Abstract
One recently identified subtype of pediatric B-precursor acute lymphoblastic leukemia (ALL) has been termed BCR-ABL1–like or Ph-like because of similarity of the gene expression profile to BCR-ABL1 positive ALL suggesting the presence of lesions activating tyrosine kinases, frequent alteration of IKZF1, and poor outcome. Prior studies demonstrated that approximately half of these patients had genomic lesions leading to CRLF2 overexpression, with half of such cases harboring somatic mutations in the Janus kinases JAK1 and JAK2. To determine whether mutations in other tyrosine kinases might also occur in ALL, we sequenced the tyrosine kinome and downstream signaling genes in 45 high-risk pediatric ALL cases with either a Ph-like gene expression profile or other alterations suggestive of activated kinase signaling. Aside from JAK mutations and 1 FLT3 mutation, no somatic mutations were found in any other tyrosine kinases, suggesting that alternative mechanisms are responsible for activated kinase signaling in high-risk ALL.
Key Points
Pediatric B-ALL cases with a gene expression signature similar to that of Philadelphia chromosome positive (Ph-like ALL) have a high risk of relapse.
Point mutations in tyrosine kinase genes are rare in pediatric Ph-like or other high-risk ALL cases except for in Janus kinase (JAK) family genes or rarely in FLT3.
Introduction
Although survival of children and adolescents with acute lymphoblastic leukemia (ALL) has improved substantially over time, 15% to 20% of patients will relapse, and most of those who experience a bone marrow relapse will die.1 Advances in understanding the genetic heterogeneity of ALL have contributed to modern risk stratification algorithms that use leukemia genotype and measurements of early treatment response to assign postinduction therapies of different intensities.2,3 However, many children who relapse have features indistinguishable from those who are cured. Furthermore, except for those few children with Philadelphia chromosome positive (Ph+) ALL, current treatments do not include molecularly targeted therapy.4
Collaborative genomic analyses performed by the Children's Oncology Group (COG) high risk (HR) childhood ALL Therapeutically Applicable Research to Generate Effective Treatments (TARGET) project have discovered novel mutations and pathway alterations that are potential therapeutic targets.5-12 We previously reported that 11.2% of patients enrolled on COG P9906 harbored mutations in JAK1 (n = 3), JAK2 (n = 17), and JAK3 (n = 1),8 the majority of which were clustered in a unique group, “R8,” when unsupervised analysis was performed of gene expression profiles (GEP) using Recognition of Outliers by Sampling Ends.9 Regardless of JAK mutation status, patients in the R8 group experienced a very poor outcome.9 JAK mutations occur almost exclusively in patients with CRLF2 genomic lesions.5,8,10,13,14 The majority of the JAK mutations in ALL are located in the JAK2 pseudokinase domain (most commonly at or near p.Arg683), are identical to those recently described in patients with Down syndrome-ALL,13,14 and are distinct from the V617F alterations catalogued in myeloproliferative disorders.15
Gene set enrichment analysis of the P9906 cohort revealed that HR IKZF1-altered B-ALL cases had a gene expression profile that was enriched in that of Ph+ ALL.7,9,12,16 Approximately half of the Ph-like cases had CRLF2 alterations, with approximately half of those cases having JAK mutations, leading us to hypothesize that mutations in other tyrosine kinase (TK) genes would be found in other cases of Ph-like and/or CRLF2 overexpressing ALL. We sequenced 126 genes encoding TKs and mediators of kinase signaling in 45 B-cell precursor HR ALL cases from COG P9906 or the successor COG HR ALL trial, AALL0232. We further sought to determine whether the Ph-like signature independently conferred a poor prognosis in AALL0232 patients, thus identifying a new marker that could be used to identify patients who might benefit from alternative or intensified therapy.
Methods
Patient selection and characteristics
Forty-five cryopreserved diagnostic bone marrow or peripheral blood specimens with at least 80% blasts from children with newly diagnosed ALL were selected for kinome sequencing (Table 1), including 23 from COG P990617 that lacked JAK mutations and 22 AALL0232 patients of unknown JAK mutation status. AALL0232 eligibility included age at least 10 years and/or initial peripheral blood white blood cell count of at least 50 000/μL. Minimal residual disease (MRD) burden was determined via flow cytometry in 1 of 2 central reference laboratories at day 29 of induction therapy.3 All P9906/AALL0232 patients or their patients/guardians provided informed consent for treatment and for banking of specimens for future research in accordance with the Declaration of Helsinki. Institutional review board approval for the laboratory studies was granted by St Jude Children's Research Hospital and the University of New Mexico.
Clinical characteristics of analyzed patients
| . | 9906 . | AALL0232 . | Total . |
|---|---|---|---|
| Age, y | |||
| 1-9 | 5 | 9 | 14 |
| > 10 | 18 | 13 | 31 |
| WBC | |||
| < 50 000/μL | 12 | 5 | 17 |
| > 50 000/μL | 11 | 17 | 28 |
| Sex | |||
| Male | 17 | 12 | 29 |
| Female | 6 | 10 | 16 |
| MRD | |||
| < 0.01% | 9 | 8 | 17 |
| > 0.01% | 14 | 14 | 28 |
| Total | 23 | 22 | 45 |
| . | 9906 . | AALL0232 . | Total . |
|---|---|---|---|
| Age, y | |||
| 1-9 | 5 | 9 | 14 |
| > 10 | 18 | 13 | 31 |
| WBC | |||
| < 50 000/μL | 12 | 5 | 17 |
| > 50 000/μL | 11 | 17 | 28 |
| Sex | |||
| Male | 17 | 12 | 29 |
| Female | 6 | 10 | 16 |
| MRD | |||
| < 0.01% | 9 | 8 | 17 |
| > 0.01% | 14 | 14 | 28 |
| Total | 23 | 22 | 45 |
GEP and sample selection
RNA extraction and GEP characterization for P9906 cases have been described previously.9,11 Affymetrix U133 Plus Version 2.0 gene expression microarray and Affymetrix SNP Version 6.0 microarray profiling were performed on 608 patients consecutively enrolled on AALL0232 with sufficient banked material available; 325 were used as a training set, after modeling the Ph-like GEP on the BCR-ABL1+ patients within this training set (n = 21).
We then applied Prediction Analysis for Microarrays (PAM),18 trained using Ph+ cases to identify all Ph-like cases (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).19 We classified patients in the test set (283 AALL0232 patients) using this Ph-like signature and assessed the outcome of all Ph-like patients enrolled on AALL0232. We further applied this Ph-like PAM algorithm to the COG P9906 samples to identify Ph-like cases in that cohort, and then assessed the prognosis of this group of ALL cases.
We selected 45 P9906 and AALL0232 cases that were either predicted to be Ph-like by PAM (31 cases; 12 of 23 from 9906 and 19 of 22 from AALL0232), or had high CRLF2 expression or other features suggestive of activated kinase signaling (n = 14) for sequence analysis of 126 genes that encode TKs or mediators of kinase signaling (supplemental Table 1). The entire coding and untranslated regions of each selected gene were subsequently amplified by PCR of whole genome amplified (QIAGEN) genomic DNA and subjected to Sanger sequencing (Beckman Coulter Genomics). A CEPH sample (NA19085) was included as a normal control. Sequence variations were detected using SNPdetector20 and novel, putative nonsilent coding mutations were selected for validation. Forty-one novel variants that failed in the validation assay or had no matching germline samples were compared with germline variants identified by the National Center for Biotechnology Information Exome Sequencing Project (http://evs.gs.washington.edu/EVS) and 1000 Genomes Project deposited in dbSNP 135 (http://www.ncbi.nlm.nih.gov/projects/SNP). For the 22 patient samples from AALL0232, we performed Sanger sequencing separately for the 5 most commonly mutated exons of JAK1 and JAK2.8 The gene expression data for COG P9906 have been deposited at the National Center for Biotechnology Information Gene Expression Omnibus (accession no. GSE11877). The gene expression data without metadata for COG AALL0232 are deposited at the National Cancer Institute caArray site, project identifier EXP-578 (https://array.nci.nih.gov/caarray/project/EXP-578).
Results and discussion
A total of 1 149 117 targeted bases from the 126 candidate genes were sequenced bidirectionally for each sample, 95% of which have high-quality sequencing data. We identified 130 755 putative variations; 179 were novel and predicted to change protein sequences after removing germline variations found in dbSNP, the Cancer Genome Atlas Project, and the normal CEPH sample NA19085. For each of the 179 putative novel variations, tumor and matching normal remission marrow DNA was resequenced to validate the original observation and to distinguish somatic from inherited variants. A total of 105 variations were germline, 20 were false positives, and 41 variations failed in the validation assay (supplemental Table 2). There were a total of 13 confirmed somatic mutations. Aside from 1 FLT3 mutation (23aainsN609) in a Ph-like case, and 12 AALL0232 patients (11 Ph-like, 1 non–Ph-like) with JAK mutations (JAK1: p.Val658Phe, n = 1; JAK2: p.Arg683Gly, n = 5; p.Arg683Ser, n = 2; p.Arg683Thr, n = 1; p.Thr875Asn, n = 2; p.Pro933Glu, n = 1), there were no confirmed somatic mutations in any other TK genes. A detailed description of mutations is tabulated in supplemental Table 3.
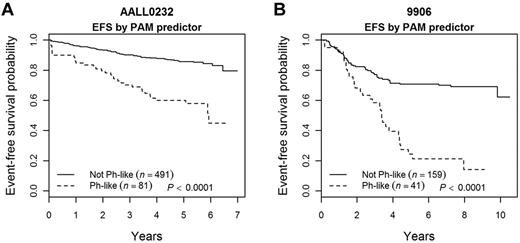
Approximately 14% (81 of 572) of AALL0232 patients studied had a Ph-like GEP by PAM (supplemental Figure 1). The 5-year event-free survival (EFS) rate for the Ph-like cases enrolled on AALL0232 was inferior to that of the non–Ph-like cases (62.6% ± 6.9% vs 85.8% ± 2.0%; P < .0001; Figure 1A). Importantly, the outcome differences were maintained regardless of randomized treatment arm, which demonstrated the superiority of high-dose methotrexate (5 g/m2) over Capizzi-style methotrexate in the first interim maintenance phase (data not shown). Similar results were seen comparing these 2 groups enrolled on P9906 (Figure 1B).
Event-free survival is poor for Ph-like patients. Using PAM clustering methodologies, NCI HR patients enrolled on AALL0232 (A) or P9906 (B) who express the Ph-like signature do poorly compared with non–Ph-like patients. Note: these EFS curves exclude BCR-ABL1+ patients.
Event-free survival is poor for Ph-like patients. Using PAM clustering methodologies, NCI HR patients enrolled on AALL0232 (A) or P9906 (B) who express the Ph-like signature do poorly compared with non–Ph-like patients. Note: these EFS curves exclude BCR-ABL1+ patients.
We also examined additional variables that could contribute to outcome in patients with the Ph-like signature. Older (at least 10 years) Ph-like patients were significantly more likely to have an initial white blood cell count ≥ 100 000/μL. Interestingly, older females with the Ph-like GEP had superior EFS compared with males (4-years EFS: 69% ± 10.2% vs 43% ± 9.4%, P = .04). Three variables were significant in a multivariable Cox regression analysis (supplemental Table 4): Ph-like GEP (hazard ratio = 1.92), end induction MRD of at least 0.01% (hazard ratio = 3.05), and hypodiploidy as defined by a DNA index < 0.81 or chromosome complement < 44 (hazard ratio = 3.17). Of note, patients who exhibited the Ph-like GEP had higher rates of MRD of at least 0.01% versus those who did not (63.8% vs 19.7%, P < .0001), concordant with our previous observations.9
In conclusion, apart from JAK family member mutations, somatic TK gene sequence mutations are rare in children with HR ALL and the Ph-like GEP or other characteristics suggestive of activated kinase signaling. Consistent with our prior findings,5,10 all of the JAK mutations identified in the AALL0232 patients occurred in patients with CRLF2 genomic lesions (7 cases with IGH@-CRLF2 and 5 cases with P2RY8-CRLF2). Regardless of the underlying genetic events that contribute to the Ph-like GEP, this subgroup, which we show is 3-4 times as common as Ph+ ALL in children, experiences a high rate of treatment failure with current therapies. In other studies,19 we have found that an important common mechanism of kinase gene activation in Ph-like HR ALL is genomic rearrangement or structural alterations that create novel fusion genes, suggesting a potential role for expanded use of TK inhibitors in selected patients with HR ALL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the National Institutes of Health Strategic Partnerships to Evaluate Cancer Gene Signatures Program (Department of Health and Human Services grant NCI U01 CA114762; C.L.W.) and the COG, including the COG Chair's grant (CA98543) and a supplement to support the TARGET Project, U10 CA98413 (COG Statistical Center), and U24 CA114766 (COG Specimen Banking), the American and Lebanese Syrian Associated Charities of St Jude Children's Research Hospital, the Frank A. Campini Foundation, the University of New Mexico Cancer Center Shared Facilities, KUGR Genomics, Biostatistics, and Bioinformatics & Computational Biology, partially supported by NCI P30 CA118100, as well as a contract from the National Cancer Institute, National Institute of Health (NCI-12400). M.L.L. is a Clinical Scholar of the Leukemia & Lymphoma Society. C.G.M. is a Pew Scholar and a St Baldrick's Scholar. K.R. is supported by a National Health and Medical Research Council (Australia) Overseas Training Fellowship and a Hematology Society of Australia and New Zealand Novartis New Investigator Scholarship; S.P.H. is the Ergen Family Chair in Pediatric Cancer.
National Institutes of Health
Authorship
Contribution: M.L.L. was the study cochair for AALL03B1, supervised research, and wrote the manuscript; J.Z. analyzed sequence data; R.C.H. facilitated sample selection and performed expression analyses; K.R., H.K., and I-M.C. performed expression analyses; D.P.-T. processed samples; G.W., X.C., J.B., M.E., and K.H.B. analyzed sequence data; W.L.C. and N.W. supervised research; M.D. performed statistics related to clinical variables; B.W. and M.J.B. performed MRD analysis; D.S.G., J.R.D., and M.S. supervised work; P.B. was the study chair for P9906; E.L. was the study chair for AALL0232; E.R. was the study cochair for AALL03B1; C.L.W. supervised gene expression analyses; C.G.M. facilitated sample selection, supervised work, performed sequencing, and wrote the manuscript; and S.P.H. supervised work and wrote the manuscript.
Conflict-of-interest disclosure: M.L.L. is a member of the Data Monitoring Committee for Bristol-Myers Squibb (trial CA 180226). The remaining authors declare no competing financial interests.
Correspondence: Cheryl L. Willman, University of New Mexico Cancer Center, 1201 Camino de Salud NE, Rm 4630, MSC07-4025, University of New Mexico, Albuquerque, NM 87131-0001; e-mail: cwillman@salud.unm.edu; Charles G. Mullighan, St Jude Children's Research Hospital, Department of Pathology, 262 Danny Thomas Pl, Mail Stop 342, Room D4047E, Memphis, TN 38105; e-mail: Charles.mullighan@stjude.org; and Stephen P. Hunger, Center for Cancer and Blood Disorders, Children's Hospital Colorado, 13123 East 16th Ave, B115, Aurora, CO 80045; e-mail: stephen.hunger@childrenscolorado.org.
References
Author notes
M.L.L. and J.Z. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal