Abstract
There is growing research interest in the mammalian Tribbles (Trib) family of serine/threonine pseudokinases and their oncogenic association with acute leukemias. This review is to understand the role of Trib genes in hematopoietic malignancies and their potential as targets for novel therapeutic strategies in acute myeloid leukemia and acute lymphoblastic leukemia. We discuss the role of Tribs as central signaling mediators in different subtypes of acute leukemia and propose that inhibition of dysregulated Trib signaling may be therapeutically beneficial.
Introduction
The mammalian Tribbles (Trib) family, Trib1, Trib2, and Trib3, is classified as pseudokinases and has important roles in many cellular processes.1 Trib1-3 are postulated to act as adaptor molecules to regulate and integrate a wide range of signaling pathways. However, the functional interpretation of the pseudokinase classification remains unclear. Here, we review the Trib family members in acute leukemias and discuss Tribs as potential therapeutic targets in acute leukemias.
We examined Trib1-3 expression in human hematopoiesis2 using the Leukaemia Gene Atlas (LGA) platform3 (Figure 1A). Statistical analyses showed that Trib1 expression significantly increased in Granulocyte/Monocyte and B-cell lineages compared with other lineages, and indeed Trib1 knockout mice have a defect in macrophage and eosinophil differentiation.4 However, Trib2 and Trib3 expressions were found to significantly increase in T-cell and Erythrocyte (ERY) lineages, respectively, and interestingly, FOG and GATA2 both bind the Trib2 promoter in oligopotent progenitors with combined erythroid/megakaryocytic lineage potential to drive Megakaryocytic/Erythroid lineages.5 Trib1 expression increased in monocyte sublineage, whereas Trib2 and Trib3 expressions increased in granulocyte sublineage (Figure 1B). Only Trib2 was differentially expressed in CD4+ve T cells (Figure 1C). We observed similarities in human hematopoiesis of Trib1-3 expression, as described above (T cells, monocytes) and dissimilarities (haematopoietic stem cell [HSC] population) using the HemaExplorer platform6 (supplemental Figure 1). As opposed to LGA platform analysis (using one study), HemaExplorer combined data sets of human hematopoiesis originated from several studies.
Distributions of Trib1-3 expression in human hematopoietic system. Expressions of Trib1-3 in (A) different hematopoietic cell lineages and among the different cell populations of (B) Granulocyte/Monocyte and (C) T-cell lineages were examined by using the LGA based on the gene expression data set from Novershtern et al.2 Statistically significant increases of Trib1-3 expressions, marked by *adjusted P < .05 and **adjusted P < .001, were determined by Welch’s t test.
Distributions of Trib1-3 expression in human hematopoietic system. Expressions of Trib1-3 in (A) different hematopoietic cell lineages and among the different cell populations of (B) Granulocyte/Monocyte and (C) T-cell lineages were examined by using the LGA based on the gene expression data set from Novershtern et al.2 Statistically significant increases of Trib1-3 expressions, marked by *adjusted P < .05 and **adjusted P < .001, were determined by Welch’s t test.
Role of Tribbles in acute myeloid leukemia (AML)
Trib1
Previous studies have identified that only Trib1 expression increases in AML due to amplification of chromosome 8q24 region as double minutes (dmin) despite other genes, including MYC, present in 8q24-dmin, and the implications of this are yet to be determined.7,8 Using the LGA, we analyzed gene expression data from 5 different studies,9-13 where samples with known AML karyotypes and/or French-American-British (FAB) subtypes were available. Trib1 expression significantly increased in AML with inv(16) or t(16;16) and in FAB M4 and M5 compared with other karyotypes and FAB subtypes (Table 1) and with healthy controls (Table 2).
Expression of Tribbles increased in specific AML karyotypes and FAB subtypes
| Studies . | Trib1 (202241_at) . | Trib2 (202478_at) . | Trib3 (218145_at) . | |||
|---|---|---|---|---|---|---|
| Karyotype . | FAB . | Karyotype . | FAB . | Karyotype . | FAB . | |
| Valk et al9 * | inv(16) or t(16; 16)† | M4† | NS | NS | t(8;21)† | M2† |
| M5‡ | t(15;17)† | M3‡ | ||||
| Gutierrez et al10 | inv(16) (p12; q13)† | M4Eo† | NS | NS | t(15;17)(q12;q21)† | M3† |
| Verhaak et al11 | idt(16)† | M4‡ | NS | RAEB† | t(6;9)† | M2‡ |
| inv(16)‡ | M5‡ | RAEB-t† | t(8;21)‡ | M3‡ | ||
| t(6;9)† | t(15;17)‡ | |||||
| Haferlach et al12 * | complex† | — | complex† | — | complex‡ | — |
| t(8;21)† | inv(16) or t(16;16)† | t(15;17)† | ||||
| Eppert et al13 | +13† | NS | inv(16)(p13;q22)† | M4Eo† | NS | NS |
| inv(16)(p13;q22)† | ||||||
| Studies . | Trib1 (202241_at) . | Trib2 (202478_at) . | Trib3 (218145_at) . | |||
|---|---|---|---|---|---|---|
| Karyotype . | FAB . | Karyotype . | FAB . | Karyotype . | FAB . | |
| Valk et al9 * | inv(16) or t(16; 16)† | M4† | NS | NS | t(8;21)† | M2† |
| M5‡ | t(15;17)† | M3‡ | ||||
| Gutierrez et al10 | inv(16) (p12; q13)† | M4Eo† | NS | NS | t(15;17)(q12;q21)† | M3† |
| Verhaak et al11 | idt(16)† | M4‡ | NS | RAEB† | t(6;9)† | M2‡ |
| inv(16)‡ | M5‡ | RAEB-t† | t(8;21)‡ | M3‡ | ||
| t(6;9)† | t(15;17)‡ | |||||
| Haferlach et al12 * | complex† | — | complex† | — | complex‡ | — |
| t(8;21)† | inv(16) or t(16;16)† | t(15;17)† | ||||
| Eppert et al13 | +13† | NS | inv(16)(p13;q22)† | M4Eo† | NS | NS |
| inv(16)(p13;q22)† | ||||||
AML samples with unknown karyotype or FAB subtype were excluded from statistical analysis provided by the LGA.
NS, no statistically significant increased expression found; –,unavailable data for statistical analysis.
AML samples with normal karyotype were not able to be included in the statistical analysis.
Statistically significant increased expression with adjusted P < .05 in Welch’s t test.
Statistically highly significant increased expression with adjusted P < .001 in Welch’s t test.
Expression of Tribbles increased in subsets of AML compared with healthy samples
| Studies . | Trib1 (202241_at) . | Trib2 (202478_at) . | Trib3 (218145_at) . | |||
|---|---|---|---|---|---|---|
| Karyotype . | FAB . | Karyotype . | FAB . | Karyotype . | FAB . | |
| Valk et al9 | inv(16) or t(16;16)* | M4* | NS | NS | t(8;21)* | M2* |
| M5* | t(15;17)† | M3† | ||||
| Haferlach et al12 | NS | — | NS | — | complex† | — |
| t(15;17)* | ||||||
| Studies . | Trib1 (202241_at) . | Trib2 (202478_at) . | Trib3 (218145_at) . | |||
|---|---|---|---|---|---|---|
| Karyotype . | FAB . | Karyotype . | FAB . | Karyotype . | FAB . | |
| Valk et al9 | inv(16) or t(16;16)* | M4* | NS | NS | t(8;21)* | M2* |
| M5* | t(15;17)† | M3† | ||||
| Haferlach et al12 | NS | — | NS | — | complex† | — |
| t(15;17)* | ||||||
Elevated expression of Tribbles in karyotypes and FAB subytypes identified in Table 1 were compared with that of healthy samples. Healthy samples were not available in Gutierrez et al,10 Verhaak et al11 and Eppert et al.13
Abbreviations are explained in Table 1.
Statistically significant increased expression with adjusted P < .05 in Welch’s t test.
Statistically highly significant increased expression with adjusted P < .001 in Welch’s t test.
Trib1 was first discovered as a cooperating gene in a murine model of HOXA9/MEIS1 myeloid leukemogenesis.14 Trib1 overexpression alone is able to induce AML in mice by promoting degradation of C/EBPα.15 Interaction between Trib1 and MEK1 leads to ERK phosphorylation and degradation of C/EBPα.16 A gain-of-function mutation, R107L in Trib1, was identified in human acute megakaryocytic leukemia,17 and its overexpression accelerates the onset of murine AML.
Trib2
Trib2 was the first family member to be identified as an oncogene, where it induced potent AML in mice through inactivation of C/EBPα.18 Trib2 induces proteasomal-dependent degradation of C/EBPα via the E3 ligase COP1,18,19 changing the C/EBPα isoform ratio in favor of the truncated oncogenic form. We did not observe elevated expression of Trib2 in any AML karyotypes or FAB subtypes (Table 1). Trib2 expression is generally low20 but up-regulated specifically in a biologically and epigenetically distinct subset of immature AML21 with silenced CEBPA and a mixed myeloid/T-lymphoid phenotype.18,22 Trib1 expression is lower in this cluster,20 which could potentially be attributed to the different roles of Trib1 and Trib2 in myeloid and lymphoid hematopoiesis.
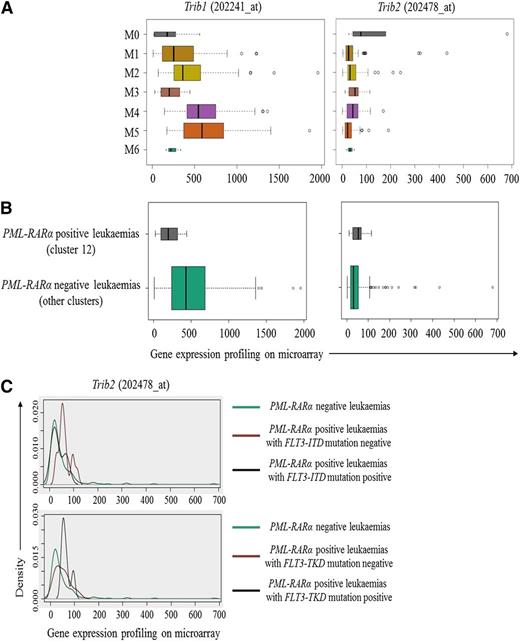
All-trans retinoic acid is a hugely successful differentiation therapy for acute promyelocytic leukemia (APL) (M3) with t(15;17) translocation (PML-RARα), and Trib2 expression levels, albeit overall low, are at higher levels in the M0 and M3 subtypes and in PML-RARα–positive leukemias (cluster 12) compared with PML-RARα–negative leukemias (Figure 2A-B). Interestingly, Trib1 levels are lower in PML-RARα–positive leukemias compared with Trib2 (Figure 2A). Approximately 37% of PML-RARα APL patients have mutations that constitutively activate FLT3.23 Cluster 12 originally identified by Valk et al9 can be divided into 2 subgroups that correspond to the FLT3-ITD mutation status. Increased Trib2 expression in this cluster was associated with FLT3-TKD but not FLT3-ITD mutations (Figure 2C). FLT3-ITD has been shown to induce a myeloproliferative disease and to cooperate with PML-RARα to induce an APL-like disease.24 However, FLT3-TKD was shown to induce a murine lymphoid disease,25 and the cooperative relationship between FLT3-TKD and PML-RARα has not been examined. In contrast to FLT3-ITD, FLT3-TKD could not induce aberrant activation of STAT5 and repression of C/EBPα and Pu.1.26 Thus, Trib1 and 2, known to inactivate C/EBPα, may be important in PML-RARα–positive leukemias that harbor FLT3-TKD mutations.
Elevated expression of Trib2 in PML-RARα–positive leukemias is associated with FLT3-TKD but not FLT3-ITD mutations. Expressions of Trib1 and Trib2 in (A) different FAB subtypes and (B) between PML-RARα– positive and –negative leukemias were examined by using the LGA based on the gene expression data set from Valk et al.9 (C) Trib2 expression in PML-RARα–positive leukemias was further stratified based on FLT3-ITD and -TKD mutation status and compared with that of PML-RARα–negative leukemias.
Elevated expression of Trib2 in PML-RARα–positive leukemias is associated with FLT3-TKD but not FLT3-ITD mutations. Expressions of Trib1 and Trib2 in (A) different FAB subtypes and (B) between PML-RARα– positive and –negative leukemias were examined by using the LGA based on the gene expression data set from Valk et al.9 (C) Trib2 expression in PML-RARα–positive leukemias was further stratified based on FLT3-ITD and -TKD mutation status and compared with that of PML-RARα–negative leukemias.
Both Trib114 and Trib227 are target genes of HOX-mediated leukemogenesis, but they are activated in a different context of murine AML. Trib1 is activated in HOXA9/MEIS1-AML,14 whereas Trib2 is activated in NUP98-HOXD13/MEIS1-AML.27 Differential activation of Trib1 and Trib2 might underlie the dissimilarities previously identified in HOXA9- and NUP98-HOXD13–mediated leukemogenesis.28,29 Trib2 was shown to also cooperate with HOXA9 and accelerate the onset of murine AML,30 indicating that both elevated Trib1 and Trib2 are cooperative events in HOXA9–positive AMLs.
Trib3
Our analysis showed that Trib3 expression significantly increased in AML with t(8;21) and t(15;17) and in FAB M2 and M3 subtypes (Tables 1 and 2). Although overexpression of Trib3 was unable to drive AML in the murine model,15 future study should determine if Trib3 is a cooperating leukemogen with AML1-ETO and PML-RARα, as it may contribute to leukemogenesis via a different mechanism.
Role of Tribbles in acute lymphoblastic leukemia (ALL)
No evidence is currently available to implicate the involvement of Trib1 and Trib3 in ALL, whereas there is growing evidence for a role of Trib2 (supplemental Figure 2).
Trib2 was differentially expressed in CD4+ve T cells (Figure 1C) and is highest in T cell-ALL (T-ALL) with normal karyotype and lowest in ALL with t(12;21).31 Trib2 was first identified in a screen for downstream effectors of NOTCH1 signaling in T-ALL18 and is associated with activating NOTCH1 mutations.31 Subsequent study showed that NOTCH1 binds to the Trib2 promoter and up-regulates its expression.22 We identified putative NOTCH1 binding sites in the promoter of Trib1, but not Trib3 (data not shown), which is interesting, as Trib3 cannot drive murine AML in the bone marrow transplant model.15 Malignant thymocytes in NOTCH1-associated T-ALL are arrested at the double positive (DP) (CD4+veCD8+ve) stage of development. In normal T-cell development, expansion and differentiation of double negative (DN) to DP thymocytes requires pre-T cell receptor (TCR) signaling. High Trib2 expression in the T-ALL subset was shown to enrich for gene sets that define TCR signaling.31 We found a gradual increase of Trib2 expression from DN to DP thymocytes using the ImmGen Data Browsers32 (supplemental Figure 3). Hence, it is likely that Trib2 is a T-cell–specific cooperative signal required by aberrant NOTCH1 signaling to drive transformation, proliferation, and survival of malignant thymocytes.
In T-ALL, Trib2 appears to be a downstream target of multiple oncogenic transcription factors. As well as NOTCH1,22,31 PITX133 and TAL134 were also found to up-regulate Trib2. PITX1 is recurrently activated in T-ALL to deregulate genes involved in T-cell development, including Trib2.33 TAL1 is aberrantly activated in 50% to 60% of human T-ALL patients,35 and 40% of these patients also develop activating mutations in NOTCH1.36 Trib2 was identified in a knockdown screen in T-ALL as one of the critical targets of the core transcriptional regulatory circuit controlled by the TAL1 complex, and, importantly, Trib2 was shown to be essential for the growth and survival of human T-ALL cell lines.34 A TAL1/LMO2 mouse model of T-ALL showed that 75% of the T-ALL mice develop spontaneous activating mutations in NOTCH1.37 Thus, further studies are warranted to examine the role of Trib2 and its potential cooperation in TAL1+ve and NOTCH1 mutant T-ALL pathogenesis.
Trib2 is also potentially involved in B cell-ALL (B-ALL) with t(1;19), as the expression level of Trib2 in this subset of ALL was higher than that in T-ALL.31 t(1;19, E2A-PBX1) is present in ∼6% of all B-ALLs, 25% of pediatric pre-B-ALL, and in rare cases of myeloid and T-cell leukemias.38 E2A-PBX1 was shown to cooperate with NOTCH1 and HOXA9, which have established relationships with Trib2 to induce T-cell lymphoma/leukaemia39 and AML40 in murine models. Given the strong link between NOTCH1 and Trib2 in T-ALL, the development of Trib inhibitors may prove to be potentially therapeutic.
Targeting Trib in AML and ALL therapy
Structurally, Trib family members have 3 clearly distinguishable regions: a C-terminal region that contains a MEK1 and COP1 E3 ligase-binding sites, a central serine/threonine kinase-like domain with an ATP binding motif, and an N-terminal region not required for oncogenicity.19 The binding of COP1 and another E3 ligase, TRIM21,41 to Trib is essential for Trib-induced proteasomal degradation of C/EBPα and AML, suggesting that potential inhibitors that may act by inhibiting the Trib-E3 ligase (COP1/TRIM21) relationship using proteasome inhibitors or molecules that interfere with the binding of the E3 ligase to the Trib C-terminal region would be effective therapeutics for Trib-induced AML. Current clinical trials using bortezomib (proteasome inhibitor) with other standard chemotherapeutics are underway in myelodysplastic syndrome and AML.
In comparison with conventional kinases, Tribs are considered pseudokinases, as they contain an N-terminal lobe in the central kinase region that contains a lysine residue critical for ATP binding and also other atypical kinase motifs.1 Although Tribs lack demonstrable serine/threonine kinase activity to date, the intact kinase domain is required for leukemogenesis.19 Thus, small molecule inhibitors that specifically target the Trib kinase domain ATP binding pocket, for example, may be good therapeutics for AML and ALL involving up-regulated Tribs. All 3 Tribs contain a conserved motif that is required for the MEK1 binding in the C terminus of the kinase domain and results in enhanced ERK phosphorylation required for the degradation of C/EBPα in AML.15,16 Thus, MEK1 inhibitors could be considered as potential chemotherapeutic agents targeting Trib function in AML and ALL. As it appears that Tribs may be central mediators of signaling pathways, they may be good therapeutic downstream targets in T-ALL and AML driven by other oncogenes (eg, NOTCH1, FLT3, HOX).
Perspectives
The specific subtypes of acute leukemia that each Trib associates with have differentiation arrest at different stages of hematopoiesis. Hence, understanding the regulation of Trib1-3 lineage-specific expression and their roles in differentiation is important. This will pave the way to understanding the molecular aberrations and cooperative signaling pathways that occur during leukemic transformation.
It is important to note the differences in the human and murine leukemias. Our analyses of human AML consist solely of mRNA expression levels, whereas in murine overexpression models, elevated protein expression would not be subject to transcriptional regulation. Indeed, the myeloid vs lymphoid differences seen in the murine vs human disease may be due to the origin of cells that overexpress Trib2. These discrepancies may be resolved through the use of conditional knockin Trib models or murine transplantations using lineage-specific donor cells. A more relevant Trib-induced leukemia murine model is necessary to gain functionally and clinically relevant data.
Though studies so far suggest Trib proteins act as adaptor molecules or decoy kinases in signaling pathways and the proteasome degradation pathway, it remains possible that Trib proteins are atypical kinases that can directly affect substrate protein activity. To date, direct substrate phosphorylation via Trib proteins has not been described, and, indeed, screens for novel Trib substrates have not been documented.
This review highlights the contribution of Trib proteins in AML and ALL signaling pathways and presents them as potential targeted therapies. Inhibitors that target activated oncogenes with essential functions in normal cells are likely to have narrow therapeutic windows and serious side effects. Available knockout mice for Trib2 and Trib3 have not shown significant phenotypic manifestations;42 however, a critical role for Trib1 in macrophage differentiation leading to adipose tissue maintenance and suppression of metabolic disorders has been demonstrated in Trib1 knockout mice.4 Future work in knockout mice is required to clarify the physiological and/or redundant roles for each Trib family member in normal and malignant hematopoiesis. Nevertheless, this indicates that selective targeting of Trib family members might be the favorable approach.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
K.L.L. is supported by Health Research Board Ireland. L.R. and K.K. are supported by the Howat Foundation and Children with Cancer, UK.
Authorship
Contribution: K.L.L. analyzed data, wrote the paper, and made the figures; L.R. wrote the paper; and K.K. designed the study and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Karen Keeshan, Paul O'Gorman leukemia research centre, University of Glasgow, 21 Shelley Rd, Glasgow, G12 0XB United Kingdom; e-mail: karen.keeshan@glasgow.ac.uk.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal