Key Points
Hematopoietic stem cell aging associates with stable transcriptional alterations.
Somatic cell reprogramming reverses functional defects associated with hematopoietic aging.
Abstract
Aging of hematopoietic stem cells (HSCs) leads to several functional changes, including alterations affecting self-renewal and differentiation. Although it is well established that many of the age-induced changes are intrinsic to HSCs, less is known regarding the stability of this state. Here, we entertained the hypothesis that HSC aging is driven by the acquisition of permanent genetic mutations. To examine this issue at a functional level in vivo, we applied induced pluripotent stem (iPS) cell reprogramming of aged hematopoietic progenitors and allowed the resulting aged-derived iPS cells to reform hematopoiesis via blastocyst complementation. Next, we functionally characterized iPS-derived HSCs in primary chimeras and after the transplantation of re-differentiated HSCs into new hosts, the gold standard to assess HSC function. Our data demonstrate remarkably similar functional properties of iPS-derived and endogenous blastocyst-derived HSCs, despite the extensive chronological and proliferative age of the former. Our results, therefore, favor a model in which an underlying, but reversible, epigenetic component is a hallmark of HSC aging.
Introduction
Many of the pathologies that could benefit from regenerative stem cell-based therapies are age-associated.1 An evident example is represented by bone marrow (BM) transplantation, a therapeutic modality that relies on the regenerative capacity of hematopoietic stem cells (HSCs) and where graft age is one of the most significant negative parameters for successful outcome.2 One key mechanism proposed to underlie HSC aging involves the acquisition of permanent genetic changes such as accumulation of DNA damage and/or telomere attrition.3 Yet, while there is a potential role for DNA damage in HSC aging, it is currently unclear if accumulation of DNA damage could explain all or even a part of the distinct transcriptional changes that associate with HSC aging and that ultimately underlie their altered function.4-7
For the purpose of regenerative medicine, alterations in the underlying genomic DNA sequence would be problematic to correct efficiently. By contrast, it might be more conceivable to alter the regulation of the transcriptome of a stem cell population, assuming permanent genomic damage does not underlie and dictate the transcriptional changes. One conceptual example is represented by induced pluripotent stem cells (iPS). These iPS cells, created by transcription factor-mediated reprogramming of somatic cells,8 progressively gain particular epigenetic features during the reprogramming process while simultaneously loosing those of the somatic starting cell,9,10 to finally reach a relatively stable embryonic stem (ES) cell-like state.11
Here, to explore the stability and potential reversibility of hematopoietic progenitor cell aging, we generated iPS cells from aged hematopoietic stem and progenitor cells (HSPCs). Next, we used these iPS cells to complement blastocysts. The iPS-derived HSCs in chimeric mice were evaluated for several parameters normally altered with age, including their telomere length, reconstitution ability, lineage competence, and production of naïve T cells. Our data suggest that a dominant feature of chronological HSC aging is an epigenetic “drift,” which can be reversed by epigenetic reprogramming.
Materials and methods
Mice
All mice were on a C57BL/6 background with the congenic marker combinations CD45.1, CD45.2, or F1CD45.1 × CD45.2. Animals were housed at animal facilities at Lund University. The Transgenic Core Facility at Lund University performed blastocyst injections. All animal experiments were performed with consent from a local ethical committee.
Retrovirus production
Retroviral vectors (pMXs-Oct3/4-IP, pMXs-Sox2-IP, pMXs-Klf4, pMXs-c-Myc-IP) were acquired from Addgene (Cambridge, MA) and were generously deposited by Shinya Yamanaka. Vector pMX-GFP was obtained from Cell Biolabs (San Diego, CA). Replication incompetent viruses were produced by transfecting the vectors into Plat-E packaging cells (Cell Biolabs, San Diego, CA) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Supernatants were harvested 48-hours posttransfection and titered using the Retro-X qRT-PCR Titration Kit (Clontech, Mountain View, CA) and fluorescence-activated cell sorter (FACS) for pMX-green flourescent protein (GFP). Viruses used in experiments had titers ranging from 4 × 106 to 4 × 108 viral particles/mL.
Hematopoietic stem and progenitor isolation
For iPS generation, mouse BM was depleted of mature cells using a cocktail of biotinylated antibodies (B220, CD4, CD8a, CD11b, Gr-1, Ly6c, and Ter119) together with anti-biotin MACS beads (Miltenyi Biotech, Bergisch Gladbach, Germany), before being further enriched for HSPCs by magnetic enrichment of CD117-positive cells (Miltenyi Biotech). For isolation and analysis of HSPCs, lineage-depleted samples were stained with streptavidin conjugated to QD605 (Invitrogen) and antibodies recognizing c-Kit, Sca-1, CD48, CD105, CD150, CD45.1, and CD45.2 (all from BioLegend, San Diego, CA), and sorted on a FACSAria (BD Biosciences, San Jose, CA). Propidium iodide (Invitrogen) was used to exclude dead cells.
The iPS generation from HSPCs
For prestimulation of primary cells prior to and during transduction, cells were cultured in OptiMEM (Invitrogen) supplemented with 10% fetal calf serum (FCS), penicillin/streptomycin (Invitrogen), 50 ng/mL stem cell factor, 5 ng/mL IL-3 (Peprotech Inc. Rocky Hill, NJ), and 5 U/mL EPO (Janssen-Cilag, Bersee, Belgium). Both candidate and established iPS cells were cultured under standard ES cell conditions; grown on irradiated mouse embryonic fibroblasts in DMEM (Invitrogen) containing 15% FCS, penicillin/streptomycin (Invitrogen), 1 mM sodium pyruvate (Invitrogen), 0.1 mM β-mercaptoethanol (Invitrogen), 1× MEM NEAA (Invitrogen), and 103 U/mL LIF (Millipore, Billerica, MA).
To generate iPS lines, 50 000 Lin-c-Kit+ cells were transduced for 2 days using Retronectin (Takara Bio Inc., Saint-Germain-en-Laye, France) coated plates preincubated with retroviruses and thereafter transferred to ES cell culture conditions. The cells were cultured for 14 days with daily media changes. On day 14, there were 25 candidate iPS cells sorted into individual wells of a 96-well plate in ES cell culture conditions based on their negativity for GFP, as iPS induction coincides with efficient silencing of retroviral long terminal repeat regions8 and CD45, a pan-hematopoietic marker lost during iPS conversion, and by being stage-specific embryonic antigen 1 and epithelial cell adhesion molecule positive.
Characterization of iPS clones
The iPS lines were assayed for viral vector integration by polymerase chain reaction (PCR) using 1 primer specific for each vector’s coding sequence and a common primer directed against the vector backbones. Chimeric mice were genotyped with a similar strategy. PCR reactions were run using Taq polymerase (Takara Bio Inc., Saint-Germain-en-Laye, France) with initial incubation at 94°C for 2 minutes followed by 35 cycles of 30 seconds at 94°C, 30 seconds at 60°C, and 30 seconds at 72°C. In vitro differentiation capacity was investigated by embryoid body formation by cultivating 250 ES or iPS cells in 20 uL hanging droplets for 48 hours in IMDM (Invitrogen) supplemented with 20% FCS, penicillin/streptomycin (Invitrogen), 1 mM sodium pyruvate (Invitrogen), 0.1 mM β-mercaptoethanol (Invitrogen) and GlutaMAX (Invitrogen). Thereafter, cells were pooled and cultured in petri dishes for 12 to 14 days before analysis. To verify iPS chimerism in adult mice, hippocampal brain sections were used to detect Y chromosome (from iPS cells) by Star fluorescence in situ hybridization protocol (Cambio Ltd, Cambridge, UK).
Bone marrow transfer and peripheral blood analysis
For bone marrow reconstitution experiments, young CD45.2 mice were lethally irradiated with 800 rad 3 to 4 hours prior to transplantation. Whole bone marrow (2 to 5 × 106 cells) or purified HSCs (200 or 1500 cells) together with 300 000 unfractionated BM support cells were injected into the tail-vein of recipient animals in a volume of 500 uL. Long-term contribution to the different peripheral blood cell lineages was followed for more than 16 weeks to assess long-term contribution of the transplanted HSCs. The analysis of the peripheral blood was done as described.12 Samples were analyzed on an LSRII (BD Biosciences). The competitive ability/transplanted HSC was measured by ((estimated frequency of transplanted competitor HSCs × % long-term myeloid chimerism)/(1 - % long-term myeloid chimerism))/actual or estimated number of test cells transplanted, with the assumption that HSCs exist at a frequency of 1/10 000.
Measurement of TRECs
T-cell receptor excision circles (TRECs) were measured in sorted CD4+/CD8+ splenic T cells by adapting a previously described protocol.13 Briefly, the sorted cells were lysed using Proteinase K (Roche, Mannheim, Germany) and they were assayed for relative TREC content using a SYBR green-based quantitative PCR (qPCR) approach where the cycle threshold value of a primer pair directed against a TREC was normalized against that of the single-copy gene Myt-1. To measure the naïve T-cell competence of HSCs in BM transfer experiments, the chimerism of the splenic CD4+/CD8+ compartment was divided by the actual, or estimated, number of transplanted HSCs and multiplied by the relative TREC incidence acquired by qPCR.
Telomere length measurement
To estimate telomere lengths in individual cells, single cells were isolated using FACS and were assayed as previously described.14-16
Affymetrix gene expression analysis
RNA was extracted from 10 000 sorted HSCs (Lin-Sca1+c-Kit+CD48-CD150+) isolated from young or aged mice, or from recipients of young and old bone marrow cells, where they in addition were selected for appropriate CD45 isoform expression using the RNeasy-micro mRNA purification kit (QIAGEN, Hilden, Germany) as previously described.12 Following a 2-round amplification protocol, biotin labeling, and hybridization to Affymetrix 430.2 gene expression arrays, probe level expression values were extracted using RMA17 and analysis performed using the dChip software18 following filtering of probes with a lower expression than 100 in either experimental group. To identify differentially expressed genes with age (Figure 1A), genes with a >1.5-fold difference in expression with a confidence interval of >90% were extracted. The microarray data used in this study can be found in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE44923 and GSE27686 (steady-state HSCs), GSE44923 (transplanted HSCs), GSE44923 (young blood-induced pluripotent stem (BiPS) cells, aged BiPS cells, and ES cells), GSE6503 (comparative dataset 1), and GSE4332 (comparative dataset 2).
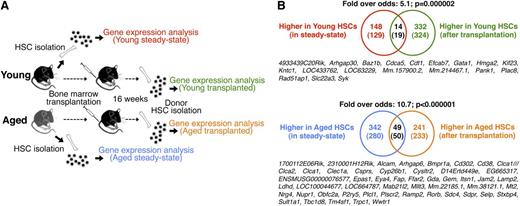
Transcriptional alterations of HSC aging persist through transplantation-mediated stress. (A) Experimental outline. (B) Venn diagrams depicting the number of genes up-regulated or down-regulated in HSCs with age, and the overlap of genes that remain differentially regulated as a consequence of HSC aging after transplantation. Numbers in Venn diagrams depict differentially regulated probe sets, with numbers in brackets indicating unique genes. The identities of common genes are shown below each diagram. The analysis was performed on gene expression arrays from 2 independent experiments totaling 6 replicate arrays for the young and aged steady-state setting and 2 replicate arrays for the young and aged transfer setting.
Transcriptional alterations of HSC aging persist through transplantation-mediated stress. (A) Experimental outline. (B) Venn diagrams depicting the number of genes up-regulated or down-regulated in HSCs with age, and the overlap of genes that remain differentially regulated as a consequence of HSC aging after transplantation. Numbers in Venn diagrams depict differentially regulated probe sets, with numbers in brackets indicating unique genes. The identities of common genes are shown below each diagram. The analysis was performed on gene expression arrays from 2 independent experiments totaling 6 replicate arrays for the young and aged steady-state setting and 2 replicate arrays for the young and aged transfer setting.
Quantitative PCR primers used for SYBR green mRNA expression analysis
Oct3/4 5′-TTCTAGCTCCTTCTGCAGGG-3′ (Fw)
5′-AGAGGGAACCTCCTCTGAGC-3′ (Rev)
Sox2 5′-CTCTGCACATGAAGGAGCAC-3′ (Fw)
5′-CCGGGAAGCGTGTACTTATC-3′ (Rev)
Klf4 5′-CAGTCGTAAGGTTTCTCGCC-3′ (Fw)
5′-GCCACCCACACTTGTGACTA-3′ (Rev)
c-Myc 5′-ACGGAGTCGTAGTCGAGGTC-3′ (Fw)
5′-AGAGCTCCTCGAGCTGTTTG-3′ (Rev)
Nanog 5′-ATGCCTGCAGTTTTTCATCC-3′ (Fw)
5′-GAGGCAGGTCTTCAGAGGAA-3′ (Rev)
β-actin 5′-CCACAGCTGAGAGGCAAATC-3′ (Fw)
5′-CTTCTCCAGGGAGGAAGAGG-3′ (Rev)
Qualitative PCR primers used for genomic DNA analysis
Oct3/4 5′-CAGTCCAACCTGAGGTCCAC-3′ (Rev)
Sox2 5′-CCGGGACCATACCATGAA-3′ (Rev)
Klf4 5′-TCCTCACGCCAACGGTTAGT-3′ (Rev)
c-Myc 5′-AGGGCTGTACGGAGTCGTAG-3′ (Rev)
Universal forward primer (viral backbone):
5′-GACGGCATCGCAGCTTGGATACAC-3′
sjTREC 5′-CCAAGCTGACGGCAGGTTT-3′ (Fw)
5′-AGCATGGCAAGCAGCACC-3′ (Rev)
Myt-1 5′-TGCCACCGCTGCTAATGAG-3′ (Fw)
5′-TGCGAACTCCTAAGCCAGCTA-3′ (Rev)
Telomeric repeats:
5′-CGGTTTGTTTGGGTTTGGGTTTGGGTTTGGGTTTGGGTT-3′ (Fw)
5′-GGCTTGCCTTACCCTTACCCTTACCCTTACCCTTACCCT-3′ (Rev)
Statistical analysis
Genome-wide gene expression microarray data were analyzed using dChip and Gene Set Enrichment Analysis, according to the manufacturer’s instructions. For fold-over-odds calculations, we applied a binomial probability calculation (Poisson distribution approximation). Otherwise, significance values were calculated by Student two-tailed t test.
Results
Genome-wide expression profiling reveals that an aging HSC signature is maintained after transplantation
It is well established that aging imposes several functional shortcomings on HSCs, including pronounced shifts in the types of mature effector cells produced and alterations in self-renewal.19 Most existing data argue that these features are autonomous to HSCs since arising phenotypes are visualized when transplanting aged HSCs into young hosts, but not vice versa.4 However, 1 concern with HSC transplantation experiments is that they tend to evaluate HSC function based on retrospective analysis of the produced progeny rather than directly at the level of HSCs. To begin to approach this issue, we transplanted young lethally irradiated mice with HSPC-enriched BM cells from young and old donors. Four-months posttransplant, we isolated highly purified linage negative, Sca-1+, ckit+, CD48-, and CD150+ HSCs from these reconstituted animals, and performed genome-wide expression analyses using Affymetrix 430.2 microarrays (“Transplanted HSCs”; two biological arrays/condition). We reasoned that this strategy would provide a mean to expose relatively permanent intrinsic gene expression changes, separate from those that associate strictly with cell division, as transplantation enforces vigorous cell cycling of the otherwise quiescent HSCs.20 By stringent analysis criteria, these experiments revealed differential expression of 573 probe sets (332 with lower and 241 with higher expression in HSCs derived from aged donors; supplemental Table 1). Next, we conducted similar analyses on HSCs from young and aged “steady-state” donors (6 biological replicates for each condition). These experiments revealed 490 probe sets with differential expression in the steady-state situation (supplemental Table 1), with the majority being up-regulated in aged HSCs (342 probe sets). The gene lists obtained from our 2 lines of experiments were next intersected to investigate a potential overlap. This revealed a highly significant correlation for both the “young-associated” (Figure 1A, odds-ratio: 5.1; P = .000002) and “aging-associated” (Figure 1B, odds-ratio: 10.7; P < .000001) probe sets. In an alternative analysis, we implemented Gene Set Enrichment Analysis.21 This independent approach confirmed a highly significant association of the steady-state young and aged HSC profiles to those obtained from young and aged HSCs subjected to transplantation (P < .001 for both up-regulated and down-regulated genes; supplemental Figure 1). A large proportion of these stably differentially expressed genes were also differentially expressed in 2 independently generated datasets4,5 of steady-state young and aged HSCs (supplemental Figure 2). These data establish that a core set of transcriptional alterations associate with HSC aging. These, in turn, persist through the multiple rounds of cell divisions that associate with HSC transplantation, and can therefore be regarded as relatively stable.
Establishment of iPS lines from young and aged hematopoietic progenitor cells
To generate iPS cells from hematopoietic progenitor cells, Lin-c-Kit+CD45.1+ HSPCs from young or aged mice were isolated and infected for 2 consecutive days with retroviruses carrying GFP, Oct3/4, Klf4, c-Myc, and Sox2. Thereafter, the cells were transferred to ES cell culture conditions. After 4 to 7 days of culture, we began to observe a vast growth of cells, with the emergence of ES-like colonies from day 14 on. However, the vigorous proliferation of other cells persisted in these cultures, complicating the isolation of emerging iPS colonies. To circumvent this problem, we isolated candidate iPS cells from other cells by FACS using antibodies against EpCAM (CD326)22 and SSEA-1 (Figure 2A). This strategy enabled effective removal of other proliferating non-iPS cells (Figure 2B and data not shown). Two iPS lines derived from young HSPCs (young BiPS1 and young BiPS2) and 2 lines derived from aged HSPCs (aged BiPS1 and aged BiPS2) were selected for further experimentation.
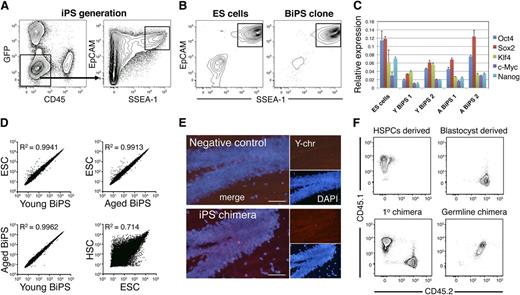
Generation of iPS cells from BM hematopoietic progenitor cells. (A) FACS strategy to isolate candidate iPS cells from HSPCs. HSPCs were transduced with retroviruses expressing GFP, Oct3/4, Klf4, c-Myc, and Sox2 for 2 days and subsequently cultured for 14 days in ES cell conditions. On day 14, candidate iPS cells were isolated by FACS based on a GFP-CD45-EpCAM+SSEA-1+ phenotype and cultured in individual wells (25 cells sorted per well) until single iPS colonies emerged. (B) Representative FACS plots of a commercially available C57BL/6 ES cell line (Primogenix B6N1, left), and an established iPS line showing the simultaneous expression of EpCAM and SSEA-1 (right). (C) Expression of pluripotency factors and induction of Nanog expression in established iPS lines (5 independent experiments, 3 replicates/cell line). (D) Correlation plots of the genome-wide expression patterns of young and aged BiPS lines to ESCs (1 experiment, relative expression values were averaged from 2 replicate arrays per condition). A correlation plot of expression patterns in ESC to HSCs is also shown to highlight the similarity in expression patterns of iPS lines and ESCs. (E) Y chromosome fluorescent in situ hybridization of a cross-section of the hippocampus from a negative control female mouse (top) and an agedBiPS primary chimera (bottom) generated from male agedBiPS cells injected into a female blastocyst. (F) FACS plots showing the congenic immunophenotype (CD45.1/CD45.2 system) of the hematopoietic cells used for cell isolation and primary chimera generation (bottom right). Peripheral blood phenotype of a germline contributed agedBiPS offspring.
Generation of iPS cells from BM hematopoietic progenitor cells. (A) FACS strategy to isolate candidate iPS cells from HSPCs. HSPCs were transduced with retroviruses expressing GFP, Oct3/4, Klf4, c-Myc, and Sox2 for 2 days and subsequently cultured for 14 days in ES cell conditions. On day 14, candidate iPS cells were isolated by FACS based on a GFP-CD45-EpCAM+SSEA-1+ phenotype and cultured in individual wells (25 cells sorted per well) until single iPS colonies emerged. (B) Representative FACS plots of a commercially available C57BL/6 ES cell line (Primogenix B6N1, left), and an established iPS line showing the simultaneous expression of EpCAM and SSEA-1 (right). (C) Expression of pluripotency factors and induction of Nanog expression in established iPS lines (5 independent experiments, 3 replicates/cell line). (D) Correlation plots of the genome-wide expression patterns of young and aged BiPS lines to ESCs (1 experiment, relative expression values were averaged from 2 replicate arrays per condition). A correlation plot of expression patterns in ESC to HSCs is also shown to highlight the similarity in expression patterns of iPS lines and ESCs. (E) Y chromosome fluorescent in situ hybridization of a cross-section of the hippocampus from a negative control female mouse (top) and an agedBiPS primary chimera (bottom) generated from male agedBiPS cells injected into a female blastocyst. (F) FACS plots showing the congenic immunophenotype (CD45.1/CD45.2 system) of the hematopoietic cells used for cell isolation and primary chimera generation (bottom right). Peripheral blood phenotype of a germline contributed agedBiPS offspring.
In initial analyses, we confirmed expression of pluripotency factors and induction of Nanog expression by quantitative reverse-transcription PCR in the newly generated iPS lines from both young and old mice (Figure 2C). To obtain a wider view of their transcriptional signatures, we performed gene expression analyses of established iPS lines using microarrays. For comparative purposes, we also included samples from 3 different C57BL/6 ES cell lines. Regardless of young or aged origin, these analyses revealed highly similar expression patterns of BiPS cells to ES cells (Figure 2D). By contrast, and as expected, large differences in expression patterns were observed when comparing iPS or ES cell lines to HSCs (Figure 2D).
To functionally characterize the generated iPS lines, we first investigated their potential to produce contracting cardiomyocytes in vitro (supplemental Movie 1). These experiments verified their potential to produce cells of an alternative lineage, but of the same germ layer (mesoderm) as the original somatic donor cells. To further investigate their lineage potential, we performed a series of blastocyst injections (supplemental Table 2). First, to investigate the potential of BiPS cells to produce a cell lineage with an alternative (ectodermal) germ layer origin than the starting blood cells, we investigated neurons in the hippocampus using Y-chromosome detection in brain sections from a female mouse chimeric with male aged BiPS cells. These analyses confirmed the presence of robust chimeric contribution to the neural lineage (Figure 2E). Finally, we set up breeding with a male-aged BiPS cell chimeric male with a wild-type C57BL/6 female, and screened 2 litters of offspring for germline transmission. Of 17 pups screened, we obtained 1 mouse with its blood system composing entirely of F1 CD45.1 × CD45.2 cells (Figure 2F), with CD45.1 representing a marker of BiPS cells (see as follows), thereby establishing totipotency of the generated BiPS cells.
Telomere length of steady state-aged and aged BiPS-derived HSCs
Somatic stem cell pools, including HSCs, are needed for the lifetime of an organism.19 However, increasing numbers of cell divisions could potentially lead to shortening of telomeres, a mechanism previously proposed to regulate stem cell behavior.23 We reasoned that this aspect could be particularly relevant in the context of our aged BiPS cells, given their proliferative history, both as a consequence of their chronological age and because of the substantial proliferation induced in these cells upon the iPS induction and maintenance. To approach this issue, we examined the telomere length in individual steady-state young (n = 251) and aged (n = 246) HSCs. These data established that aged HSCs harbored approximately 11% shorter telomeres in comparison with their young counterparts (P < .0234; Figure 3A), which represents the baseline effect of aging on the telomere length of HSCs.
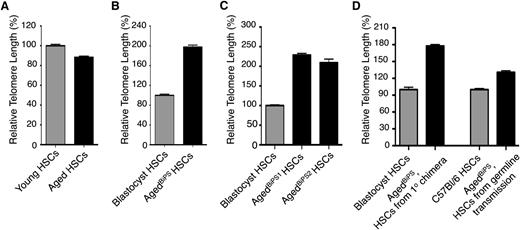
Single-cell telomere length analysis of HSCs from various sources. (A) The relative telomere length was measured by qPCR in single sorted HSCs in 2 independent experiments from 3 individual young and aged mice (n = 251 and 246 cells in each group, respectively). (B) Single cell telomere length measurements of blastocyst- and agedBiPS2-derived HSCs from an agedBiPS2 primary chimera (n = 86 and 66 cells in each group, respectively). (C) Relative telomere length in blastocyst, agedBiPS1- and agedBiPS2-derived HSCs isolated from repopulated recipient mice (2 independent isolations, n = 200, 130, and 53 cells, respectively). (D) Relative telomere length in (left) blastocyst and agedBiPS derived HSCs from a germline founder (agedBiPS2 primary chimera, n = 43 and 41 cells in each group, respectively) and (right) from a germline offspring mouse compared with an age-matched C57BL/6 control (n = 84 cells in each group). Error bars indicate mean ± SEM.
Single-cell telomere length analysis of HSCs from various sources. (A) The relative telomere length was measured by qPCR in single sorted HSCs in 2 independent experiments from 3 individual young and aged mice (n = 251 and 246 cells in each group, respectively). (B) Single cell telomere length measurements of blastocyst- and agedBiPS2-derived HSCs from an agedBiPS2 primary chimera (n = 86 and 66 cells in each group, respectively). (C) Relative telomere length in blastocyst, agedBiPS1- and agedBiPS2-derived HSCs isolated from repopulated recipient mice (2 independent isolations, n = 200, 130, and 53 cells, respectively). (D) Relative telomere length in (left) blastocyst and agedBiPS derived HSCs from a germline founder (agedBiPS2 primary chimera, n = 43 and 41 cells in each group, respectively) and (right) from a germline offspring mouse compared with an age-matched C57BL/6 control (n = 84 cells in each group). Error bars indicate mean ± SEM.
Although a few previous studies have reported a significant elongation of telomeres in established iPS cell lines compared with the telomere length of the somatic donor cell type,24,25 it has remained unclear whether telomere length is appropriately maintained following re-differentiation of iPS cells. Furthermore, it remained a formal possibility that the somatic cell type itself might influence on the telomere length dynamics associated with iPS formation and maintenance. This was relevant to our work, as we used hematopoietic cells, whereas most previous studies on iPS cells have used fibroblasts as somatic donor cells. When investigating aged BiPS-derived HSCs in the primary chimeric setting, which due to cell availability was restricted to the aged BiPS2 clone, we found a striking (twofold, P < .0001) elongation of telomeres compared with the blastocyst control HSCs (Figure 3B). Next, we conducted similar experiments on aged BiPS-derived HSCs after transplantation. These experiments demonstrated that aged BiPS cells maintained their approximately twofold elongated telomeres (P < .0001 for both aged BiPS1 and aged BiPS2 cells), despite the exposure of these cells to severe hematopoietic stress/proliferation (Figure 3C). Finally, we investigated the telomere length of HSCs derived from a germline offspring mouse (Figure 2F). From this mouse, we could observe a significant reduction in telomere length as compared with the telomere length of HSCs from the primary chimera (parent), although telomere length still remained significantly longer in such offspring (approximately 31% longer compared with that of the somatic starting cells) (P < .0001; Figure 3D). Taken together, these experiments demonstrate that iPS formation from HSPCs associates with elongation of telomeres in a similar manner as previously described for iPS cells with a fibroblast origin.24
Reprogramming of aged hematopoietic progenitors into totipotency alleviates the functional alterations that associate with chronological hematopoietic aging
As a last and main aim of our work, we set out to functionally characterize the hematopoietic differentiation capacity of BiPS from aged HSPCs. Despite extensive research, the exact cues guiding in vitro differentiation of ES/iPS cells into adult-type HSCs have not yet been established. Therefore, we evaluated the hematopoietic differentiation potential of young BiPS and aged BiPS cells in chimeric mice generated via blastocyst injections (supplemental Table 2). Because we had a specific interest in the hematopoietic system, we had initially devised our experiments such that we could evaluate the BiPS-derived contribution using the congenic CD45.1 (expressed on BiPS cells) and CD45.2 (blastocyst-derived cells) markers. As CD45 is expressed on all nucleated blood cells, this made it possible to detail hematopoiesis derived from young BiPS and aged BiPS in chimeric mice in a direct manner.
As lineage skewing is a dominant feature of HSC aging, we first evaluated the lineage distribution of BiPS-derived cells in the primary chimeric mice.26-29 These analyses revealed that the hematopoietic lineage distributions of iPS-derived cells, regardless of origin, were remarkably similar to those of the endogenous blastocyst origin (Figure 4A). Another hallmark of HSC aging is a dramatic increase in numbers of phenotypically defined HSCs.4,5,27,28 We therefore evaluated this aspect in mice chimeric with aged BiPS cells, but failed to observe any differences when comparing to the blastocyst-derived HSCs (Figure 4B). By contrast, and in agreement with previous data,4,5,27,28 aged steady-state mice displayed dramatically elevated frequencies of HSCs (12.9-fold, P < .0001; Figure 4B).
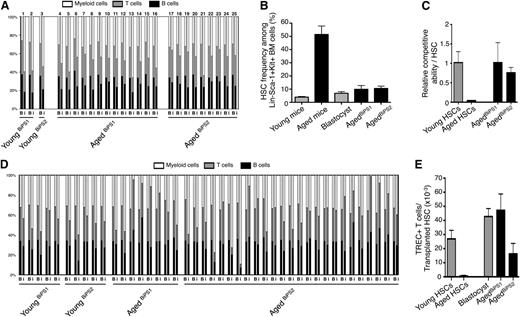
Hematopoiesis derived from iPS lines with an aged hematopoietic progenitor origin display multiple young functional characteristics. (A) Ratios of the peripheral lineage distribution in individual chimeric animals derived from young and aged BiPS lines. Chimeric ratios are depicted as previously described26,27 for both blastocyst derived (B) and iPS derived (i) hematopoiesis. Numbers above bars denote individual mice, whose total blood cell chimerism can be found in supplemental Table 2. (B) The frequency of HSCs among lineage negative Sca-1 positive c-kit positive BM cells in steady-state young and aged mice (3 independent experiments, 6 and 9 mice, respectively), and among blastocyst- and agedBiPS-derived lineage negative Sca-1 positive c-kit positive BM cells in primary chimeras (3 independent experiments, totaling 8, 3, and 5 mice, respectively). (C) Competitive repopulating ability of young, aged, agedBiPS1, and agedBiPS2 HSCs assessed as the ability of each transplanted HSC to repopulate the peripheral myeloid compartment of lethally irradiated young recipients 16 weeks posttransfer (3 independent experiments, n = 5, 25, 12, and 25 mice per group, respectively). (D) In vivo lineage distribution after transplantation of young and aged BiPS HSCs into lethally irradiated young recipients. Analysis was performed (as in A). (E) The ability of transplanted young, aged, blastocyst, agedBiPS1, and agedBiPS2-derived HSCs to generate naïve TREC+ T cells in repopulated recipient mice (from 2 independent experiments, totaling 4, 4, 10, 3, and 7 mice, respectively). Bars depict mean ± SEM.
Hematopoiesis derived from iPS lines with an aged hematopoietic progenitor origin display multiple young functional characteristics. (A) Ratios of the peripheral lineage distribution in individual chimeric animals derived from young and aged BiPS lines. Chimeric ratios are depicted as previously described26,27 for both blastocyst derived (B) and iPS derived (i) hematopoiesis. Numbers above bars denote individual mice, whose total blood cell chimerism can be found in supplemental Table 2. (B) The frequency of HSCs among lineage negative Sca-1 positive c-kit positive BM cells in steady-state young and aged mice (3 independent experiments, 6 and 9 mice, respectively), and among blastocyst- and agedBiPS-derived lineage negative Sca-1 positive c-kit positive BM cells in primary chimeras (3 independent experiments, totaling 8, 3, and 5 mice, respectively). (C) Competitive repopulating ability of young, aged, agedBiPS1, and agedBiPS2 HSCs assessed as the ability of each transplanted HSC to repopulate the peripheral myeloid compartment of lethally irradiated young recipients 16 weeks posttransfer (3 independent experiments, n = 5, 25, 12, and 25 mice per group, respectively). (D) In vivo lineage distribution after transplantation of young and aged BiPS HSCs into lethally irradiated young recipients. Analysis was performed (as in A). (E) The ability of transplanted young, aged, blastocyst, agedBiPS1, and agedBiPS2-derived HSCs to generate naïve TREC+ T cells in repopulated recipient mice (from 2 independent experiments, totaling 4, 4, 10, 3, and 7 mice, respectively). Bars depict mean ± SEM.
Next, we explored the capacity of aged BiPS cells to regenerate a hematopoietic system in a competitive transfer setting. To this end, we transplanted BM mononuclear cells from primary aged BiPS chimeric mice into lethally irradiated CD45.2 recipient hosts (2-5 × 106 cells/mouse). 16 weeks after transplantation, we evaluated the aged BiPS and blastocyst-derived hematopoiesis in the peripheral blood of these hosts. These experiments revealed the aged BiPS reconstitution to be of similar magnitude and quality when compared with the reconstitution patterns obtained from young HSCs in steady state (Figure 4C-D), as well as to the reconstitution pattern observed from the blastocyst control (Figure 4D). This was in sharp contrast to chronologically aged HSCs, which were characterized by a dramatic reduction in repopulation capacity (23.8-fold, P < .0001; Figure 4C). Finally, we performed more detailed investigations to explore the intrinsic ability of aged BiPS-derived HSCs to generate naïve T cells. We were particularly interested in this aspect as this parameter is normally difficult to probe in steady-state aged mice due to the thymic atrophy that accompanies aging. To this end, we estimated the frequencies of T cells containing TRECs, a molecular marker of recent thymic emigrants,13 per transplanted HSC. These analyses revealed that steady-state aged HSCs have a strongly reduced T-cell potential when compared with their young counterparts (38.4-fold reduction, P = .005; Figure 4E). By contrast, aged BiPS-derived HSCs generated naïve T cells at similar levels as both young HSCs and the endogenous blastocyst control-derived HSCs (Figure 4E).
Discussion
Many parameters act together and complicate both the study and generalizations that can be made about the aging of multicellular organisms. These include not only the highly varying onset in both time and degree, whereby aging affects different organs and tissues, but also its pleiotropic outcomes. Such considerations aside, 1 common feature of aging involves what seems as an inevitable loss of overall fitness. At a cellular level, this could result in either the generation of functionally defective cells, a reduced ability in the generation of appropriate cells, or a combination of both scenarios. The recognition that most organs and tissues are maintained by primitive self-renewing stem cells, whose sole function is to provide organisms with appropriate cellular offspring for a lifetime, has in recent years fueled the emergence of theories of stem cell origins to the aging process.3,19 The blood system is particularly amenable to study the autonomous aspects of these processes because of its extensively well-established character and the ability to regenerate an entire blood system from HSCs via transplantation.
An initial striking observation made in these studies was the dramatic elongation of telomeres that associated with the reprogramming process. These telomeres were appropriately maintained during the transition of iPS cells to differentiated progeny, and required germline dilution for substantial shortening, which agree with recent data showing that the combined telomere lengths of the parents determine telomere lengths of offspring.30 Although telomere extension may be a necessity for the generation of functional iPS cells, the long telomeres of C57BL/6 mice14 combined with the fact that chronologically aged HSCs show only modest telomere shortenings, argue perhaps against telomere attrition as the primary driver of murine HSC aging.
We interpret our results on the “young-like” functional capacity of aged-derived BiPS cells to support a view in which reversible epigenetic (and transcriptional) alterations are central for the development and/or maintenance of the phenotypes associated with HSC aging. Although a few reports have previously suggested that iPS cells themselves can associate with varying degrees of molecular “resemblance,” or “memory” of its somatic donor origin,31,32 we could find little evidence for an epigenetic memory with functional consequences in the BiPS lines studied here. Some data argue that this central question perhaps can be explained from the perspective of an incomplete reprogramming process.33 In an alternative interpretation, it might be envisioned that the strategy used in our work could favor selection of a subset of HSPCs unaffected by age and/or DNA damage. Although we cannot formally rule out this possibility, we consider it less likely. This is foremost based on the similarities in efficiencies of iPS formation of young and aged HSPCs (unpublished observations), and the very strong dominance of myeloid-biased HSCs in the aging setting.4 It might also be conceivable that aging HSPCs have acquired mutations that by themselves result in altered epigenetic features. An extension of this view would be that initial causative mutations would be compensated for during the iPS process and thus redundant for subsequent re-differentiation. Our experiments did not address this issue directly, but we did find that young BiPS, aged BiPS, and ES cells had remarkably similar genome-wide expression profiles and thus showed no evidence of transcriptional compensation.
Previously, sequencing of murine iPS lines revealed a mutational spectra correlating distinctly to that observed in the somatic cell donor cells.34 Based on such data, it can be anticipated that if murine HSPC aging do associate with an increase DNA mutation frequency, similar to the aging of human healthy HSPC,35 then aged BiPS lines should have significantly more DNA mutations. However, even if this could be established, its consequence would require extensive sequencing combined with loss and/or gain of function studies; a line of experiments beyond the scope of the work presented here.
We interpret our data to show that the epigenetic reset coinciding with iPS induction has immediate functional benefits for subsequent differentiation. This should be the result of normalized regulation of certain defined loci dysregulated with age, and thus a careful evaluation of the epigenetic properties of aged HSPCs might provide both candidate genes and more general regulators. For instance, the key epigenetic regulator Ezh2 is down-regulated in aged HSCs.36 Although we previously found little evidence of epigenetic changes at the p16INK4a locus in aged HSCs,36 a well-established Ezh2 target loci in other cells,37 the broad and potent roles of Ezh2 might well underlie other epigenetic alterations governing the HSC aging process.38
In conclusion, our experiments demonstrate that reprogrammed aged HSPCs have functional differentiation potential similar to those of vastly younger cells. Whether the aging state can be subjected to reversal in a more physiological or less invasive setting, and not merely in a highly artificial reprogramming scenario, remains a key outstanding question.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was generously supported by project grants (to D.B.) from the Swedish Cancer Society, the Swedish Medical Research Council (project grants and consortia grants Hemato-Linné and Stemtherapy), the Swedish Pediatric Leukemia Foundation, Ingabritt och Arne Lundbergs Forskningsstiftelse, and a grant for research on regenerative medicine from AFA Insurance.
Authorship
Contribution: M.W. and D.B. planned the study. M.W., G.L.N., G.S., and A.U. performed most experiments. M.-A.M.F. and R.M. undertook the blastocyst injections. T.D. performed the fluorescent in situ hybridization experiments. M.S. helped with microarray hybridizations. M.W., and D.B. analyzed the data and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: David Bryder, Immunology Section, Institution for Experimental Medical Science, BMC D14, Lund, 221 84, Sweden; e-mail: david.bryder@med.lu.se.