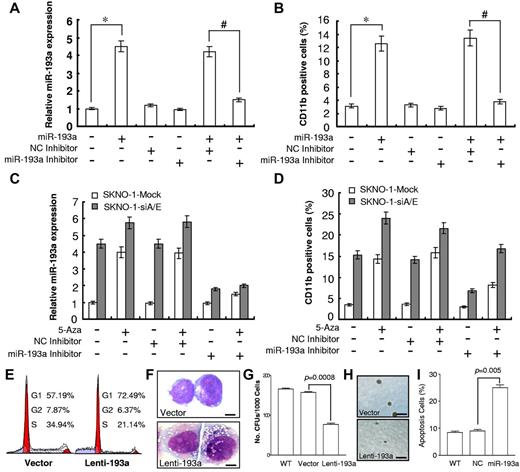
Ectopic expression of a synthetic miR-193a in SKNO-1 cells resulted in a 4.5-fold increase in the miR-193a expression and induced myelomonocytic differentiation marker CD11b from 3.1% to 12.6% in these cells. Cotransfection with a miR-193a inhibitor to sequester the miRNA in an inactive form inhibited any increased in the expression of miR-193a and CD11b (Figure 6A-B). When SKNO-1 and SKNO-1-siA/E cells transfected with either miR-193a inhibitor or negative control were treated with the DNMTs inhibitor 5-Aza, it resulted in approximately a 4-fold increase in miR-193a expression in SKNO-1-mock cells and 30% increase in miR-193a levels in SKNO-1-siA/E cells (Figure 6C) with a decrease in the methylation status of the endogenous pre-miR-193a CpG island (Figure 2E). Flow cytometry showed that the expression levels of CD11b were increased by approximately 3.5-fold and 1.6-fold in SKNO-1-mock and SKNO-1-siA/E cells, respectively (Figure 6D). Both SKNO-1-mock and SKNO-1-siA/E cells transfected with miR-193a inhibitor and treated with 5-Aza showed a decrease in miR-193a levels compared with 5-Aza treatment alone (Figure 6C). Flow cytometry showed that in the presence of 5-Aza, the knockdown of miR-193a produced a 30% to 40% reduction of CD11b compared with 5-Aza treatment alone. However, it maintained the cells at higher differentiation levels compared with control (Figure 6D).
Enhancement of miR-193a levels by siAML1/ETO, 5-Aza treatment and ectopic miR-193a restores myeloid differentiation in SKNO-1 cells. (A) Relative qRT-PCR quantification of miR-193a level (*P = .004, #P = .006) and (B) flow cytometric analysis of CD11b expression in SKNO-1 cells at 144 hours after transfection with 100nM synthetic miR-193a, NC inhibitor or miR-193a inhibitor. NC, Negative Control. Histograms show 3 independent evaluations ± SD (*P = .005, #P = .008). (C) Relative qRT-PCR quantification of miR-193a level and (D) flow cytometric analysis of CD11b expression in SKNO-1 and SKNO-1-siA/E cells at 144 hours in the presence or absence of 2.5μM 5-aza after transfection with 100nM synthetic NC inhibitor or miR-193a inhibitor. Histograms show 3 independent evaluations ± SD. (E) Representative flow cytometric analysis of cell-cycle distribution of SKNO-1 cells transfected with vector or lenti-193a at 72 hours. (F) Morphologic analysis of SKNO-1 cells at 4 days after transfection with vector or Lenti-193a. Scale bar = 10 μm. (G) Analysis of CFU and (H) morphology in SKNO-1 cells during lenti-193a infection using methylcellulose culture system. WT indicates wild-type. Scale bar = 1 mm. Data are the representative of 3 independent experiments. (I) Flow cytometry analysis of apoptosis in SKNO-1 cells at 72 hours after transfection with 100nM synthetic miR-193a or NC. Histograms show 3 independent evaluations ± SD.
Enhancement of miR-193a levels by siAML1/ETO, 5-Aza treatment and ectopic miR-193a restores myeloid differentiation in SKNO-1 cells. (A) Relative qRT-PCR quantification of miR-193a level (*P = .004, #P = .006) and (B) flow cytometric analysis of CD11b expression in SKNO-1 cells at 144 hours after transfection with 100nM synthetic miR-193a, NC inhibitor or miR-193a inhibitor. NC, Negative Control. Histograms show 3 independent evaluations ± SD (*P = .005, #P = .008). (C) Relative qRT-PCR quantification of miR-193a level and (D) flow cytometric analysis of CD11b expression in SKNO-1 and SKNO-1-siA/E cells at 144 hours in the presence or absence of 2.5μM 5-aza after transfection with 100nM synthetic NC inhibitor or miR-193a inhibitor. Histograms show 3 independent evaluations ± SD. (E) Representative flow cytometric analysis of cell-cycle distribution of SKNO-1 cells transfected with vector or lenti-193a at 72 hours. (F) Morphologic analysis of SKNO-1 cells at 4 days after transfection with vector or Lenti-193a. Scale bar = 10 μm. (G) Analysis of CFU and (H) morphology in SKNO-1 cells during lenti-193a infection using methylcellulose culture system. WT indicates wild-type. Scale bar = 1 mm. Data are the representative of 3 independent experiments. (I) Flow cytometry analysis of apoptosis in SKNO-1 cells at 72 hours after transfection with 100nM synthetic miR-193a or NC. Histograms show 3 independent evaluations ± SD.
We also showed that ectopic expression of miR-193a increased the fraction of cells in the G1 phase from 57.19% to 72.49%, whereas it decreased the fraction of cells in the S phase from 34.94% to 21.14% (Figure 6E). Ectopic expression of miR-193a also reduced the number and the size of CFU, induced apoptosis, and brought about morphologic changes in SKNO-1 cells, such as chromatin condensation, decreased nuclear/cytoplasmic ratio, and appearance of primary granules, which are consistent with their maturation into granulocytes (Figure 6F-I).
All these data indicated that miR-193a plays a significant role in both cell cycle progression and differentiation fate of myeloid precursors.
In vivo study
Having demonstrated the relevance of the AML1/ETO/DNMTs/HDACs/miR-193a feedback loop on miR-193a regulation, we tested whether this loop represented a potentially viable therapeutic target to overcome AML1/ETO leukemia in vivo. Approximately 20 million viable SKNO-1 cells were inoculated subcutaneously in the right flank of immunocompromised “nude” mice. When tumors reached 50 mm3, synthetic miR-193a or scramble oligonucleotides were injected directly into the tumors every 3 or 4 days for a total of 4 times (Figure 7A). At day 7 after the first treatment, tumors injected with synthetic miR-193a were significantly smaller than those injected with scramble oligonucleotide (P = .001) and wild type controls (P = .001; Figure 7B). At day 14, the average tumor weights in mice treated with scramble oligonucleotides were 2.83g and those treated with synthetic miR-193a were 0.48g (P = .001, Figure 7C-E). Treatment with synthetic miR-193a was also associated with down-regulation of AML1/ETO, DNMT3a, HDAC3, CCND1, and MDM2, and up-regulation of miR-193a and PTEN levels (Figure 7F-G). We also found that there was a down-regulation of KIT and inhibition of KIT-dependent downstream signaling effectors in miR-193a transfected mouse xenografts (Figure 7G). All these results indicated that miR-193a overexpression significantly contributes to the blockage of malignant-cell proliferation, and targeting of AML1/ETO/DNMTs/HDACs through a miR-193a–induced feedback network. This represents a promising therapeutic approach to treat t(8;21) leukemia.
miR-193a inhibits leukemic growth in vivo. (A) Diagram illustrating the experimental design of the mice xenograft experiment. (B) Tumor volumes measured at the indicated days during the experiment. WT (n = 3), scramble (n = 6), and synthetic miR-193a (n = 6). (C) Tumor weight averages and (D) photographs of 2 mice between scramble and synthetic miR-193a–treated mice groups at the end of the experiment (day 14; P = .001). Bars represent SD. (E) Ectopic miR-193a expression significantly inhibits tumor growth in mice engrafted with SKNO-1 cells. (F) Relative quantification of indicated gene expression in xenografts injected with synthetic miR-193a or scramble. (G) Immunoblot analysis demonstrated the down-regulation of AML1/ETO, KIT and indicated downstream effectors in xenografts injected with synthetic miR-193a or scramble. (H) Schematic model for the epigenetic silencing of miR-193a and PTEN genes by AML1/ETO and the interplay among AML1/ETO corepressor complex, miR-193a, PTEN, KIT, and CCND1 in myeloid differentiation.
miR-193a inhibits leukemic growth in vivo. (A) Diagram illustrating the experimental design of the mice xenograft experiment. (B) Tumor volumes measured at the indicated days during the experiment. WT (n = 3), scramble (n = 6), and synthetic miR-193a (n = 6). (C) Tumor weight averages and (D) photographs of 2 mice between scramble and synthetic miR-193a–treated mice groups at the end of the experiment (day 14; P = .001). Bars represent SD. (E) Ectopic miR-193a expression significantly inhibits tumor growth in mice engrafted with SKNO-1 cells. (F) Relative quantification of indicated gene expression in xenografts injected with synthetic miR-193a or scramble. (G) Immunoblot analysis demonstrated the down-regulation of AML1/ETO, KIT and indicated downstream effectors in xenografts injected with synthetic miR-193a or scramble. (H) Schematic model for the epigenetic silencing of miR-193a and PTEN genes by AML1/ETO and the interplay among AML1/ETO corepressor complex, miR-193a, PTEN, KIT, and CCND1 in myeloid differentiation.
Discussion
Recent evidence suggests that miRNAs are novel epigenetic targets in cancer.21 Their altered regulation by oncogenic proteins or transcriptional factors, which can control each other function in negative feedback loops, is potentially of great relevance in the pathogenesis of leukemia. Tumor specific down-regulation of subsets of miRNAs has generally been observed in various types of human cancer.22 DNA methylation-based regulation of miRNAs was also recently described.5,23 In this study, we showed that the heterochromatic silent state of miR-193a and PTEN genes contribute to t(8;21) leukemogenesis by activating the PTEN/PI3K signal pathway. Furthermore, AML1/ETO targets miR-193a and PTEN through its interaction with the AML1 binding sites at the pre–miR-193a and PTEN upstream regions where they recruit HDACs and DNMTs. The AML1/ETO-associated complex resets the miR-193a and PTEN genes through changes in chromatin conformation and DNA methylation to a repressed ground state and thus contributes to the differentiation block of myeloid precursors (Figure 7H). Study of miR-193a expression disclosed a feedback circuitry comprising of AML1/ETO/DNMTs/HDACs complex and tumor suppressor miR-193a, which together play a crucial role in the differentiation process by suppressing the miR-193a, and thereby expanding the oncogenic activity of the AML1/ETO fusion protein. Of note, ectopic miR-193a expression, down-regulation of AML1/ETO protein levels, or the use of demethylating agents reactivate miR-193a expression and restore myeloid differentiation in t(8;21)-AML blasts. Our findings suggest that a regulatory network exists between miR-193a and the transcription factor AML1/ETO during myeloid cell proliferation and differentiation. Changing the balance from one to the other may result in the shift from differentiation to proliferation, which results in AML.
PTEN is a tumor suppressor gene and its role in tumor biology is well characterized.24 In this study, we showed that PTEN is negatively regulated by the oncoprotein AML/ETO at the mRNA level, which is suggested that the down-regulation of PTEN by AML1/ETO may play a crucial role in accelerating cancer progression. Our study, for the first time, describes the mechanism for the deregulation of PTEN by AML1/ETO-mediated gene silencing and indicates that the epigenetic inactivation of the PTEN gene may represent an important step in AML progression.
Our findings provide new insights into the miRNA-mediated inhibition of myeloid cell-cycle progression, a key step that is disrupted in different AML subtypes. We showed that AML1/ETO regulates miR-193a, and miR-193a blocks the myeloid cell-cycle progression by targeting AML1/ETO, KIT, CCND1, and MDM2. We speculate the possibility of miR-193a targeting other cell-cycle regulators during granulopoiesis. Although different miRNAs including miR-193b have been reported to target CCND1,25 these miRNAs have not been reported to be expressed in granulopoiesis. Because miR-193a is most expressed in granulopoiesis, we propose that miR-193a and PTEN inhibition by AML1/ETO is the major pathway through which AML1/ETO mediates cell-cycle inhibition in t(8;21) AML. However, there is also a possibility that other miRNAs regulated by AML1/ETO can target CCND1 during myelogenesis.
The regulatory circuitry described in this study provides a model for understanding the complexity of the epigenetic/transcriptional silencing of expression of a single miRNA by a leukemia-associated fusion product and of the multiple direct and indirect posttranscriptional consequences of this oncogenic event in leukemogenesis. It is also interesting to note that the fine-tuning of miR-193a expression is in part because of an autoregulatory loop wherein the translational repression of the miRNA targets allows the establishment of miRNA sustained expression. Notably, the relevant function of miR-193a up-regulation would not only be a decrease in AML1/ETO, KIT, CCND1 and MDM2 mediated functions, but also an inhibition of other putative substrates not yet tested, and possibly important for cell-lineage specification. In conclusion, our study provides new insight into understanding the molecular processes required for myeloid differentiation and offers the possibility of identifying new targets for the development of novel therapeutic approaches to leukemia.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Shruti Shah (SIRO Clinpharm Pvt Ltd) for providing editorial assistance and the funding for the editorial support provided by Xi′an Janssen Pharmaceutical Ltd. The authors also thank Dr Xiaofei Zheng for the miRNA expression library, Prof Qishou Xu and Dr Weidong Han for experimental support, and M. S. Li Cheng for technical assistance.
This work was supported by grants from the National Basic Research Program of China (2005CB522400), National Natural Science Foundation of China (90919044, 30971297, 81000221, and 81170518), High and New Technology Program of PLA (2010gxjs091), and Capital Medical Development Scientific Research Fund (No. 2007-2040).
Authorship
Contribution: Y.L., L.G., and X.L. performed experiments and analyzed the data; L.G. and Lili Wang provided AML patient samples and clinical data; X.G., W.W., J.S., L.D., J.L., C.X., and Lixin Wang performed experiments; M.Z., M.J., and J.Z. performed statistical analysis; M.A.C., C.N., C.D.B. and G.M. commented on the paper; Y.L. wrote the paper; Y.L. and L.Y. designed the research; and L.Y. supervised the work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Li Yu, Deptartment of Hematology and BMT Center, Chinese PLA General Hospital, 28 Fuxing Road, Beijing 100853, China; e-mail: chunhuiliyu@yahoo.com; or Guido Marcucci, The Ohio State University Comprehensive Cancer Center, Biomedical Research Tower, 460 West 12th Ave, Columbus, OH 43210; e-mail: guido.marcucci@osumc.edu.
References
Author notes
Y.L., L.G., and X.L. contributed equally to this work.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal