Key Points
Multiple myeloma cells inhibit myeloma-specific T cells through expression of carcinoembryonic antigen-related cell adhesion molecule-6.
Abstract
Although functionally competent cytotoxic, T cells are frequently observed in malignant diseases, they possess little ability to react against tumor cells. This phenomenon is particularly apparent in multiple myeloma. We here demonstrate that cytotoxic T cells reacted against myeloma antigens when presented by autologous dendritic cells, but not by myeloma cells. We further show by gene expression profiling and flow cytometry that, similar to many other malignant tumors, freshly isolated myeloma cells expressed several carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) at varying proportions. Binding and crosslinking of CEACAM-6 by cytotoxic T cells inhibited their activation and resulted in T-cell unresponsiveness. Blocking of CEACAM-6 on the surface of myeloma cells by specific monoclonal antibodies or CEACAM-6 gene knock down by short interfering RNA restored T-cell reactivity against malignant plasma cells. These findings suggest that CEACAM-6 plays an important role in the regulation of CD8+ T-cell responses against multiple myeloma; therefore, therapeutic targeting of CEACAM-6 may be a promising strategy to improve myeloma immunotherapy.
Introduction
T-cell responses against tumor-associated antigens have been described in many tumors1-6 and often cause an accumulation of tumor-specific memory T cells in lymphoid organs or in the blood.2,7,8 However, the capacity of these T cells to react against autologous tumor cells is generally low.9,10 T-cell unresponsiveness against tumor cells has been demonstrated for a broad variety of tumors, and especially multiple myeloma (MM). Despite remarkable recent advances in its treatment, MM remains incurable in most patients using conventional therapy.11 Immunotherapy in the form of allogeneic hematopoietic stem cell transplantation is regarded a potential cure in a subset of patients because of its well-documented graft-versus-myeloma effect.12-15 Unfortunately, this procedure is associated with high mortality and morbidity and remains an option only for a few selected patients.15-18 In the autologous setting, the potential of a patient’s own immune system to recognize tumor cells has become a focus of many investigations.19 Patients with monoclonal gammopathy of undetermined significance exerted strong T-cell responses against plasma cells in the bone marrow (BM).20 In some patients with MM, clonal expansions of cytotoxic CD8+ T cells have been described, and this correlated with a better prognosis, suggesting the presence of protective tumor-specific T-cell clones.21-23 Similarly, T cells reactive against idiotype antibodies,24 MUC1,22 or unknown antigens derived from plasma cell lysates25 could be detected either in freshly isolated T cells from patient BM or after repetitive stimulations in vitro. Other studies reported an inability of autologous T cells to respond to autologous tumor cells or shared myeloma-associated antigens,20,26 suggesting a tumor-specific or generalized state of T-cell unresponsiveness.27
Previous reports suggested that the expression of carcinoembryonic antigen-related cell adhesion molecules (CEACAM)-1 on malignant melanoma cells is responsible for the inhibition of melanoma-specific T-cell responses.28 Although CEACAM-1 expression has not been reported for MM, a case report described increased concentrations of CEACAM-5 in the plasma from a myeloma patient.29
The CEACAM gene family is part of the immunoglobulin superfamily.30 CEACAM receptors consist of a variable immunoglobulin domain–like region followed by up to 6 constant immunoglobulin domain–like structures that are attached to the cell membrane by glycosylphosphatidylinositol anchors.31 The best characterized biological function of CEACAMs is the support of cell–cell adhesion through their homophilic interactions. Multiple cellular activities have been attributed to CEACAM (CD66) proteins, including a role in the differentiation and arrangement of 3-dimensional tissue structure, angiogenesis, apoptosis, tumor suppression, and metastasis.31 A role in the modulation of innate and adaptive immune responses was suggested for CEACAM-1 (CD66a), which possesses a cytoplasmic tail containing an immunoreceptor tyrosine-based inhibitory motif. CEACAM-1 is induced on T cells during T-cell receptor (TCR)-specific activation and mediates a rapid blocking of T-cell effector functions upon homophilic binding with CEACAMs expressed on target cells.32 However, CEACAMs can also exert heterophilic interactions because different members of this superfamily are known to bind to each other and even to other types of molecules, including components of the extracellular matrix, growth-factor receptors, integrins, and cadherins.33
Here we addressed the question of whether myeloma cells might escape recognition by autologous, myeloma-specific CD8+ T cells through CEACAM expression.
Materials and methods
Patients and healthy donors
Patients with previously untreated MM presenting at the University Hospitals of Heidelberg and Montpellier as well as 25 healthy donors have been included into the study after providing written informed consent in accordance with the Declaration of Helsinki. The study was approved by the local ethics committee (#229/2003 and S-152/2010). Diagnosis, staging, and assessment of response to treatment followed standard criteria.28-30 Clinical parameters according to 332 samples subjected to gene expression profiling have been previously published.34 Of those, 247 patients underwent frontline high-dose chemotherapy with 200 mg/m2 melphalan and autologous stem cell transplantation. Clinical characteristics related to samples from 77 MM patients subjected to immune testing are shown in supplemental Table 1.
Samples and tumor cell lines
Healthy BM plasma cells and myeloma cells were purified as previously described.35-38 Isolated myeloma cells were kept in RPMI 1640 medium (PAA), with 10% fetal calf serum (Biochrom AG) and used within 24 hours for the stimulation of autologous T cells. The myeloma cell lines RPMI8226 and SKMM-2 (German Collection of Microorganisms and Cell Cultures), the breast cancer cell lines MCF739 and KS,40 the pro-monocytic leukemia cell line U937,39 and the melanoma cell line MeWo41 (all from American Type Cell Culture) were cultured as recommended.
Gene expression profiling
Gene expression profiling was performed as previously published.35-38 In brief, after RNA extraction, labeled complementary RNA was generated using the small sample labeling protocol vII (Affymetrix) and hybridized U133 2.0 plus arrays according to the manufacturer’s instructions. When different probe sets were available for the same gene, we chose the probe set yielding the maximal variance and the highest signal expression data are deposited in ArrayExpress (accession number E-MTAB-317). Gene expression analyses were performed on GeneChip Robust Multiarray Averaging36 preprocessed data sets of the B-cell lineage. Because 2 different in vitro transcription amplification labeling kits have been used, batch correction was performed using ComBat.37 To assess the presence or absence of gene expression, the Presence-Absence calls with Negative Probesets (PANP) algorithm38 based on probes with no known hybridization partners was used, indicating genes assessed as either “marginal” or “present” as being expressed.
Flow cytometry
For the analysis of CEACAM expression, freshly isolated plasma cells and myeloma cells, purified T cells and dendritic cells (DCs) from healthy individuals (see the following section) and myeloma cell lines were incubated on ice with fluorescein isothiocyanate–labeled mouse anti-human CEACAM-5 monoclonal antibody (mAb) (clone: CI-P83-1; Dianova) or with the following mouse mAbs: anti-CEACAM-8 (clone: G10F5; BD Pharmingen), anti-CEACAM-1 (clone: GM8G5; Axxora or Santa Cruz), or anti-CEACAM-6 (clone: 9A6; Axxora). This incubation was followed by incubation with polyclonal fluorescein isothiocyanate–conjugated goat anti-mouse immunoglobulin (Ig)G/IgM (BD Pharmingen). Staining of myeloma or plasma cells, T cells, and DCs was conducted by labeling with phycoerythrin-conjugated mouse anti-human CD138 mAb (clone B-B4; Miltenyi Biotech), mouse anti-human CD3 (clone: UCHT1; BD Pharmingen), -CD8 (G42-8; BD Pharmingen), -CD57 (NK-1; BD Pharmingen), or mouse anti-human CD11c (clone: B-ly6; BD Pharmingen). Control labeling was evaluated with isotype-matched mAb (BD Pharmingen). All antibodies were used at dilutions of 1:100. Dead cells were excluded from the analysis using propidium iodide or 7-AAD (BD Pharmingen) staining. Only propidium iodide– or 7-AAD-negative cells were recorded and a minimum of 2 × 105 cells were acquired per test sample on a FACSCANTO (BD). All analyses were performed using FlowJo software (Tree Star).42
CEACAM inhibition
For the inhibition of CEACAM-mediated signaling with CEACAM-specific mAb, 105 myeloma cells or breast cancer cells were incubated for 30 minutes on ice with 30 μg/mL mAbs against CEACAM-1 (clone: GM8G5; Axxora), CEACAM-5 (clone: CI-P83-1; Dianova), CEACAM-6 (clone: 9A6, Axxora or Genovac or clone 5E506, Biozol), or CEACAM-8 (clone: G10F5; BD Pharmingen). As a control, tumor cells were incubated with the same concentration of isotype-matched mAb (BD Pharmingen). Afterward, tumor cells were washed 3 times and cocultured with T cells for 40 hours for interferon-γ (IFN-γ) enzyme-linked immunospot (Elispot) assays. In some cases, concentrations of blocking antibodies were modified, as stated in the figure legends. For inhibition of CEACAM-6–mediated signaling with CEACAM-6–specific short interfering RNA (siRNA), RPMI8226, and myeloma cells were transfected with stealth select CEACAM-6 siRNA (National Center for Biotechnology Information reference sequence NM_002483.3; Invitrogen) using oligofectamine reagent (Invitrogen) diluted 1:4 in OptiMEM medium (Invitrogen) according to the manufacturer’s instructions. A monolayer of 3 × 105 cells was grown in a 6-well plate (TPP and Biochrom AG) and transfected with 0.04 μM siRNA for 72 hours. siGFP RNA was used as a negative control. Reverse transcription-polymerase chain reaction (PCR) (forward primer: 5′GGA GGA AGG ACA GCA GGG CCA 3′; reverse primer: 5′AGT GAG GCT GTG AGC AGG ACC3′; Eurofins MWG Operon) was used to verify CEACAM-6 gene knockdown. Transfected cells were cultured for 24 hours in culture media and subsequently used in functional assays.
Isolation of T cells
For use in functional assays, T cells were cultured as previously described.43,44 In brief, cells were cultured for 7 days in RPMI 1640 medium containing 10% human AB serum, 100 U/mL interleukin (IL)-2, and 60 U/mL IL-4, followed by overnight incubation without ILs, and separation from contaminating cells by anti-CD19, anti-CD15, and anti-CD56 mAb-conjugated magnetic beads (Dynal). Afterward, CD8+ T cells were isolated using anti-CD8–coated magnetic microbeads (MACS beads; Miltenyi Biotec). In some experiments, regulatory T cells were depleted from CD8+ T cells using anti-CD25 MACS beads according to the manufacturer’s protocol (Miltenyi Biotec).
Isolation of myeloma cells
BM plasma cells were purified from BM aspirates using CD138-microbeads (Miltenyi Biotec) as previously described.36 Purity of isolated myeloma cells was determined to be ≥80% by flow cytometry (FACSCalibur; Becton Dickinson). Isolated myeloma cells were kept in RPMI 1640 medium (PAA) with 10% fetal calf serum (Biochrome AG) and used within 24 hours for the stimulation of autologous T cells.
Generation of DCs
DCs were generated as previously described.39 In brief, adherent cells from peripheral blood samples were cultured for 7 days in serum-free XVIVO 20 containing 50 ng/mL recombinant human granulocyte-macrophage colony stimulating factor and 1000 U/mL IL-4. DCs were enriched by depleting CD3 and CD19 cells with anti-CD3 and anti-CD19-coupled magnetic beads, and pulsed for 2 hours with respective test or control antigens.
Functional T cell assays
IFN-γ or perforin-producing T lymphocytes were detected by Elispot assays according to the manufacturer’s protocol (Mabtech) as previously described.2 For analysis of T-cell responses to antigen-pulsed autologous DCs, DCs were pulsed with 20 μg/mL of HLA-A2-restricted peptides, or 200 μg/mL long synthetic peptides or protein from cell lysates. The following test antigens were used: the HLA-A0201–restricted peptide MUC112-202 long synthetic peptide MUC11-100; tetanus toxoid (Merck Biosciences); or cell lysate–derived from the RPMI8226 myeloma cell line. Negative control antigens included HLA-A0201–binding control peptide HIVgag77-85,2 hu IgG,39 and cell lysates derived from the breast cancer cell line MCF739 or the promonocytic leukemia cell line U937.39 Pulsed DCs were incubated with autologous T cells at a ratio of 1:5 for 40 hours. IFN-γ or perforin spots were measured using KS ELISPOT software (Zeiss or CTL Europe). Spots induced by control antigens were considered to be background. Individuals were designated as responders if spot numbers in wells containing test antigen–pulsed DCs exceeded a total of 10 spots and were significantly higher (P ≤ .05, 2-sided Student t test) than in control wells containing DCs pulsed with negative control antigens. Cell lysates were generated as previously described by 5 cycles of freezing and thawing, followed by passage through a 0.22-μm filter.39 In some cases, T cells were directly cocultured together with viable autologous or allogeneic tumor cells in Elispot wells at a ratio of 5:1 instead of antigen-pulsed DCs. Each test well contained 105 T cells. Statistical comparison was performed in triplicates per group.
For transwell experiments, 5 × 105 purified CD8+ T cells/well from MM patients in 500 µL X-vivo 20 medium were plated into the lower chambers of transwell plates using 3 µm pore inserts (ThinCerts; Greiner) and activated with Staphylococcus aureus enterotoxin B (SEB, Baxter; 10 µg/mL medium). A total of 1 × 105 CEACAM-6–positive RPMI8226 myeloma cells was added into the upper or lower chambers as indicated in the figure legends at a ratio of TC:MM cells of 5:1. In some test groups, CEACAM-6 on MM cells was blocked by preincubation with anti CEACAM-6 mAb (clone 9A6). IFN-γ concentrations in the lower chambers were assessed after 11 hours by enzyme-linked immunosorbent assay (OptEIA human IFN-γ ELISA set; BD Biosciences) according to the manufacturer`s instructions.
Phosphoprotein analysis
The 106 purified CD8+ T cells from myeloma patients per well were plated in 96-well plates in 100 µL of cytokine free X vivo 20 medium. Afterward, 2 × 105 RPMI 8226 myeloma cells in 50 µL of cytokine-free X vivo 20 medium per well were added. These were either untreated or pretreated with 30 µg/mL anti-CEACAM6 mAb for 30 minutes on ice followed by careful washing. After 5 minutes of coculture, cells were harvested, lysed, and further processed to assess the concentrations of TCR-associated signaling molecules using 7-plex TCR signaling Kit-phosphoprotein (Millipore) with one replicate/test group according to the manufacturer’s protocol. Measurements were performed using Luminex100 Bio-Plex System (Luminex). For all analyses, we used Bio-Rad Bio-Plex Manager software version 4.1.1. (BioRad Life Science Research). The total protein concentrations were measured using BSA Protein Assay Kit (Thermo Scientific).
Statistical evaluation
Differences between test groups were analyzed using Student t test. Gene expression data were analyzed as described previously. In all tests, results were considered statistically significant if the P value was ≤ .05. All statistical computations for gene expression data were performed using R version 2.7.045 and Bioconductor46 version 2.2.
Results
Functional inhibition of MM cell–reactive T cells upon interaction with myeloma cells
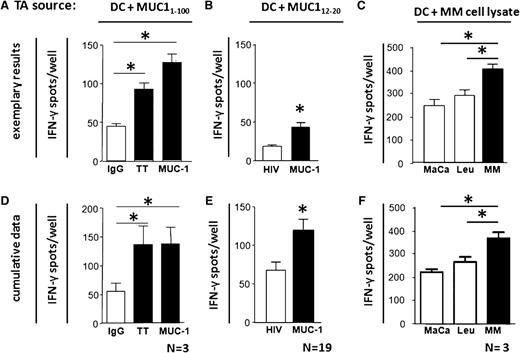
To evaluate whether MM patients possess T cells that can react against myeloma-associated antigens, we analyzed by short-term ex vivo IFN-γ Elispot assays patients’ IFN-γ secretion upon coculture with MUC1-pulsed autologous DCs. All 3 tested MM patients exhibited significant CD8+ T-cell responses against a long synthetic MUC1 peptide, which were comparable to their response against the recall antigen tetanus toxoid, which we used as a positive control. Significant T-cell responses against the HLA-A0201–restricted MUC-112-20 epitope were detectable in 8 of 19 MM patients (42%). The numbers of IFN-γ-secreting T cells were also significantly increased in four MM patients whose test wells contained DCs loaded with RPMI8226 myeloma cell line lysates, compared with control wells containing lysates from unrelated MFC7 or U937 cells. Thus, functionally competent and myeloma antigen-reactive T cells are present in the BM of patients with MM and are capable to recognize myeloma cell–derived antigens when presented by autologous DCs. Representative experiments for individual patients are shown in Figure 1A-C, and cumulative data of all tested patients are shown in Figure 1D-F.
Presence of myeloma antigen–specific type 1 T cells in BM of myeloma patients. (A) Isolated BM T cells from MM patients were tested by short-term IFN-g Elispot assay for reactivity against autologous DCs pulsed with either MUC11-100 long peptide, tetanus toxoid (TT) as positive control, or hu IgG as negative control. (B) Ex vivo isolated BM T cells from HLA0201-positive MM patients were tested for reactivity against autologous DCs pulsed with HLA-A0201–restricted epitope MUC1 12-20 or HIVgag77-85 (negative control antigen). (C) Isolated, purified CD81 BM T cells from MM patients were tested by short-term IFN-g Elispot assay for reactivity against autologous DCs pulsed with lysate derived from myeloma cell line RPMI-8226 (MM), or lysates derived from the myeloma-unrelated tumor breast cancer cell line MCF7 (MaCa) or from the promonocytic leukemia cell line U937 (Leu), which served as negative control antigens. Representative experiments of 4 different MM patients are shown in (A-C). Respective cumulative data are shown in (D-F). N: number of patients. *Significant difference compared with respective control group (P < .05, 2-sided Student t test). TA, tumor antigen.
Presence of myeloma antigen–specific type 1 T cells in BM of myeloma patients. (A) Isolated BM T cells from MM patients were tested by short-term IFN-g Elispot assay for reactivity against autologous DCs pulsed with either MUC11-100 long peptide, tetanus toxoid (TT) as positive control, or hu IgG as negative control. (B) Ex vivo isolated BM T cells from HLA0201-positive MM patients were tested for reactivity against autologous DCs pulsed with HLA-A0201–restricted epitope MUC1 12-20 or HIVgag77-85 (negative control antigen). (C) Isolated, purified CD81 BM T cells from MM patients were tested by short-term IFN-g Elispot assay for reactivity against autologous DCs pulsed with lysate derived from myeloma cell line RPMI-8226 (MM), or lysates derived from the myeloma-unrelated tumor breast cancer cell line MCF7 (MaCa) or from the promonocytic leukemia cell line U937 (Leu), which served as negative control antigens. Representative experiments of 4 different MM patients are shown in (A-C). Respective cumulative data are shown in (D-F). N: number of patients. *Significant difference compared with respective control group (P < .05, 2-sided Student t test). TA, tumor antigen.
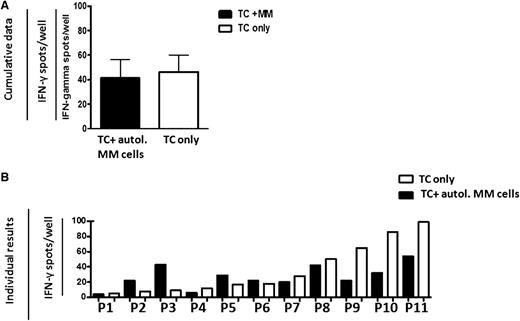
We next assessed whether autologous T cells can recognize myeloma antigens when presented by autologous myeloma cells instead of professional antigen-presenting cells. To this end, BM T cells from 11 MM patients were cocultured with sorted CD138+ autologous myeloma cells in a short-term IFN-γ Elispot assay. Addition of MM cells to ex vivo isolated BM-derived autologous CD8+ T cells did not increase IFN-γ spots above background levels (Figure 2A). Individual patient results are shown in Figure 2B; in 7 cases, T-cell activity was reduced upon coculture with myeloma cells, whereas increased IFN-γ secretion was only observed in 4 cases. In no cases did T-cell reactivity in myeloma cocultures exceed the T-cell reactivity in cocultures with the T cell– and natural killer cell–depleted, CD138-negative fraction of sorted BM cells (data not shown). Therefore, although major histocompatibility complex class I–restricted myeloma reactive T cells are present in the BM of myeloma patients, in most patients they do not exert reactivity toward autologous myeloma cells. This corresponds with the previously reported lack of T-cell cytotoxicity against autologous myeloma cells in MM patients.20
Reactivity of BM T cells from MM patients is inhibited upon coculture with myeloma cells. Cumulative mean ± standard error of the mean (SEM) numbers of IFN-γ–secreting, freshly isolated BM T cells from 11 MM patients (A) and mean values of individual results (B) from these patients with (black bars) or without (white bars) coculture with sorted CD138+ autologous (autol.) myeloma cells, as analyzed by short-term IFN-γ Elispot assay.
Reactivity of BM T cells from MM patients is inhibited upon coculture with myeloma cells. Cumulative mean ± standard error of the mean (SEM) numbers of IFN-γ–secreting, freshly isolated BM T cells from 11 MM patients (A) and mean values of individual results (B) from these patients with (black bars) or without (white bars) coculture with sorted CD138+ autologous (autol.) myeloma cells, as analyzed by short-term IFN-γ Elispot assay.
Aberrant CEACAM expression on myeloma cells
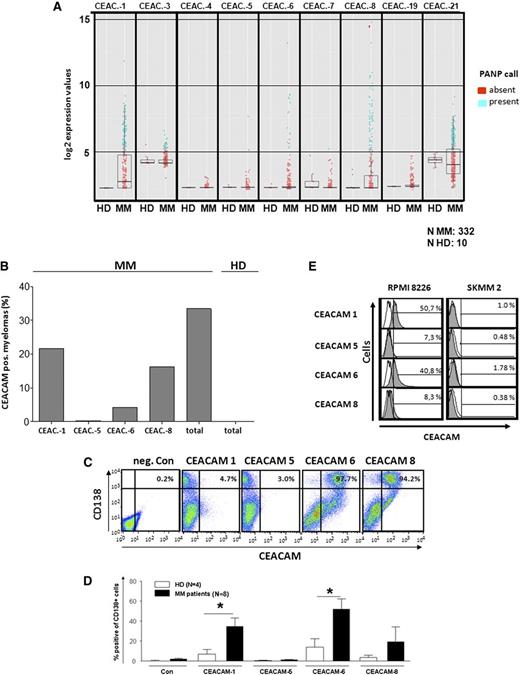
One of the major aims of this study was to assess the potential expression of CEACAM family members in MM. Therefore, expression of CEACAMs 1, 3-8, 19, and 21 was first analyzed in CD138+-purified myeloma cells from a large cohort of 332 previously untreated, therapy-requiring myeloma patients, and in purified BM plasma cells from 10 healthy donors by gene expression profiling. Using the PANP algorithm, we unexpectedly detected frequent expression of a broad variety of CEACAMs, namely CEACAM-1, CEACAM-3, CEACAM-6, CEACAM-8, and CEACAM-21 in malignant plasma cells, unlike normal BM plasma cells. The other CEACAMs were not (CEACAM-4, CEACAM-7, and CEACAM-19) or hardly (CEACAM-5) expressed (Figure 3A). In positive samples, CEACAMs−1, −6 and, −8 showed the highest relative expression levels. They have been associated with progression of human malignant disorderspreviously45-47 and therefore we concentrated our further analysis on these CEACAMs. The proportions of myeloma patients and healthy individuals expressing messenger (mRNA) of these CEACAMs (using the PANP algorithm) are shown in Figure 3B. In summary, 33.4% of myeloma patients expressed at least 1, 6.3% 2, and 2.7% 3 of these CEACAMs.
CEACAM expression in MM. (A) Expression of CEACAM-mRNA in CD138+ purified plasma cells from BM of myeloma patients (MM, n = 332) or healthy subjects (HD, n = 10), as determined by Affymetrix GeneChip arrays. Blue dots indicate expression; red indicates lack of expression as determined by the PANP-algorithm (see “Materials and methods”). (B) Percentages of samples expressing the respective CEACAMs in (A). (C-E) Expression of CEACAM protein on CD138+ myeloma cells from BM of MM patients on plasma cells from healthy subjects and on myeloma cell lines RPMI8226 and SKMM2, as determined by flow cytometry. (C) Dot plots showing expressions of CD138 and CEACAMs on BM cells from 1 MM patient. (D) Cumulative proportions of CEACAM-expressing CD138+ cells from BM of MM patients (black bars) or healthy individuals (white bars). (E) Gray histograms demonstrate CEACAM expression on gated CD138+ cells from myeloma cell lines RPMI8226 and SKMM2. Proportions of CEACAM-positive cells are shown as percentages (%). Negative controls are shown as white histograms. *Significant difference between patients and healthy donors (P < .05, 2-sided Student t test).
CEACAM expression in MM. (A) Expression of CEACAM-mRNA in CD138+ purified plasma cells from BM of myeloma patients (MM, n = 332) or healthy subjects (HD, n = 10), as determined by Affymetrix GeneChip arrays. Blue dots indicate expression; red indicates lack of expression as determined by the PANP-algorithm (see “Materials and methods”). (B) Percentages of samples expressing the respective CEACAMs in (A). (C-E) Expression of CEACAM protein on CD138+ myeloma cells from BM of MM patients on plasma cells from healthy subjects and on myeloma cell lines RPMI8226 and SKMM2, as determined by flow cytometry. (C) Dot plots showing expressions of CD138 and CEACAMs on BM cells from 1 MM patient. (D) Cumulative proportions of CEACAM-expressing CD138+ cells from BM of MM patients (black bars) or healthy individuals (white bars). (E) Gray histograms demonstrate CEACAM expression on gated CD138+ cells from myeloma cell lines RPMI8226 and SKMM2. Proportions of CEACAM-positive cells are shown as percentages (%). Negative controls are shown as white histograms. *Significant difference between patients and healthy donors (P < .05, 2-sided Student t test).
Then, protein expression of CEACAM-1, CEACAM-6, CEACAM-8, and CEACAM-5 was analyzed by flow cytometry using specific mAbs in CD138+ primary myeloma cells of 8 MM patients, in normal BM plasma cells, and in the RPMI-8226 and SKMM2 myeloma cell lines. Figure 3C shows a representative CEACAM expression profile of 1 patient; Figure 3D contains cumulative data. Compared with plasma cells from healthy donor, myeloma cells showed overall significantly increased expression of CEACAM-1 and CEACAM-6: 5 to 31% were CEACAM-1+ and 11 to 98% were CEACAM-6+. CEACAM-8 expression was detectable in subfractions of some myelomas. RPMI8226 myeloma cells expressed CEACAM-1 and CEACAM-6 weakly but homogeneously, as assessed by fluorescence-activated cell sorter analysis (Figure 3E), and confirmed by PCR, western blot, and immune precipitation (data not shown). The SKMM2 cell line was CEACAM-negative (Figure 3E). Healthy donor plasma cells showed low CEACAM expression, with ≤5% CEACAM-1+, ≤10% CEACAM-6+, and ≤3% CEACAM-8+ (Figure 3D). Taken together, in myeloma patients, the proportions of CEACAM-positive plasma cells among all plasma cells were heterogeneous but significantly increased compared with healthy individuals. Because CEACAMs were expressed only on subpopulations of myeloma cells, gene array analysis measuring to integral median of expression in the whole population in relation to the PANP-based threshold tends to underestimate the presence of fractions of CEACAM-positive myeloma cells in patients.
CEACAM-6 inhibits T-cell reactivity against myeloma cells
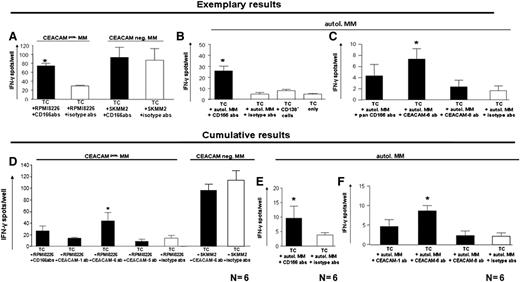
To evaluate whether myeloma cell CEACAM expression was involved in T-cell immune tolerance, BM T cells from MM patients were cultured with either CEACAM-positive (RPMI8226) or CEACAM-negative (SKMM2) (Figure 4A,D) allogeneic or purified CD138+ autologous myeloma cells (Figure 4B,C,E,F). These were pretreated with blocking anti-CEACAM (–1, −6, and −8) or respective isotype mAb. In the allogeneic setting, T-cell reactivity is based on alloreactivity against foreign HLA molecules; in the autologous setting, myeloma TCRs are directed against myeloma-associated antigens. Anti-CEACAM mAbs were removed from culture media before T-cell addition. Because of limitations in sorted patient-derived myeloma cells, analyses in the autologous setting included 5 × 104 T cells and 1 × 104 myeloma cells per well instead of 1 × 105 T cells and 2 × 104 myeloma cells, which were used for experiments with allogeneic cocultures. This may possibly explain overall spot number differences between the 2 experimental setups. T-cell reactivity was low against the CEACAM+ RPMI8226 myeloma cell line or autologous MM cells, but significantly increased after combined inhibition of CEACAM-1, CEACAM-6, and CEACAM-8 (Figure 4A-B). T cells strongly reacted against the CEACAM-negative SKMM2 myeloma cell line, and this reaction was not further increased by CEACAM blocking (Figure 4A).
Blocking of CEACAM on myeloma cells increases T-cell reactivity against myeloma cells. (A-C) Mean ± SEM IFN-γ spot numbers of 3 wells per test group from 3 different patients. IFN-γ secretion by T cells cocultured with the CEACAM-1+ and CEACAM-6+ myeloma cell line RPMI-8226 (A, left), the CEACAM-negative myeloma cell line SKMM2 (A, right) or with autologous myeloma cells (B-C). Compared with pretreatment with respective isotype antibodies, a significant increase of IFN-γ secretion was observed when CEACAM-positive myeloma cells were pretreated with mixed anti-CEACAM (–1, –6, and –8; CD166) antibodies (A,C), or with anti-CEACAM-6 mAb, but not anti-CEACAM-8 mAb (C). Further negative control groups contained T cells cocultured with CD138− BM cells or T cells only. (D) Cumulative results from 6 patients showing mean + SEM numbers of IFN-γ secreting cells after coculture with CEACAM-positive RPMI8226 myeloma cells or with CEACAM-negative SKMM2 myeloma cells pretreated with anti-CEACAM (–1, –6, or –8) mAb (black bars) or with respective isotype antibodies (white bars). (E-F) Cumulative results from six patients showing mean + SEM numbers of IFN-γ-secreting cells after coculture with autologous myeloma cells pretreated with mixed anti-CEACAM–1, –6, or –8 mAb (E, black bar), with single antibodies against CEACAM-1, CEACAM-6, or CEACAM-8 (F, black bars) or with respective isotype antibodies (white bars). Significant increase of IFN-γ secretion was observed only upon blocking of CEACAMs on myeloma cells. *Significant differences between test and respective control groups (P < .05, 2-sided Student t test).
Blocking of CEACAM on myeloma cells increases T-cell reactivity against myeloma cells. (A-C) Mean ± SEM IFN-γ spot numbers of 3 wells per test group from 3 different patients. IFN-γ secretion by T cells cocultured with the CEACAM-1+ and CEACAM-6+ myeloma cell line RPMI-8226 (A, left), the CEACAM-negative myeloma cell line SKMM2 (A, right) or with autologous myeloma cells (B-C). Compared with pretreatment with respective isotype antibodies, a significant increase of IFN-γ secretion was observed when CEACAM-positive myeloma cells were pretreated with mixed anti-CEACAM (–1, –6, and –8; CD166) antibodies (A,C), or with anti-CEACAM-6 mAb, but not anti-CEACAM-8 mAb (C). Further negative control groups contained T cells cocultured with CD138− BM cells or T cells only. (D) Cumulative results from 6 patients showing mean + SEM numbers of IFN-γ secreting cells after coculture with CEACAM-positive RPMI8226 myeloma cells or with CEACAM-negative SKMM2 myeloma cells pretreated with anti-CEACAM (–1, –6, or –8) mAb (black bars) or with respective isotype antibodies (white bars). (E-F) Cumulative results from six patients showing mean + SEM numbers of IFN-γ-secreting cells after coculture with autologous myeloma cells pretreated with mixed anti-CEACAM–1, –6, or –8 mAb (E, black bar), with single antibodies against CEACAM-1, CEACAM-6, or CEACAM-8 (F, black bars) or with respective isotype antibodies (white bars). Significant increase of IFN-γ secretion was observed only upon blocking of CEACAMs on myeloma cells. *Significant differences between test and respective control groups (P < .05, 2-sided Student t test).
Next, we blocked CEACAMs separately; increased TCRs were observed with anti-CEACAM-1 or anti-CEACAM-8 mAb in some patients, but only CEACAM-6 blockade induced significant increases against both allogeneic RPMI8226 cells and autologous MM cells in all 6 patients (Figure 4C-F). Because the anti-CEACAM-6 antibody did not recognize the CEACAM-1–positive melanoma cell line MeWo (data not shown), we can exclude potential crossreactivity to CEACAM-1, which can be expressed on activated T cells. Moreover, we could exclude binding of the CEACAM-6 antibody to resting or activated T cells or DCs (supplemental Figure 1), which is in line with our observation that the addition of CEACAM-6 blocking antibodies had no significant effect on T-cell reactivity against myeloma antigen–pulsed DCs (supplemental Figure 2).
CEACAM-6 gene knock down in RPMI8226 cells by specific siRNAs strongly increased their ability to elicit IFN-γ secretion by CD8+ T cells from MM patients to a comparable extent as antibody blockade, confirming that the observed increase in T-cell reactivity after antibody blockade was due to CEACAM-6 inhibition and not to unspecific effects of the anti-CEACAM-6 antibody (supplemental Figure 3). Taken together, these data demonstrate that CEACAM-6 on myeloma cells suppresses the reactivity of myeloma-reactive T cells.
CEACAM-6 inhibition on tumor cells increases TCR activation and cytotoxic T lymphocyte (CTL) activity
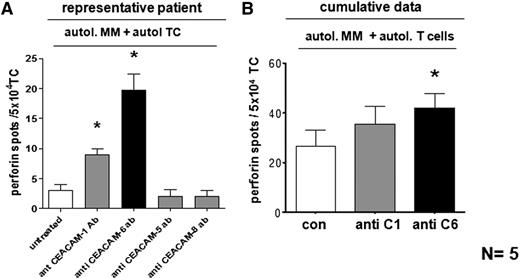
To assess the impact of CEACAM-6 on the cytotoxic T-cell response to tumor cells, we analyzed the secretion of perforin—a major cytotoxic effector molecule of CTL— from CD8+ T cells of 5 MM patients upon coculture with sorted autologous MM cells. Perforin secretion significantly increased upon CEACAM-6 inhibition, and to some extent upon CEACAM-1 inhibition (Figure 5), but not after blockade of CEACAM-5 or CEACAM-8 (data not shown). Figure 5A shows results of 1 myeloma patient; Figure 5B contains cumulative data from all 5 patients.
Blocking of CEACAM-6 on myeloma cells increases cytotoxic activity of tumor-reactive CTL. (A,B) Perforin secretion of ex vivo isolated, purified CD8+ T cells from a representative (A) or 5 (cumulative) MM patients (B), cocultured with sorted autologous myeloma cells in a perforin Elispot assay. Inhibition of CEACAM-1, CEACAM-5, and CEACAM-8 on myeloma cells was conducted by specific mAbs. A significant increase of perforin-secreting CTL was achieved upon CEACAM-6 inhibition (black bar). Unblocked myeloma cells (con; white bar) served for comparison. Data are shown as mean ± SEM numbers of perforin spots. TC, T cells only. *P < .05 or (*)P < .1, statistically significant differences between test and respective control groups as calculated by two-sided Student t test.
Blocking of CEACAM-6 on myeloma cells increases cytotoxic activity of tumor-reactive CTL. (A,B) Perforin secretion of ex vivo isolated, purified CD8+ T cells from a representative (A) or 5 (cumulative) MM patients (B), cocultured with sorted autologous myeloma cells in a perforin Elispot assay. Inhibition of CEACAM-1, CEACAM-5, and CEACAM-8 on myeloma cells was conducted by specific mAbs. A significant increase of perforin-secreting CTL was achieved upon CEACAM-6 inhibition (black bar). Unblocked myeloma cells (con; white bar) served for comparison. Data are shown as mean ± SEM numbers of perforin spots. TC, T cells only. *P < .05 or (*)P < .1, statistically significant differences between test and respective control groups as calculated by two-sided Student t test.
So far, our experiments indicated that CEACAM-6 can inhibit the reactivation of resting myeloma reactive T cells during their contact with myeloma cells. We next analyzed whether encounter of myeloma cells would also suppress the activity of preactivated CTL and whether this could be abolished by CEACAM-6 blockade.
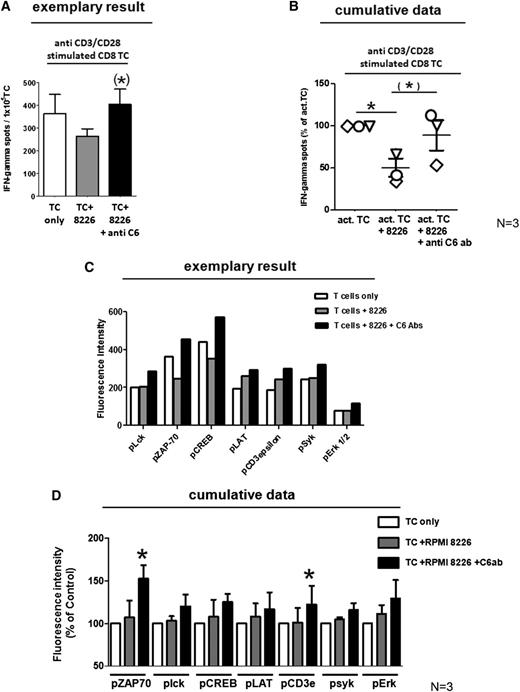
We therefore activated CD8+ T cells from MM patients by polyclonal TCR stimulation with anti-CD3 and anti-CD28 mAbs, cocultured aliquots with RPMI8226 myeloma cells in the presence or absence of CEACAM-6 blocking antibodies, and assessed their activity by IFN-γ Elispot assay. As shown in Figure 6A-B, IFN-γ secretion was significantly reduced in the presence of CEACAM-6–positive myeloma cells but restored through CEACAM-6 inhibition. Suppression of cytokine secretion required direct cell–cell contact between T cells and myeloma cells because the presence of CEACAM-6–positive myeloma cells did not affect IFN-γ release from cocultured activated CD8+ T cells when they were separated from each other by semipermeable membranes in transwell experiments (supplemental Figure 4).
Impact of CEACAM-6 on TCR signaling. (A) Polyclonally activated CD8+ T cells show reduced secretion of IFN-γ in response upon coculture with CEACAM-6–positive RPMI8226 myeloma cells assessed by IFN-γ Elispot assay of 1 representative of 3 MM patients. Mean ± SEM numbers of IFN-γ spots are shown for test wells containing only activated T cells (white bar), activated T cells in coculture with RPMI8226 myeloma cells (gray bar), or activated T cells in coculture with RPMI8226 myeloma cells on which CEACAM-6 was blocked before by CEACAM-6–specific mAbs (black bar). Cumulative data from all 3 tested patients are shown as % of IFN-γ secretion relative to corresponding wells containing TC only in (B). Each patient is represented by a different symbol. *P < .05 or (*)P < .1, statistically significant differences between test and respective control groups as calculated by 1-sided Student t test. (C) TCR signaling is reduced by CEACAM-6 on myeloma cells. The amount of different phosphorylated TCR-associated signaling molecules was assessed on CD8+ T cells from MM patients either cultured alone directly after ex vivo isolation (white bars) or after 5 minutes of coculture with untreated RPMI8226 myeloma cells (gray bars) or with RPMI8226 myeloma cells on which CEACAM-6 had been blocked by anti–CEACAM-6 mAb (black bars). Quantification of protein phosphorylation was conducted with phosphoprotein-specific antibodies by Luminex technology using 1 replicate per test group. One representative experiment of 3 is shown. Cumulative data from 3 tested patients are shown in (D) as % of fluorescence intensity relative to corresponding wells containing activated TC only. Mean ± SEM values from 3 independent experiments with different myeloma patients are shown. *Significant difference (P < .05) between TC treated with myeloma cells and T-cells treated with CEACAM-6 blocked myeloma cells (2-sided Student t test).
Impact of CEACAM-6 on TCR signaling. (A) Polyclonally activated CD8+ T cells show reduced secretion of IFN-γ in response upon coculture with CEACAM-6–positive RPMI8226 myeloma cells assessed by IFN-γ Elispot assay of 1 representative of 3 MM patients. Mean ± SEM numbers of IFN-γ spots are shown for test wells containing only activated T cells (white bar), activated T cells in coculture with RPMI8226 myeloma cells (gray bar), or activated T cells in coculture with RPMI8226 myeloma cells on which CEACAM-6 was blocked before by CEACAM-6–specific mAbs (black bar). Cumulative data from all 3 tested patients are shown as % of IFN-γ secretion relative to corresponding wells containing TC only in (B). Each patient is represented by a different symbol. *P < .05 or (*)P < .1, statistically significant differences between test and respective control groups as calculated by 1-sided Student t test. (C) TCR signaling is reduced by CEACAM-6 on myeloma cells. The amount of different phosphorylated TCR-associated signaling molecules was assessed on CD8+ T cells from MM patients either cultured alone directly after ex vivo isolation (white bars) or after 5 minutes of coculture with untreated RPMI8226 myeloma cells (gray bars) or with RPMI8226 myeloma cells on which CEACAM-6 had been blocked by anti–CEACAM-6 mAb (black bars). Quantification of protein phosphorylation was conducted with phosphoprotein-specific antibodies by Luminex technology using 1 replicate per test group. One representative experiment of 3 is shown. Cumulative data from 3 tested patients are shown in (D) as % of fluorescence intensity relative to corresponding wells containing activated TC only. Mean ± SEM values from 3 independent experiments with different myeloma patients are shown. *Significant difference (P < .05) between TC treated with myeloma cells and T-cells treated with CEACAM-6 blocked myeloma cells (2-sided Student t test).
In myeloma patients, clonally expanded CTL populations often express immune regulatory molecules associated with T-cell exhaustion and anergy, a prominent one being CD57, which may contribute to their dysfunctional phenotype.48 To assess whether interactions with CEACAM-6–positive myeloma cells may be involved in the induction of such a phenotype, we cocultured CD8+ T cells from 3 myeloma patients with RPMI8226 myeloma cells. CD57 expression was detectable on 28.7+19.8% of ex vivo analyzed CTL but was not influenced by coculture with CEACAM-6–positive myeloma cells (supplemental Figure 5).
We now assessed whether T-cell suppression may be mediated by direct influences of CEACAM-6 on the TCR signaling cascade. To this end, we explored the activation of major TCR-associated signaling molecules in patient-derived T cells upon contact with RPMI 8226 cells with or without CEACAM-6 blockade. To this end, we harvested CD8+ T cells after 2 to 5 minutes of coculture with RPMI tumor cells at a ratio of 5:1 and quantified in T-cell lysates the amount of activated signaling molecules by the Luminex method using phosphoprotein-specific antibodies. Although contact with myeloma cells did not induce the activation of lck, ZAP70, CREB, LAT, CD3ε, syk, or ERK, coculture with RPMI8226 cells that had been pretreated with anti-CEACAM-6 mAb to inhibit CEACAM-6 interactions caused the activation of all tested TCR-associated signaling molecules, particularly of ZAP70 (Figure 6C-D).
Discussion
Over the past few years, profound advances in the field of tumor immunotherapy have facilitated the development of tumor vaccines that efficiently elicit tumor antigen–specific CTL responses.49 However, the clinical efficiency of current tumor immunotherapies remains low.49 Many tumors have the capacity to block effector functions of T cells, and it is hypothesized that this is 1 of the reasons for the limited activity of tumor immunotherapy.50 Therefore, the identification of factors that regulate the activity of tumor-specific T cells during their interaction with tumor cells is of major clinical importance. We demonstrate here the role of CEACAM-6 in regulating effector T cell responses and its exploitation by malignant plasma cells.
In the current study, we detected T-cell reactivity against myeloma-associated antigens in myeloma patients. This reactivity was abrogated upon coculture with autologous myeloma cells and could be fully restored by blocking CEACAMs on the surface of myeloma cells. Using gene expression profiling, we demonstrate for the first time the expression of CEACAMs−1, –6, and –8 on the surface of primary myeloma cells and on a myeloma cell line, which significantly exceeded those of normal plasma cells from healthy donors in the case of CEACAM–1 and −6; this was validated by flow cytometry. Considerable expression as determined by the PANP algorithm of at least 1 of the tested CEACAMs at the mRNA level was detectable in 33.4% of patients. Although CEACAM-6 mRNA was detectable only in a small proportion of patients, CEACAM-6 protein was frequently expressed by subpopulations of myeloma cells. This might be explained by the fact that in some cases, the CEACAM-6–positive subpopulations or the amount of CEACAM-6 mRNA expression might be too small to reveal differential signals on a gene chip array. In addition, protein expression may not always be indicated by gene transcription because CEACAMs can be stored in vesicles until their release to the cell surface.32 The probability of an underestimation of CEACAM-6 expression by the gene array analysis was further supported by our observation of a weak, homogeneous CEACAM-6 protein expression in RPMI-8226 cells that was detectable by flow cytometry, reverse transcription PCR, and immune precipitation and also clearly indicated by the applied functional CEACAM-6 blocking analyses, but failed to be significantly detected by gene array analysis.
So far, a role of CEACAMs in the inhibition of T-cell responses has been demonstrated for CEACAM-1 but not for other CEACAMs. Only CEACAM-1 and −20 possess isoforms with immunoreceptor tyrosine-based inhibitory motif–containing cytoplasmic tails32 that generally transmit inhibitory signals. Although we could demonstrate that CEACAM-1 and CEACAM-8 blocking improved myeloma-specific T-cell reactivity in some patients, CEACAM-6 exerted a stronger and more consistent inhibitory effect on T-cell responses. Because CEACAM-6 has no intrinsic signaling capacity, its inhibitory activity is presumably mediated by binding to ligands on the T-cell surface.
CEACAMs can bind to a variety of molecules such as extracellular matrix components, integrins, or growth factor receptors. Besides, the homologous nature of the N-terminal IgV domains of the CEACAM family members and their structural composition of an immunoglobulin variable domain-like region followed by up to 6 immunoglobulin constant domain-like regions predisposes them for trans and cis homophilic and heterophilic interactions with other CEACAMs, including CEACAM-1 and such interactions have indeed been described, for example between CEACAM-1 and −5 or CEACAM-6 and −8.33 Therefore, recruitment and crosslinking of CEACAM-1 at the immunological synapse and subsequent suppression of T-cell effector function could potentially be mediated by heterophilic interactions as well. CEACAM-1 is not expressed on the surface of resting CD8+ T cells32 but stored in intracellular vesicles and rapidly externalized within 24 to 72 hours upon activation by cytokines or antigen recognition. These kinetics are similar to that of CD69 and therefore precede the activation-induced expression of the co-inhibitory receptor CTLA4.32 CEACAM-1 ligation and phosphorylation recruits SH2-domain-containing protein tyrosine phosphatase 1 (SHP1). SHP1 dephosphorylates ZAP70, which results in the inhibition of TCR signaling. Thereby, CEACAM-1 ligation leads to an early inhibition of T-cell activation within 10 minutes after activation, as demonstrated by reduced expression of CD69 and reduced proliferation.51
We here demonstrate that CEACAM-6 on myeloma cells not only inhibited IFN-γ secretion of ex vivo isolated and resting, but also of polyclonally activated CD8+ T cells from MM patients. Coculture with CEACAM-6–positive myeloma cells resulted in a CEACAM-6–mediated dephosphorylation of ZAP70 and inhibition of phosphorylation of other molecules important for TCR signaling such as CREB, lck, LAT, CD3ε, syk, and Erk.52-54 Although the identification of the CEACAM-6 ligand was not addressed in our study, these findings are consistent with an activation of SHP phosphatases by CEACAM-6 ligation. A direct or indirect role of CEACAM-1 cannot be excluded although direct interactions between CEACAM-1 and −6 have not been reported so far. Moreover, we cannot rule out that CEACAMs that were not specifically analyzed in this study, namely CEACAM-3 and −21, might also possess the potential to modulate T-cell reactivity in myeloma patients.
Although the recently observed expression of CEACAM-1 on some malignant melanomas was interpreted as a mechanism to escape recognition by tumor antigen–specific effector T cells, many tumors do not express CEACAM-1 but a variety of other CEACAMs. In several malignancies, including breast and colorectal cancer, CEACAM-6 expression was correlated with increased risk of relapse and reduced survival.47,55,56 However, CEACAM-6 modulated immune responses in malignant diseases have not been examined before. It is tempting to speculate that mechanisms underlying the adverse prognosis of CEACAM-6 in breast and colon cancer and potentially other tumors might be mediated by impaired T-cell–mediated immune response. Our findings are derived from a rather small group of patients and from a T-cell population that was functionally skewed in the BM microenvironment in a myeloma-specific manner. Therefore, they need to be interpreted with caution as regards to the relevance of CEACAM-6 for anti-tumor immune responses in general.
The present study is the first demonstration of CEACAM-1, CEACAM-6, and CEACAM-8 expression on a subfraction of purified myeloma cell samples. Our data indicate that CEACAMs on myeloma cells in these patients might inhibit the reactivation of tumor-reactive CD8+ T cells upon interaction with myeloma cells, possibly contributing as 1 of the mechanisms to the well-characterized immune resistance of MM.
Targeting the interactions of CEACAM-6 and −1 between T cells and their target cells might contribute to strengthen therapeutic T-cell responses against MM and have therefore therapeutic implications.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dieter Zopf and Wolf Doecke, both of Bayer-Health Care, for valuable experimental advice and thank Dr Fabienne McClanahan for language revision.
This study was supported in part by the Deutsches Krebsforschungszentrum-Bayer Health Care Alliance and by the Deutsche Forschungsgemeinschaft, Bonn, Germany, including the SFB/TRR79; the Dietmar Hopp Foundation, St. Leon-Rot, Germany; the University of Heidelberg, Germany; the Ligue Nationale Contre Le Cancer, Paris, France; and the 7th framework program of the European Union (OverMyR).
Authorship
Contribution: M.W.-H., D.H., M.H., A.S., T.M., K.N., H.G., and A.D.H. collected and prepared patient material; N.K., C.P., S.J., and L.U. conducted experiments; H.C., B.B., B.G., and H.B. generated CTL clones; T.R., J.-F.R., B.K., D.H., and H.G. conducted and analyzed gene array analysis; B.B. generated and contributed breast cancer cell lines; and P.B. designed the experiments and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for H.B. is Department of Radiology, University Hospital of Tübingen, Germany.
Correspondence: Philipp Beckhove, Translational Immunology Division, German Cancer Research Center, DKFZ, INF 280, 69120 Heidelberg, Germany; e-mail: p.beckhove@dkfz.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal