Key Points
Bone marrow mesenchymal stromal cells preserve CML stem cells from elimination following tyrosine kinase inhibitor treatment.
N-cadherin and Wnt signaling contribute to protection of CML stem cells by mesenchymal cells and may represent new treatment targets.
Abstract
Tyrosine kinase inhibitors (TKIs) are highly effective in treatment of chronic myeloid leukemia (CML) but do not eliminate leukemia stem cells (LSCs), which remain a potential source of relapse. TKI treatment effectively inhibits BCR-ABL kinase activity in CML LSCs, suggesting that additional kinase-independent mechanisms contribute to LSC preservation. We investigated whether signals from the bone marrow (BM) microenvironment protect CML LSCs from TKI treatment. Coculture with human BM mesenchymal stromal cells (MSCs) significantly inhibited apoptosis and preserved CML stem/progenitor cells following TKI exposure, maintaining colony-forming ability and engraftment potential in immunodeficient mice. We found that the N-cadherin receptor plays an important role in MSC-mediated protection of CML progenitors from TKI. N-cadherin–mediated adhesion to MSCs was associated with increased cytoplasmic N-cadherin–β-catenin complex formation as well as enhanced β-catenin nuclear translocation and transcriptional activity. Increased exogenous Wnt-mediated β-catenin signaling played an important role in MSC-mediated protection of CML progenitors from TKI treatment. Our results reveal a close interplay between N-cadherin and the Wnt–β-catenin pathway in protecting CML LSCs during TKI treatment. Importantly, these results reveal novel mechanisms of resistance of CML LSCs to TKI treatment and suggest new targets for treatment designed to eradicate residual LSCs in CML patients.
Introduction
Chronic myeloid leukemia (CML) originates from a primitive hematopoietic cell that is transformed by the BCR-ABL oncogene.1 The deregulated tyrosine kinase activity of the BCR-ABL protein leads to increased proliferation, reduced apoptosis, and disturbed interaction with the extracellular matrix, resulting in abnormal expansion of myeloid progenitors and differentiated myeloid cells. The BCR-ABL tyrosine kinase inhibitors (TKIs) imatinib (IM), nilotinib, and dasatinib are highly effective in treatment of CML, resulting in complete cytogenetic responses and major reductions in BCR-ABL transcript levels in most chronic phase (CP) CML patients.2,3 Conversely, cessation of drug treatment leads to disease recurrence in most CML patients, and the fraction of patients in whom TKI treatment can be discontinued remains low.4 A persistent population of leukemia stem cells (LSCs) is the likely source of relapse after TKI discontinuation.5 Quiescent CML LSCs are especially resistant to TKI-induced apoptosis. Because TKI treatment effectively inhibits BCR-ABL kinase activity in primitive CML LSCs, there is increasing interest in identifying BCR-ABL kinase-independent mechanisms that may contribute to preservation of LSCs during TKI treatment.6-9
Normal hematopoietic stem cells (HSCs) localize to specialized microenvironmental niches in the bone marrow (BM) that provide critical signals to regulate HSC numbers and quiescence and support HSC preservation.10 Although less well studied, there is some evidence that LSC generation and propagation may also be supported by microenvironmental cells.11 In addition, BM stromal cells or extracellular matrix may protect leukemia cells from the effects of chemotherapy and small molecule–targeted therapies.12-14 For example, the VLA-4 and CXCR4 receptors (required for normal HSC homing and retention to the hematopoietic niche) have been found to be important for acute myeloid leukemia LSC homing and growth,12,15 and blockade of these receptors can enhance acute myeloid leukemia LSC sensitivity to chemotherapy. Moreover, antibody-mediated targeting of the CD44 adhesion receptor can prevent trafficking of CML LSCs to the BM and promote LSC differentiation.16,17 Culture of CML cell lines with stromal cell–conditioned medium or with fibronectin is reported to result in resistance to IM.18,19 In turn, IM treatment increases CXCR4-mediated migration of CML cell lines to BM mesenchymal stromal cells (MSCs) and results in increased cell cycle arrest and survival of quiescent cells.20 Although these results with cell lines and murine models suggest that microenvironmental interactions could protect CML cells from TKI treatment, the role of the BM stromal microenvironment in protecting primary human CML LSCs from TKI treatment and the underlying molecular interactions are not well studied. Two recent studies have indicated that culture of primary CML CD34+ cells with conditioned media from or in direct contact with BM stroma can protect CML cells from TKI-induced cell death or inhibition of colony formation.21,22 However, these studies have not evaluated microenvironmental interactions with phenotypically or functionally defined primitive CML stem/progenitor cells.
Previous studies have indicated a potential role for the cell–cell adhesion receptor N-cadherin in HSC regulation by BM osteoblasts and resistance of leukemia cells to therapy.23,24 Although recent reports indicate that N-cadherin is not required for regulation of murine HSCs,25,26 the role of N-cadherin in human hematopoiesis is not well studied.14,24,27 Cadherins mediate adhesion through homotypic interaction between cell surface receptors. Cadherins can associate with β-catenin leading to their stabilization, linkage to the actin cytoskeleton, and enhanced cell–cell adhesion.28,29 Wnt ligands in the microenvironment can activate β-catenin signaling through engagement of Frizzled and LRP6 receptors. Canonical Wnt signaling results in stabilization and nuclear translocation of β-catenin, complex formation with tissue-coding factor/leukokinesis-enhancing factor (TCF/LEF), and transcription of target genes.30 β-catenin signaling may regulate normal HSC self-renewal and hematopoietic reconstituting ability.31 Enhanced Wnt–β-catenin activity is reported to contribute to LSC transformation in blast crisis CML, although its role in CP CML is less clear.32,33 In a BCR-ABL transduction-transplantation model, expression of BCR-ABL in β-catenin–deleted HSCs resulted in reduced LSC self-renewal and leukemia development.34,35 Deletion of β-catenin in established leukemia synergized with IM to eliminate CML stem cells.36 Potential mechanisms underlying increased β-catenin in CML cells include BCR-ABL–mediated β-catenin phosphorylation leading to protein stabilization and activation of nuclear signaling37 and reduced β-catenin degradation related to GSK3β inactivation downstream of BCR-ABL,38 or missplicing of GSK3β as reported in blast crisis CML LSCs.37 However, the role of exogenous microenvironmental Wnt in activating β-catenin signaling in CML LSCs has not been evaluated. In this study, we determined whether human BM MSCs affected the survival, proliferation, and in vitro and in vivo growth of primitive CML stem/progenitor cells exposed to TKI treatment. In this context, we evaluated the roles of N-cadherin and exogenous Wnt–β-catenin signaling in modulating response to TKI treatment.
Methods
Subjects
CML samples were obtained from patients in CP without prior IM treatment from the City of Hope Cancer Center and the University of Glasgow. Leukopheresis samples were processed for CD34+ cell selection with CliniMACS (Miltenyi Biotech, Teterow, Germany). Cord blood samples were provided by StemCyte (Arcadia, CA). Normal BM cells were obtained from healthy donors at City of Hope Cancer Center. Mononuclear cells were isolated from cord blood and BM by Ficoll-Hypaque (Sigma Diagnostics, St. Louis, MO) density gradient centrifugation for 30 minutes at 400 g. CD34+ cells were isolated by using magnetic bead selection (Miltenyi Biotech, Auburn, CA). All subjects signed an informed consent form. Sample acquisition was approved by the Institutional Review Boards at the City of Hope National Medical Center, in accordance with an assurance filed with and approved by the Department of Health and Human Services, and the North Glasgow University Hospital Division of the National Health Service Greater Glasgow and Clyde, and met all requirements of the Declaration of Helsinki.
Cell culture
Human reverse transcriptase (hTERT) –immortalized primary human marrow MSCs were maintained in RPMI 1640 with 10% fetal bovine serum for no more than 5 passages. MSCs were subcultured in tissue culture plates (Falcon; BD Biosciences, San Jose, CA) until confluent followed by irradiation at 20 Gy. CD34+ cells were cultured in the presence or absence of irradiated MSCs at 37°C with 5% CO2 in serum-free expansion medium (Stem Cell Technologies, Vancouver, BC, Canada) supplemented with growth factors (GFs) at concentrations similar to those found in stroma-conditioned medium from long-term BM cultures (granulocyte-macrophage colony-stimulating factor 200 pg/mL, granulocyte colony-stimulating factor 1 ng/mL, stem cell factor 200 pg/mL, leukemia inhibitory factor 50 pg/mL, macrophage inflammatory protein-1 200 pg/mL, and interleukin-6 1 ng/mL). Stock solutions of the TKIs IM, nilotinib (Novartis Pharmaceuticals, East Hanover, NJ), and dasatinib (Bristol-Myers Squibb, New York, NY) were prepared in dimethylsulfoxide and stored at –20°C. IM, nilotinib, and dasatinib were added at concentrations of 5 µM, 5 µM, and 0.15 µM, respectively.
Apoptosis, proliferation, cell cycle, and colony-forming cell assays
To assess apoptosis, cells from CML patients and healthy controls (n = 3 each) were cultured in the presence or absence of MSCs and TKIs for 96 hours, then labeled with Annexin V-PE (BD Biosciences PharMingen, San Diego, CA), and analyzed by flow cytometry (FACScalibur; BD Biosciences). Apoptotic cells were defined as Annexin V-PE+. Proliferation was assessed by labeling cells with 5- (and 6-) carboxyfluorescein diacetate succinimidyl ester (CFSE; Molecular Probes, Eugene, OR), followed by culture for 96 hours with or without MSCs and TKIs. At the end of the culture period, CFSE levels were analyzed by flow cytometry. ModFit software (Verity, Topsham, ME) was used to fit the data, determine the percentage of cells in each generation, and generate a proliferation index. The position of the parent generation was set on the basis of an aliquot of cells treated with 4% paraformaldehyde immediately after sorting. For cell cycle analysis, cells were labeled with Ki67 and 4,6 diamidino-2-phenylindole and analyzed by flow cytometry. For colony-forming capacity assays, CD34+CD38− and CD34+CD38+ cells cultured with or without TKIs and MSCs were plated in methylcellulose progenitor culture, and burst-forming unit-erythroid and colony-forming unit-granulocyte and macrophage cells were counted after 14 days.
Engraftment of human cells in immunodeficient mice
CML CD34+ cells (2 × 106 cells per mouse) from 3 newly diagnosed CP CML patients were cultured for 96 hours with and without MSCs and IM. Subsequently, all cells were harvested and transplanted via tail vein injection into 8-week-old sublethally irradiated (3 Gy) NSG mice (The Jackson Laboratory, Bar Harbor, ME). Mice were euthanized after 4 or 10 weeks, and marrow contents of femurs, spleen cells, and blood cells were obtained. Human cell engraftment was assessed by labeling with anti-human CD45 antibody and analyzed by flow cytometry. Specific cell subsets were detected by using antibodies to human CD34, CD33, CD11b, CD71, CD3, and CD19. BCR-ABL messenger RNA (mRNA) levels in marrow cells obtained from mice were evaluated by quantitative polymerase chain reaction (Q-PCR).
Evaluation of adhesion and N-cadherin
Adherent and nonadherent cells were separated by using a standardized wash procedure. Plates were shaken well, and nonadherent cells were collected, followed by addition of phosphate-buffered saline, flushing and collection of adherent MSCs, and checking with the microscope to make sure all cells were detached from MSCs. Adherent and nonadherent cells were counted, and the fraction of adherent cells as a proportion of total cells was calculated. N-cadherin expression was evaluated by staining CD34+ cells with Alexa Fluor 488 conjugated anti-human N-cadherin antibody (cat:FAB6426G; R&D Systems, Minneapolis, MN). To block N-cadherin, CML CD34+ cells were incubated with a blocking anti–N-cadherin monoclonal antibody (GC-4 40 μg/mL, Sigma; St. Louis, MO), control immunoglobulin G, or with N-cadherin blocking short cyclic peptides with HAV sequences (N-Ac-CHAVDIC-NH2; NCDH 0.15 mM) or control peptides with HGV sequences (NCDH control at 0.15 mM) for 3 days on MSCs. The cyclic HAV peptide is an effective N-cadherin antagonist.39 Cells were analyzed for apoptosis, intracellular signaling, and Q-PCR.
Evaluation of Wnt–β-catenin reporter activity
A β-catenin–activated reporter (BAR) Wnt–β-catenin reporter system was used. The reporter construct contains a concatemer of 12 TCF response elements separated by distinct 5-base linkers that drive Wnt-responsive transcription of a Venus enhanced yellow fluorescent protein variant.40 This reporter was inserted into a lentivirus plasmid expressing red fluorescent protein (RFP) via a spleen focus-forming virus promoter (pBARvr). A control reporter (pfuBARvr) has a 2-base substitution in each TCF binding site. Infectious lentivirus vector stocks were used to transduce CML CD34+ cells. Transduced CML CD34+ cells were cultured with MSCs with or without IM in serum-free expansion medium with low GFs for 96 hours and analyzed by flow cytometry. Transduced cells were identified on the basis of RFP expression, and target gene activation was measured by assessing median channel fluorescence of Venus within RFP-positive cells. Wnt activity was also analyzed after culture with conditioned medium from Wnt1-transduced cells and FKWnt1-transduced cells as controls. Wnt1 and FKWnt1 lentivirus plasmids were transfected into HT1080, together with packaging and envelope plasmids, and conditioned medium was collected at 48 hours.
Confocal microscopy
CML CD34+ cells were cultured in 8-chamber polystyrene vessels with or without MSCs and treated with IM, DKK1 (R&D systems), NCDH, or control for 48 hours. Cells were fixed, permeabilized, and incubated with mouse anti–β-catenin antibodies (14/β-catenin; BD Biosciences) and sheep anti-human/mouse/rat N-cadherin antibody (cat:AF6426; R&D Systems). Cells were washed and incubated with Texas Red–conjugated anti-mouse and fluorescein isothiocyanate–conjugated anti-sheep secondary antibodies. Slides were examined by using a Zeiss upright LSM 510 2-photon confocal microscope.
Real-time Q-PCR analysis
Total RNA was extracted by using the RNAeasy Mini Kit (Qiagen, Valencia, CA). First-strand complementary DNA was synthesized by using Superscript III First-Strand Kit (Invitrogen, Grand Island, NY). Q-PCR analysis for detection of BCR-ABL transcripts was performed in triplicate by using a real-time TaqMan assay and the ABI Prism 7700 sequence detector (Applied Biosystems, Foster City, CA), as previously described.5 BCR levels were measured as internal controls. Results were expressed as a BCR-ABL:BCR ratio. Q-PCR analysis was also performed for detection of N-cadherin, cyclin D1, c-Myc, and peroxisome proliferator-activated receptor delta transcripts, and glyceraldehyde 3-phosphate dehydrogenase and β-actin were measured as internal controls (supplemental Table 1). Results were expressed as a ratio to glyceraldehyde 3-phosphate dehydrogenase or β-actin.
Microarray analysis
RNA was obtained from CML CD34+ cells treated with or without IM (5 μM) and MSCs for 96 hours (n = 3 each), aand then amplified, labeled, and hybridized to GeneChip 1.0 arrays (Affymetrix, Santa Clara, CA). Microarray data analysis was performed by using R (version 2.9; www.r-project.org) with genomic analysis packages from Bioconductor (version 2.4; www.bioconductor.org). The 33 297 probes represented on the microarray were filtered by cross-sample mean and for a standard deviation of greater than the 25% quantile, yielding 18 624 probes representing 12 553 genes. Linear regression was used to model the gene expression with a 2 × 2 factorial design and matched samples. Differentially expressed genes were identified by calculating the empirical Bayes-moderated t statistic, and P values were adjusted by false discovery rate (FDR) using the LIMMA package (Bioconductor). Gene set enrichment analysis (GSEA) was performed by using GSEA software version 2.04 (http://www.broadinstitute.org/gsea/) to detect enrichment of predetermined gene sets using t scores from all genes for 1263 gene sets in the C2 (curated gene sets) category from the Molecular Signature Database (MsigDB).41 For significant gene sets related to the Wnt–β-catenin and cadherin pathways, leading-edge genes commonly enriched in these sets were analyzed. Heatmaps were plotted to show contrasts among the different treatment groups.
Western blotting
CD34+ cells cultured with or without IM and MSCs for 48 to 96 hours were lysed in buffer containing 0.5% Nonidet P-40 (Sigma Diagnostics) and 0.5% sodium deoxycholate supplemented with phenylmethylsulfonyl fluoride (1 mM/L), protease inhibitor mixture, and phosphatase inhibitors (50 mM/L sodium fluoride, 1 mM/L sodium vanadate; all from Sigma Diagnostics). For nuclear cytoplasmic fractionation, cells were lysed in hypotonic buffer for 5 minutes, gently agitated for 1 minute on ice, and centrifuged at 13 000 rpm at 4°C for 10 seconds. Supernatants were collected as the cytoplasmic extract. After washing with hypotonic buffer, nuclear pellets were incubated in high-salt buffer at 4°C for 30 minutes, and supernatants were collected as nuclear extracts after centrifugation at 13 000 rpm at 4°C for 5 minutes.42 Proteins were resolved on 4% to 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and transferred to nitrocellulose membrane. Primary antibodies used were anti–phospho-Crkl (Cell Signaling, Danvers, MA), anti–N-cadherin (cat:AF6426; R&D Systems), anti–N-cadherin (3B9; Invitrogen), anti–β-catenin (14/β-catenin; BD Biosciences), anti–p-GSK3β (S9), anti–p-AKT, anti-actin (Cell Signaling), and anti-tubulin and anti-Sp1 (Santa Cruz Biotechnology, Santa Cruz, CA). Horseradish peroxidase–conjugated secondary antibodies were from Jackson ImmunoResearch Laboratories (West Grove, PA). Antibody detection was performed by using the SuperFemto kit (Pierce Biotechnology, Rockford, IL).
Duolink in situ proximity ligation assay
In situ proximity ligation assay (PLA) technology was used to locate and quantify interaction of N-cadherin and β-catenin, using the Duolink kit (Olink Biosciences, Uppsala, Sweden).43 CML CD34+ cells cultured with or without MSCs and IM for 96 hours were dropped on poly-L-lysine–coated slides, fixed, permeabilized, and incubated with sheep anti-human/anti-mouse/anti-rat N-cadherin (cat:AF6426; R&D Systems) and mouse anti-human β-catenin (14/β-catenin; BD Biosciences) antibodies. After washing, cells were incubated with species-specific secondary antibodies, PLA probe anti-goat plus, and PLA probe anti-mouse minus, each with attached unique short DNA strands that can interact when in close proximity (<40 nm) through subsequent addition of 2 circle-forming DNA oligonucleotides. The oligonucleotides were joined by enzymatic ligation, amplified via rolling circle amplification using a polymerase, which resulted in several-hundredfold replication of the DNA circle, and labeled with complementary oligonucleotide probes to highlight the product. The resulting amplification product was seen as a bright dot by using fluorescence microscopy.
Statistical analysis
Data obtained from independent experiments were reported as the mean plus or minus standard error of the mean. Significance levels were determined by Student t test analysis.
Results
BM MSCs inhibit apoptosis and enhance maintenance of CML stem/progenitor cells following TKI treatment
We studied the effect of primary human BM MSCs, immortalized by enforced expression of hTERT, on survival and proliferation of CML and normal CD34+CD38– stem cells/primitive progenitors and CD34+CD38+ committed progenitors treated with IM, nilotinib, or dasatinib. As previously described in detail, hTERT-immortalized MSCs have a normal karyotype, do not form foci in soft agar, can differentiate into osteoblasts and chondrocytes, and are indistinguishable from nonimmortalized primary BM MSCs in their capacity to support normal CD34+ stem/progenitor cells and leukemic lymphoblasts.44
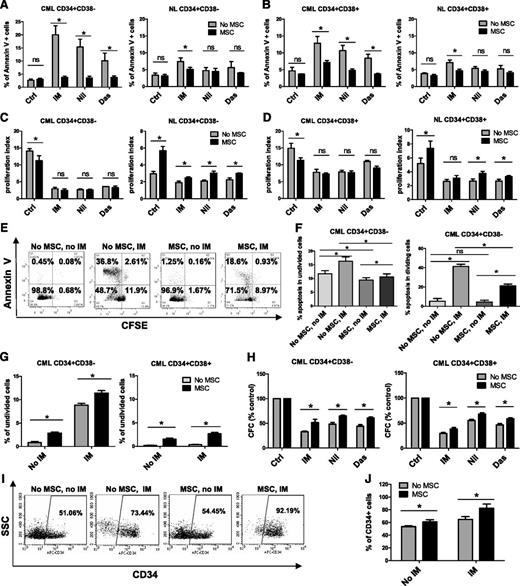
CD34+ cells were labeled with CFSE, and CFSE+ CD34+CD38− as well as CD34+CD38+ cells were selected by using flow cytometry and incubated with therapeutically relevant doses of IM (5 µM), nilotinib (5 µM), or dasatinib (0.15 µM) under low GF conditions for 96 hours, with or without coculture with MSCs. GFs were added to allow direct comparison between cells cultured with or without MSCs, since culture without MSCs requires addition of GFs. Following culture, cells were labeled with Annexin V-PE and analyzed for apoptosis and cell division (reduction in CFSE fluorescence) by flow cytometry. MSCs did not significantly affect apoptosis of CML CD34+CD38− or CD34+CD38+ cells in the absence of TKI treatment but did protect them from apoptosis following TKI treatment (Figure 1A-B). MSC coculture resulted in significantly greater reduction of IM-induced apoptosis of CML CD34+CD38− compared with CD34+CD38+ cells but resulted in similar reduction of apoptosis of both cell types after treatment with other TKIs (supplemental Figure 1A). Culture with MSCs did not significantly affect apoptosis of normal CD34+CD38– and CD34+CD38+ cells regardless of TKI treatment (Figure 1A-B). Untreated CML CD34+CD38– and CD34+CD38+ cells had a significantly higher proliferation rate than their normal counterparts (Figure 1C-D). Proliferation was reduced by TKIs in both CML subpopulations in the absence of MSCs. Culture with MSCs slightly reduced CML stem/progenitor cell proliferation without TKI treatment but did not significantly change proliferation of cells treated with TKIs (Figure 1C-D). Proliferation of untreated CML CD34+CD38− and CD34+CD38+ cells was similar with and without MSCs. However, proliferation of CML CD34+CD38+ cells was significantly greater than that of CML CD34+CD38– cells, both with and without MSCs, in the presence of TKIs (supplemental Figure 1B). MSC coculture resulted in a modest increase in normal progenitor proliferation (Figure 1C-D). Proliferation of untreated normal CD34+CD38+ cells was significantly greater than that of CD34+CD38− cells without MSCs but was not significantly different when cocultured with MSCs. There was no significant difference in proliferation between two cell types in the presence of TKIs both with and without MSCs (supplemental Figure 1B). Coculture with MSCs significantly reduced apoptosis of both nondividing (CFSE bright) and dividing (CFSE low) CML CD34+CD38− cells after IM treatment (Figure 1E-F) and increased the proportion of nondividing cells with or without IM treatment (Figure 1G). MSC coculture also resulted in a significantly enhanced colony-forming capacity of TKI-treated CML CD34+CD38+ and CD34+CD38− cells compared with TKI treatment without MSCs (Figure 1H). MSC coculture did not result in significantly different effects on colony formation from CML CD34+CD38− cells compared with CD34+CD38+ cells (supplemental Figure 1C). We observed an increased frequency of CML cells that expressed CD34 following MSC coculture with or without IM compared with culture without MSCs (Figure 1I-J). These results suggest that MSCs enhance preservation of CML stem/progenitor cells during TKI treatment by reducing apoptosis and increasing the number of undivided stem/progenitor cells.
MSCs protect CML CD34+CD38− and CD34+CD38+ cells from TKI treatment. Primary CML and normal (NL) progenitor (CD34+) cells were stained with CFSE. CFSE+ primitive cells (Lin−CD34+CD38−) and committed cells (Lin−CD34+CD38+) were sorted by flow cytometry and cultured for 96 hours with or without MSCs and were treated with IM (5 μm), nilotinib (Nil; 5 μm), or dasatinib (Das; 0.15 μm) or were left untreated. The percentages of apoptotic primitive cells (A) and committed cells (B) were assessed on the basis of Annexin V+ labeling. A proliferation index was calculated based on reduction in CFSE levels for CML and NL CD34+CD38− cells (C) and CD34+CD38+ cells (D) treated with IM, nilotinib, or dasatinib compared with controls (Ctrls) with and without MSCs. (E) Representative flow cytometry plots and (F) graph showing apoptosis (Annexin V+ cells) within undivided (CFSE bright) and dividing (CFSE dim) CML CD34+CD38− cells treated with IM with and without MSCs. (G) Percentages of undivided (CFSE bright) CML CD34+CD38− and CD34+CD38+ cells after culture with or without IM treatment and with or without MSCs. (H) CML CD34+CD38− and CD34+CD38+ cells cultured for 96 hours with or without MSCs with or without treatment with IM (5 µm), nilotinib (5 µm), or dasatinib (0.15 µm) were plated in methylcellulose progenitor assays, and colony-forming capacity (CFC) frequencies were determined. (I) Representative flow cytometry plot and (J) graph showing CD34+ expression after culture of CML CD34+ cells with or without IM treatment and with or without MSCs. ns, not significant. n = 3. *P < .05.
MSCs protect CML CD34+CD38− and CD34+CD38+ cells from TKI treatment. Primary CML and normal (NL) progenitor (CD34+) cells were stained with CFSE. CFSE+ primitive cells (Lin−CD34+CD38−) and committed cells (Lin−CD34+CD38+) were sorted by flow cytometry and cultured for 96 hours with or without MSCs and were treated with IM (5 μm), nilotinib (Nil; 5 μm), or dasatinib (Das; 0.15 μm) or were left untreated. The percentages of apoptotic primitive cells (A) and committed cells (B) were assessed on the basis of Annexin V+ labeling. A proliferation index was calculated based on reduction in CFSE levels for CML and NL CD34+CD38− cells (C) and CD34+CD38+ cells (D) treated with IM, nilotinib, or dasatinib compared with controls (Ctrls) with and without MSCs. (E) Representative flow cytometry plots and (F) graph showing apoptosis (Annexin V+ cells) within undivided (CFSE bright) and dividing (CFSE dim) CML CD34+CD38− cells treated with IM with and without MSCs. (G) Percentages of undivided (CFSE bright) CML CD34+CD38− and CD34+CD38+ cells after culture with or without IM treatment and with or without MSCs. (H) CML CD34+CD38− and CD34+CD38+ cells cultured for 96 hours with or without MSCs with or without treatment with IM (5 µm), nilotinib (5 µm), or dasatinib (0.15 µm) were plated in methylcellulose progenitor assays, and colony-forming capacity (CFC) frequencies were determined. (I) Representative flow cytometry plot and (J) graph showing CD34+ expression after culture of CML CD34+ cells with or without IM treatment and with or without MSCs. ns, not significant. n = 3. *P < .05.
BM MSCs enhance maintenance of CML LSCs with in vivo repopulating capacity
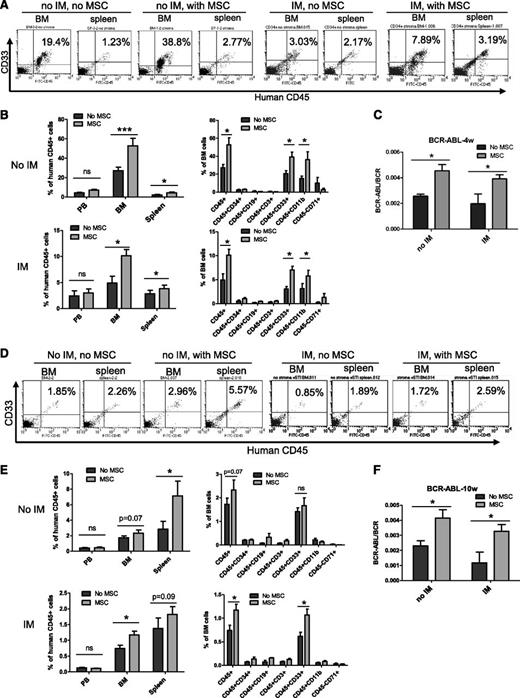
We next evaluated the effect of MSCs on maintenance of CML LSCs with in vivo repopulating capacity. CML CD34+ cells were cultured with and without MSCs and IM for 96 hours and transplanted into irradiated immunodeficient NOD.Cg-PrkdcscidIL2rgtm1Wjl/SzJ (NSG) mice. Cells cultured with MSCs demonstrated significantly increased engraftment in the BM and spleen of NSG mice at 4 weeks (short-term engraftment) and 10 weeks (longer-term engraftment) after transplantation, both with and without IM treatment. Engraftment of human cells was higher in the BM compared with the spleen at 4 weeks but was higher in the spleen compared with BM at 10 weeks after transplantation (Figures 2A-B,D-E). The majority of human cells engrafted in the BM and spleen were CD33- and/or CD11b-expressing myeloid cells (Figures 2B and 2E). Q-PCR analysis of BM cells confirmed engraftment of BCR−ABL+ cells and showed increased engraftment of BCR−ABL+ cells from CML CD34+ cells cocultured with MSCs at 4 (Figure 2C) and 10 weeks (Figure 2F). These results indicate that BM MSCs enhance maintenance of CML LSCs with in vivo repopulating capacity and protect them from TKI treatment.
MSCs protect CML LSCs capable of engrafting NSG mice from TKI treatment. CML CD34+ cells (2 × 106 cells per mouse) from 3 patients were cultured for 96 hours with and without MSCs and with or without IM and transplanted into NSG mice. Mice were euthanized after 4 and 10 weeks, and marrow contents of femurs, spleen cells, and blood cells were obtained. (A) Flow cytometry plots and (B) graphs showing human cell engraftment in peripheral blood (PB), BM, and spleen at 4 weeks (4w) posttransplantation. (C) BCR-ABL mRNA levels in marrow cells obtained from mice at 4 weeks posttransplantation. (D) Flow cytometry plots and (E) graphs showing human cell engraftment in PB, BM, and spleen at 10 weeks posttransplantation. (F) BCR-ABL mRNA levels in marrow cells obtained from mice at 10 weeks (10w) posttransplantation. ns, not significant. n = 5. *P < .05; ***P < .001.
MSCs protect CML LSCs capable of engrafting NSG mice from TKI treatment. CML CD34+ cells (2 × 106 cells per mouse) from 3 patients were cultured for 96 hours with and without MSCs and with or without IM and transplanted into NSG mice. Mice were euthanized after 4 and 10 weeks, and marrow contents of femurs, spleen cells, and blood cells were obtained. (A) Flow cytometry plots and (B) graphs showing human cell engraftment in peripheral blood (PB), BM, and spleen at 4 weeks (4w) posttransplantation. (C) BCR-ABL mRNA levels in marrow cells obtained from mice at 4 weeks posttransplantation. (D) Flow cytometry plots and (E) graphs showing human cell engraftment in PB, BM, and spleen at 10 weeks posttransplantation. (F) BCR-ABL mRNA levels in marrow cells obtained from mice at 10 weeks (10w) posttransplantation. ns, not significant. n = 5. *P < .05; ***P < .001.
N-cadherin contributes to MSC-mediated protection of CML stem/progenitor cells from IM
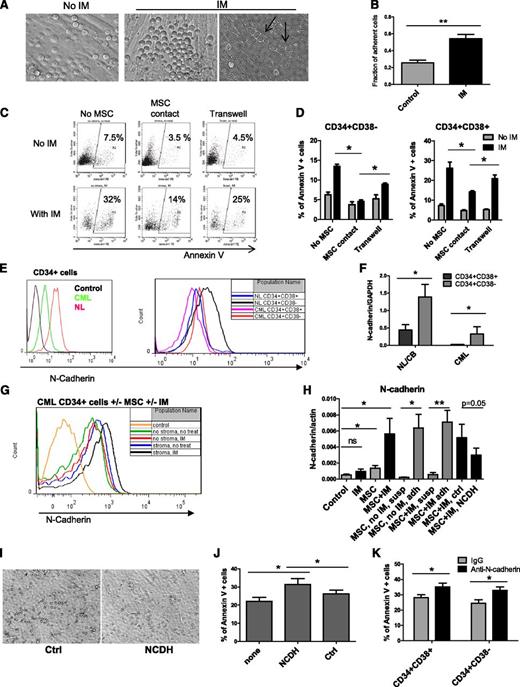
IM treatment enhanced adhesion of CML CD34+ cells to MSCs (Figure 3A-B). Some adherent CML CD34+ cells migrated underneath MSC layers (Figure 3A, arrowhead). Cultured CML CD34+CD38+ and CD34+CD38− cells separated from MSCs by a Transwell insert with 0.45-μm pores showed increased apoptosis with IM compared with cells grown in direct contact with MSCs but were less sensitive than cells grown without any exposure to MSCs (Figure 3D). CML CD34+CD38+ cells demonstrated significantly higher apoptosis compared with CD34+CD38− cells with or without MSC coculture (supplemental Figure 2A). Representative plots for CD34+ cells are shown in Figure 3C. Since N-cadherin has been shown to contribute to resistance of leukemia cells to therapy, we evaluated its role in MSC-mediated protection of CML stem/progenitor cells.23,24 In both normal and CML samples, N-cadherin was expressed at higher levels in CD34+CD38− compared with CD34+CD38+ cells (Figure 3E-F). However, cell surface N-cadherin expression and N-cadherin mRNA were lower in CML compared with normal CD34+ cells, CD34+CD38− cells, and CD34+CD38+ cells (Figure 3E-F). We observed increased cell surface N-cadherin expression and N-cadherin mRNA expression in CML cells treated with IM in the presence of MSCs compared with untreated controls, IM alone, or MSCs alone (Figure 3G-H). Increased N-cadherin mRNA expression was restricted to MSC-adherent cells (Figure 3H). We used anti–N-cadherin blocking antibodies or anti–N-cadherin short cyclic HAV peptide (NCDH) to block N-cadherin interactions.39 CML CD34+ cells incubated with NCDH peptides had reduced adhesion to MSCs compared with controls (Figure 3I). The NCDH peptide reduced N-cadherin mRNA expression compared with control scrambled peptide (Figure 3H). Western blotting also showed enhanced N-cadherin expression in IM-treated CML CD34+ cells cultured with MSCs (Figure 4A). Increased N-cadherin protein expression was limited to MSC-adherent cells in both the absence and the presence of IM (Figure 4B). Treatment with NCDH peptide reduced N-cadherin protein expression compared with control peptide treatment (Figure 4B). CML CD34+ cells incubated with NCDH peptides also showed enhanced apoptosis following IM treatment compared with cells exposed to control peptides (Figure 3J). Likewise, CML CD34+CD38− and CD34+CD38+ cells incubated with anti–N-cadherin antibodies showed significantly reduced adhesion to MSCs (data not shown) and significantly increased apoptosis following IM treatment compared with cells incubated with control immunoglobulin G (Figure 3K).
Role of N-cadherin in MSC-mediated protection of CML stem/progenitor cells from TKI treatment. (A) CML CD34+ cells cocultured on MSCs without IM (left) and with IM (right) adhered to and migrated underneath (right, arrowhead) MSCs. (B) The fraction of MSC-adherent CML CD34+ cells with or without IM. (C) Representative flow cytometry plots and (D) graphs showing apoptosis of CML CD34+ cells (C) and CD34+CD38+ and CD34+CD38− cells (D) cultured without MSCs in a Transwell insert above MSC layers or in contact with MSCs in the absence or presence of IM (n = 3). (E) Representative flow cytometry plots showing N-cadherin expression on CML and normal CD34+, CD34+CD38−, and CD34+CD38+ cells. (F) N-cadherin mRNA levels in CML and normal CD34+CD38− and CD34+CD38+ cells (n = 3 CML patients and 3 healthy controls). (G) Representative flow cytometry plots showing N-cadherin expression on CML CD34+ cells cultured with and without IM and with or without MSCs. (H) N-cadherin mRNA levels in CML CD34+ cells cultured with and without IM in the presence or absence of MSCs (left), in adherent (adh) and nonadherent suspension (susp) cells, and in cells treated with an N-cadherin blocking (NCDH) and NCDH control peptide (right) (n = 3). (I) Reduced adhesion of CML CD34+ cells to MSCs in the presence of NCDH peptides. (J) Apoptosis of IM-treated CML CD34+ cells cocultured with MSCs in the presence of NCDH or control peptides (n = 3). (K) Apoptosis of IM-treated CML CD34+CD38− and CD34+CD38+ cells cocultured with MSCs and N-cadherin blocking or isotype control antibodies (n = 3). ns, not significant. *P < .05; **P < .01.
Role of N-cadherin in MSC-mediated protection of CML stem/progenitor cells from TKI treatment. (A) CML CD34+ cells cocultured on MSCs without IM (left) and with IM (right) adhered to and migrated underneath (right, arrowhead) MSCs. (B) The fraction of MSC-adherent CML CD34+ cells with or without IM. (C) Representative flow cytometry plots and (D) graphs showing apoptosis of CML CD34+ cells (C) and CD34+CD38+ and CD34+CD38− cells (D) cultured without MSCs in a Transwell insert above MSC layers or in contact with MSCs in the absence or presence of IM (n = 3). (E) Representative flow cytometry plots showing N-cadherin expression on CML and normal CD34+, CD34+CD38−, and CD34+CD38+ cells. (F) N-cadherin mRNA levels in CML and normal CD34+CD38− and CD34+CD38+ cells (n = 3 CML patients and 3 healthy controls). (G) Representative flow cytometry plots showing N-cadherin expression on CML CD34+ cells cultured with and without IM and with or without MSCs. (H) N-cadherin mRNA levels in CML CD34+ cells cultured with and without IM in the presence or absence of MSCs (left), in adherent (adh) and nonadherent suspension (susp) cells, and in cells treated with an N-cadherin blocking (NCDH) and NCDH control peptide (right) (n = 3). (I) Reduced adhesion of CML CD34+ cells to MSCs in the presence of NCDH peptides. (J) Apoptosis of IM-treated CML CD34+ cells cocultured with MSCs in the presence of NCDH or control peptides (n = 3). (K) Apoptosis of IM-treated CML CD34+CD38− and CD34+CD38+ cells cocultured with MSCs and N-cadherin blocking or isotype control antibodies (n = 3). ns, not significant. *P < .05; **P < .01.
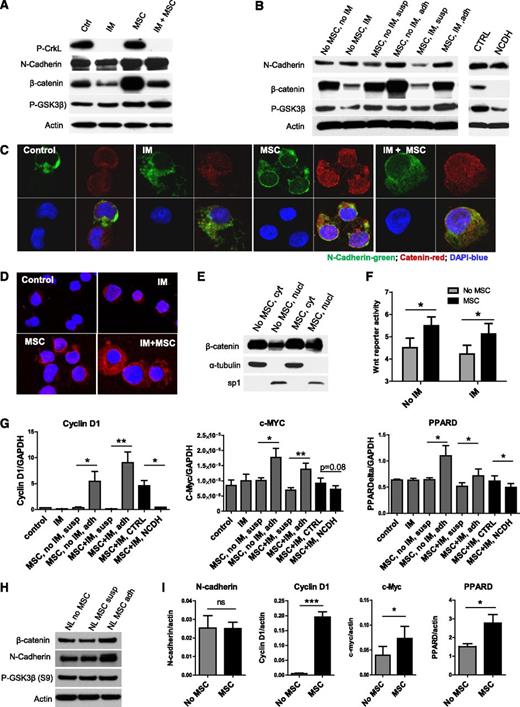
Enhanced Wnt–β-catenin signaling in TKI-treated CML stem/progenitor cells cocultured with MSCs. (A) Western blotting for P-Crkl, N-cadherin, β-catenin, P-GSK3β (S9), and actin in CML CD34+ cells cultured with and without MSCs and with and without IM treatment. (B) Western blotting for N-cadherin, β-catenin, P-GSK3β (S9), and actin in CML CD34+ cells and MSC-adherent and MSC-nonadherent CML CD34+ cells with and without IM treatment, and in CML CD34+ cells cocultured with MSCs and IM and NCDH or control peptide as shown. (C) Immunofluorescence microscopy of CML CD34+ cells labeled with antibodies to N-cadherin (green) and β-catenin (red) following culture as shown. Nuclei were labeled with 4,6 diamidino-2-phenylindole (DAPI; blue). Results shown are representative of 100 cells analyzed per slide. (D) N-cadherin and β-catenin protein–protein interactions were evaluated by using Duolink in situ PLA technology. Protein interactions are visualized as red dots. (E) Cytoplasm (cyt) and nuclear (nucl) fractions from CML CD34+ cells cultured with and without MSCs were analyzed by western blotting for β-catenin, α-tubulin, and sp1. (F) Wnt–β-catenin–related transcriptional activity was evaluated by using an improved TOPFlash reporter system in CML CD34+ cells cultured as shown. (G) Q-PCR analysis for mRNA expression of the Wnt–β-catenin target genes cyclin-D1, c-Myc, and peroxisome proliferator-activated receptor delta (PPARD) in CML CD34+ cells cultured as shown. (H) Western blotting for β-catenin, N-cadherin, P-GSK3β (S9), and actin in normal CD34+ cells cultured with and without MSCs. (I) Q-PCR analysis for mRNA expression of N-cadherin, β-catenin and Wnt–β-catenin target genes cyclin-D1, c-Myc, and PPARD in normal CD34+ cells cultured as shown. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; ns, not significant. n = 3. *P < .05; **P < .01; ***P < .001.
Enhanced Wnt–β-catenin signaling in TKI-treated CML stem/progenitor cells cocultured with MSCs. (A) Western blotting for P-Crkl, N-cadherin, β-catenin, P-GSK3β (S9), and actin in CML CD34+ cells cultured with and without MSCs and with and without IM treatment. (B) Western blotting for N-cadherin, β-catenin, P-GSK3β (S9), and actin in CML CD34+ cells and MSC-adherent and MSC-nonadherent CML CD34+ cells with and without IM treatment, and in CML CD34+ cells cocultured with MSCs and IM and NCDH or control peptide as shown. (C) Immunofluorescence microscopy of CML CD34+ cells labeled with antibodies to N-cadherin (green) and β-catenin (red) following culture as shown. Nuclei were labeled with 4,6 diamidino-2-phenylindole (DAPI; blue). Results shown are representative of 100 cells analyzed per slide. (D) N-cadherin and β-catenin protein–protein interactions were evaluated by using Duolink in situ PLA technology. Protein interactions are visualized as red dots. (E) Cytoplasm (cyt) and nuclear (nucl) fractions from CML CD34+ cells cultured with and without MSCs were analyzed by western blotting for β-catenin, α-tubulin, and sp1. (F) Wnt–β-catenin–related transcriptional activity was evaluated by using an improved TOPFlash reporter system in CML CD34+ cells cultured as shown. (G) Q-PCR analysis for mRNA expression of the Wnt–β-catenin target genes cyclin-D1, c-Myc, and peroxisome proliferator-activated receptor delta (PPARD) in CML CD34+ cells cultured as shown. (H) Western blotting for β-catenin, N-cadherin, P-GSK3β (S9), and actin in normal CD34+ cells cultured with and without MSCs. (I) Q-PCR analysis for mRNA expression of N-cadherin, β-catenin and Wnt–β-catenin target genes cyclin-D1, c-Myc, and PPARD in normal CD34+ cells cultured as shown. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; ns, not significant. n = 3. *P < .05; **P < .01; ***P < .001.
N-cadherin mediated interaction with MSCs enhances Wnt–β-catenin signaling in CML CD34+ cells
Canonical Wnt signaling results from engagement of Frizzled and LRP6 receptors on HSCs by Wnt proteins, leading to activation and nuclear translocation of β-catenin, complex formation with TCF, and transcription of target genes.30 Immortalized BM MSCs express several Wnt genes, including Wnt1, Wnt2B, Wnt3, Wnt5A, Wnt 5B, Wnt6, Wnt8B, and Wnt16 (data not shown). IM treatment resulted in reduction in total β-catenin levels in CML CD34+ cells on western blotting (Figure 4A). CML CD34+ cells showed increased expression of β-catenin after coculture with MSCs with and without IM treatment. β-catenin was increased in MSC-adherent CML CD34+ cells compared with nonadherent cells (Figure 4B). NCDH blocking peptides inhibited β-catenin expression in CML CD34+ cells cultured with MSCs (Figure 4B). We also observed increased β-catenin labeling by using immunofluorescence microscopy in CML CD34+ cells cultured with MSCs compared with cells cultured without MSCs with or without IM treatment (Figure 4C). Increased β-catenin labeling was seen in the nucleus as well as the cytoplasm of MSC-cocultured cells. Cytoplasmic β-catenin colocalized with N-cadherin. By using the Duolink PLA assay to detect intermolecular interactions between N-cadherin and β-catenin, we observed increased N-cadherin–β-catenin interactions in CML CD34+ cells cocultured with MSCs, which were increased after IM treatment (Figure 4D). Nuclear cytoplasmic fractionation confirmed an increase in nuclear β-catenin in CML CD34+ cells following MSC coculture (Figure 4E; supplemental Figure 3A). These results indicate that MSC coculture results in increased β-catenin levels in CML CD34+ cells, which includes increased cytoplasmic β-catenin in complex with N-cadherin as well as increased nuclear β-catenin.45,46 Wnt transcriptional activity, evaluated by using an improved BAR,40 was enhanced in CML CD34+ cells following MSC coculture with or without IM (Figure 4F). Q-PCR confirmed upregulation of Wnt target genes, cyclin D1, c-Myc, and peroxisome proliferator-activated receptor delta in MSC-adherent but not nonadherent CML CD34+ cells (Figure 4G). Treatment with NCDH, but not control peptides, inhibited Wnt target gene expression. These results support a role for N-cadherin–mediated adhesion to MSCs in enhanced transcription of β-catenin target genes in CML CD34+ cells. As with CML CD34+ cells, we also saw increased expression of β-catenin and N-cadherin (Figure 4H; supplemental Figure 3B) and of Wnt target genes in normal CD34+ cells cocultured with MSCs (Figure 4I).
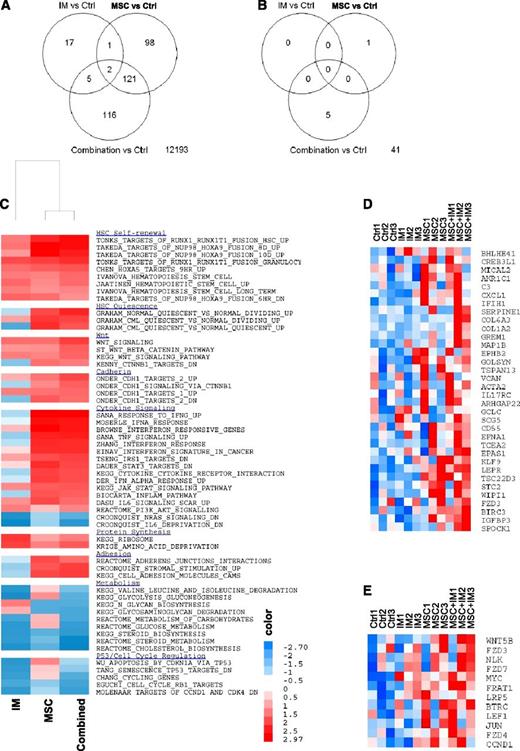
Microarray analysis
We evaluated the effect of IM and MSC treatment on gene expression in CML CD34+ cells. The number of differentially expressed genes [log2(fold change [FC]) > 0.585 (FC > 1.5), and FDR ≤ 0.25] when comparing the different treatments is shown in Figure 5A. The interaction between MSCs and IM in determining gene expression in CML CD34+ cells was analyzed by comparing changes in gene expression induced by the combination of IM and MSCs versus the sum of gene expression changes induced by IM alone and MSCs alone [interaction = (combination − MSCs) − (IM − control)]. Only 47 of 12 553 genes demonstrated significant interaction [log2(FC) > 0.585; P ≤ .05] (Figure 5B), indicating that the additive effects of IM and MSC treatment can explain the bulk of changes in gene expression. GSEA to determine enrichment of gene sets in IM and MSC-treated cells confirmed enrichment of Wnt–β-catenin and cadherin gene sets. GSEA also showed enrichment of gene sets related to HSC self-renewal and quiescence, cytokine signaling, adhesion, metabolism, and cell cycle regulation in MSC-treated CML CD34+ cells with or without IM (Figure 5C; Table 1). We identified the leading edge genes that contributed to enrichment of cadherin and catenin gene sets in CML CD34+ cells cultured on MSCs. Expression of these genes with different treatments is shown in Figure 5D and Figure 5E, respectively. N-cadherin–related genes upregulated by MSC coculture included extracellular matrix proteins VCAN, SPOCK1, COL6A3, and COL1A2; regulators of cell development EPNA1 and EPHB2; cytoskeleton regulators ACTA2, MICAL2, and MAP1B; apoptosis regulators, including TSC22D, BIRC3, and GREM1; and transcription factors KLF9, CREB3L1, BHLHE41, TSC22D3, and TCEA2. Wnt–β-catenin pathway genes upregulated by MSC coculture included WNT5B; the Wnt receptors FZD3, FZD4, FZD7, and LRP5; regulators of Wnt signaling BTRC, NLK1, FRAT1; the Wnt effector LEF1; and the Wnt target genes MYC and CCND1.
Microarray assay of gene expression in CML CD34+ cells cocultured with and without MSCs and with or without IM. (A) The number of differentially expressed genes when comparing the different treatments and (B) the interactions between MSCs and IM in determining gene expression in CML CD34+ cells are shown. GSEA showed increased expression of gene sets related to HSC self-renewal and quiescence, cytokine signaling, adhesion, metabolism, and cell cycle regulation in MSC-cocultured CML CD34+ cells with or without IM (C) and enrichment of cadherin (D) and Wnt–β-catenin (E) –related gene sets.
Microarray assay of gene expression in CML CD34+ cells cocultured with and without MSCs and with or without IM. (A) The number of differentially expressed genes when comparing the different treatments and (B) the interactions between MSCs and IM in determining gene expression in CML CD34+ cells are shown. GSEA showed increased expression of gene sets related to HSC self-renewal and quiescence, cytokine signaling, adhesion, metabolism, and cell cycle regulation in MSC-cocultured CML CD34+ cells with or without IM (C) and enrichment of cadherin (D) and Wnt–β-catenin (E) –related gene sets.
Gene sets enriched in CML CD34+ cells following exposure to MSCs and IM
| Gene sets . | NES . | FDR . | ||||
|---|---|---|---|---|---|---|
| IM . | MSC . | Combined . | IM . | MSC . | Combined . | |
| HSC self-renewal | ||||||
| TONKS_TARGETS_OF_RUNX1_RUNX1T1_FUSION_HSC_UP | 1.5031 | 2.389541 | 2.668802 | 0.200 | <0.001 | <0.001 |
| TAKEDA_TARGETS_OF_NUP98_HOXA9_FUSION_8D_UP | 1.539169 | 2.792789 | 2.660975 | 0.180 | <0.001 | <0.001 |
| TAKEDA_TARGETS_OF_NUP98_HOXA9_FUSION_10D_UP | 1.172108 | 2.766266 | 2.500532 | 0.508 | <0.001 | <0.001 |
| TONKS_TARGETS_OF_RUNX1_RUNX1T1_FUSION_GRANULOCYTE_UP | 1.993256 | 2.144978 | 2.329874 | 0.013 | <0.001 | <0.001 |
| CHEN_HOXA5_TARGETS_9HR_UP | 1.612002 | 1.801589 | 1.919964 | 0.127 | 0.014 | 0.002 |
| IVANOVA_HEMATOPOIESIS_STEM_CELL | 1.48788 | 1.386235 | 1.660943 | 0.206 | 0.151 | 0.033 |
| JAATINEN_HEMATOPOIETIC_STEM_CELL_UP | 1.690402 | 1.326127 | 1.658947 | 0.097 | 0.151 | 0.033 |
| IVANOVA_HEMATOPOIESIS_STEM_CELL_LONG_TERM | 1.456441 | 1.241877 | 1.370937 | 0.234 | 0.268 | 0.143 |
| TAKEDA_TARGETS_OF_NUP98_HOXA9_FUSION_6HR_DN | 1.714095 | 1.645097 | 1.249897 | 0.086 | 0.041 | 0.243 |
| HSC quiescence | ||||||
| GRAHAM_NORMAL_QUIESCENT_VS_NORMAL_DIVIDING_UP | −0.85093 | 2.016533 | 2.410049 | 0.866 | 0.001 | <0.001 |
| GRAHAM_CML_QUIESCENT_VS_NORMAL_DIVIDING_UP | 1.078994 | 2.199812 | 2.085968 | 0.599 | <0.001 | <0.001 |
| GRAHAM_CML_QUIESCENT_VS_NORMAL_QUIESCENT_UP | −1.95649 | 2.334091 | 1.929717 | |||
| Wnt | ||||||
| WNT_SIGNALING | 1.644355 | 1.602941 | 2.137335 | 0.111 | 0.054 | <0.001 |
| ST_WNT_BETA_CATENIN_PATHWAY | 1.186699 | 1.251462 | 1.795597 | 0.492 | 0.260 | 0.012 |
| KEGG_WNT_SIGNALING_PATHWAY | 1.45817 | 1.374894 | 1.707158 | 0.233 | 0.157 | 0.023 |
| KENNY_CTNNB1_TARGETS_DN | −1.36828 | 1.599336 | 1.469737 | 0.211 | 0.055 | 0.094 |
| Cadherin | ||||||
| ONDER_CDH1_TARGETS_2_UP | 0.921706 | 1.535746 | 2.065623 | 0.822 | 0.074 | 0.001 |
| ONDER_CDH1_SIGNALING_VIA_CTNNB1 | −1.01896 | 1.318553 | 1.775083 | 0.627 | 0.195 | 0.014 |
| ONDER_CDH1_TARGETS_1_UP | 1.462469 | 1.571189 | 1.665949 | 0.229 | 0.064 | 0.032 |
| ONDER_CDH1_TARGETS_2_DN | −1.44575 | 1.831754 | 1.64301 | 0.156 | 0.011 | 0.036 |
| Cytokine signaling | ||||||
| SANA_RESPONSE_TO_IFNG_UP | −0.95045 | 2.965142 | 2.836384 | 0.726 | <0.001 | <0.001 |
| MOSERLE_IFNA_RESPONSE | −0.39523 | 2.628911 | 2.578175 | 1.000 | <0.001 | <0.001 |
| BROWNE_INTERFERON_RESPONSIVE_GENES | 0.795554 | 2.824429 | 2.561327 | 0.944 | <0.001 | <0.001 |
| SANA_TNF_SIGNALING_UP | −1.19179 | 2.440201 | 2.483553 | 0.383 | <0.001 | <0.001 |
| ZHANG_INTERFERON_RESPONSE | −0.82431 | 2.363861 | 2.330738 | 0.893 | <0.001 | <0.001 |
| EINAV_INTERFERON_SIGNATURE_IN_CANCER | 0.452984 | 2.327512 | 2.303409 | 1.000 | <0.001 | <0.001 |
| TSENG_IRS1_TARGETS_DN | 1.086487 | 1.9622 | 2.110712 | 0.595 | 0.002 | <0.001 |
| DAUER_STAT3_TARGETS_DN | 0.49214 | 2.332711 | 2.044684 | 1.000 | <0.001 | 0.001 |
| KEGG_CYTOKINE_CYTOKINE_RECEPTOR_INTERACTION | −1.05504 | 2.092004 | 1.973682 | 0.575 | <0.001 | 0.001 |
| DER_IFN_ALPHA_RESPONSE_UP | −0.61103 | 2.152106 | 1.965927 | 0.997 | <0.001 | 0.002 |
| KEGG_JAK_STAT_SIGNALING_PATHWAY | 1.323564 | 1.822952 | 1.868087 | 0.360 | 0.012 | 0.004 |
| BIOCARTA_INFLAM_PATHWAY | −1.03839 | 1.934039 | 1.792361 | 0.600 | 0.004 | 0.012 |
| DASU_IL6_SIGNALING_SCAR_UP | 1.249695 | 2.034135 | 1.658946 | 0.426 | 0.001 | 0.033 |
| REACTOME_PI3K_AKT_SIGNALING | 1.451631 | 0.752397 | 1.376669 | 0.238 | 0.922 | 0.139 |
| CROONQUIST_NRAS_SIGNALING_DN | −2.47053 | −1.06302 | −1.86165 | <0.001 | 0.667 | 0.020 |
| CROONQUIST_IL6_DEPRIVATION_DN | −2.64986 | −1.10654 | −2.33675 | <0.001 | 0.586 | <0.001 |
| Protein synthesis | ||||||
| KEGG_RIBOSOME | 2.226323 | 1.02579 | 1.438408 | 0.005 | 0.552 | 0.106 |
| KRIGE_AMINO_ACID_DEPRIVATION | 2.124014 | 1.422338 | 1.829972 | 0.007 | 0.129 | 0.008 |
| Adhesion | ||||||
| REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | −0.6271 | 1.994029 | 1.614459 | 0.995 | 0.002 | 0.044 |
| CROONQUIST_STROMAL_STIMULATION_UP | −1.17825 | 1.87128 | 2.138987 | 0.400 | 0.007 | <0.001 |
| KEGG_CELL_ADHESION_MOLECULES_CAMS | −1.02396 | 2.055671 | 2.157459 | 0.621 | 0.001 | <0.001 |
| Metabolism | ||||||
| KEGG_VALINE_LEUCINE_AND_ISOLEUCINE_DEGRADATION | −2.35516 | 0.562928 | −1.52169 | <0.001 | 0.996 | 0.121 |
| KEGG_GLYCOLYSIS_GLUCONEOGENESIS | −2.53665 | 1.257924 | −1.82849 | <0.001 | 0.252 | 0.025 |
| KEGG_N_GLYCAN_BIOSYNTHESIS | 1.491404 | −1.90203 | −1.87135 | 0.206 | 0.042 | 0.019 |
| KEGG_GLYCOSAMINOGLYCAN_DEGRADATION | 0.943999 | −1.89651 | −2.00295 | 0.801 | 0.040 | 0.007 |
| REACTOME_METABOLISM_OF_CARBOHYDRATES | −2.04296 | 0.833918 | −2.01312 | 0.001 | 0.835 | 0.007 |
| REACTOME_GLUCOSE_METABOLISM | −2.31217 | 0.88233 | −2.23553 | <0.001 | 0.771 | 0.002 |
| KEGG_STEROID_BIOSYNTHESIS | −2.28708 | −1.82998 | −2.25745 | <0.001 | 0.052 | 0.003 |
| REACTOME_STEROID_METABOLISM | −2.27821 | −1.58039 | −2.67395 | <0.001 | 0.156 | <0.001 |
| REACTOME_CHOLESTEROL_BIOSYNTHESIS | −2.44969 | −1.7996 | −2.7018 | <0.001 | 0.063 | <0.001 |
| Cell cycle regulation | ||||||
| WU_APOPTOSIS_BY_CDKN1A_VIA_TP53 | −2.30721 | 1.345716 | −1.0242 | <0.001 | 0.176 | 0.607 |
| TANG_SENESCENCE_TP53_TARGETS_DN | −2.26777 | 0.669011 | −1.25232 | <0.001 | 0.973 | 0.322 |
| CHANG_CYCLING_GENES | −2.21082 | 0.580398 | −1.90182 | <0.001 | 0.993 | 0.014 |
| EGUCHI_CELL_CYCLE_RB1_TARGETS | −2.00287 | −1.28341 | −2.06781 | 0.001 | 0.375 | 0.005 |
| MOLENAAR_TARGETS_OF_CCND1_AND_CDK4_DN | −2.2664 | −0.99657 | −2.17995 | 0.000 | 0.764 | 0.002 |
| Gene sets . | NES . | FDR . | ||||
|---|---|---|---|---|---|---|
| IM . | MSC . | Combined . | IM . | MSC . | Combined . | |
| HSC self-renewal | ||||||
| TONKS_TARGETS_OF_RUNX1_RUNX1T1_FUSION_HSC_UP | 1.5031 | 2.389541 | 2.668802 | 0.200 | <0.001 | <0.001 |
| TAKEDA_TARGETS_OF_NUP98_HOXA9_FUSION_8D_UP | 1.539169 | 2.792789 | 2.660975 | 0.180 | <0.001 | <0.001 |
| TAKEDA_TARGETS_OF_NUP98_HOXA9_FUSION_10D_UP | 1.172108 | 2.766266 | 2.500532 | 0.508 | <0.001 | <0.001 |
| TONKS_TARGETS_OF_RUNX1_RUNX1T1_FUSION_GRANULOCYTE_UP | 1.993256 | 2.144978 | 2.329874 | 0.013 | <0.001 | <0.001 |
| CHEN_HOXA5_TARGETS_9HR_UP | 1.612002 | 1.801589 | 1.919964 | 0.127 | 0.014 | 0.002 |
| IVANOVA_HEMATOPOIESIS_STEM_CELL | 1.48788 | 1.386235 | 1.660943 | 0.206 | 0.151 | 0.033 |
| JAATINEN_HEMATOPOIETIC_STEM_CELL_UP | 1.690402 | 1.326127 | 1.658947 | 0.097 | 0.151 | 0.033 |
| IVANOVA_HEMATOPOIESIS_STEM_CELL_LONG_TERM | 1.456441 | 1.241877 | 1.370937 | 0.234 | 0.268 | 0.143 |
| TAKEDA_TARGETS_OF_NUP98_HOXA9_FUSION_6HR_DN | 1.714095 | 1.645097 | 1.249897 | 0.086 | 0.041 | 0.243 |
| HSC quiescence | ||||||
| GRAHAM_NORMAL_QUIESCENT_VS_NORMAL_DIVIDING_UP | −0.85093 | 2.016533 | 2.410049 | 0.866 | 0.001 | <0.001 |
| GRAHAM_CML_QUIESCENT_VS_NORMAL_DIVIDING_UP | 1.078994 | 2.199812 | 2.085968 | 0.599 | <0.001 | <0.001 |
| GRAHAM_CML_QUIESCENT_VS_NORMAL_QUIESCENT_UP | −1.95649 | 2.334091 | 1.929717 | |||
| Wnt | ||||||
| WNT_SIGNALING | 1.644355 | 1.602941 | 2.137335 | 0.111 | 0.054 | <0.001 |
| ST_WNT_BETA_CATENIN_PATHWAY | 1.186699 | 1.251462 | 1.795597 | 0.492 | 0.260 | 0.012 |
| KEGG_WNT_SIGNALING_PATHWAY | 1.45817 | 1.374894 | 1.707158 | 0.233 | 0.157 | 0.023 |
| KENNY_CTNNB1_TARGETS_DN | −1.36828 | 1.599336 | 1.469737 | 0.211 | 0.055 | 0.094 |
| Cadherin | ||||||
| ONDER_CDH1_TARGETS_2_UP | 0.921706 | 1.535746 | 2.065623 | 0.822 | 0.074 | 0.001 |
| ONDER_CDH1_SIGNALING_VIA_CTNNB1 | −1.01896 | 1.318553 | 1.775083 | 0.627 | 0.195 | 0.014 |
| ONDER_CDH1_TARGETS_1_UP | 1.462469 | 1.571189 | 1.665949 | 0.229 | 0.064 | 0.032 |
| ONDER_CDH1_TARGETS_2_DN | −1.44575 | 1.831754 | 1.64301 | 0.156 | 0.011 | 0.036 |
| Cytokine signaling | ||||||
| SANA_RESPONSE_TO_IFNG_UP | −0.95045 | 2.965142 | 2.836384 | 0.726 | <0.001 | <0.001 |
| MOSERLE_IFNA_RESPONSE | −0.39523 | 2.628911 | 2.578175 | 1.000 | <0.001 | <0.001 |
| BROWNE_INTERFERON_RESPONSIVE_GENES | 0.795554 | 2.824429 | 2.561327 | 0.944 | <0.001 | <0.001 |
| SANA_TNF_SIGNALING_UP | −1.19179 | 2.440201 | 2.483553 | 0.383 | <0.001 | <0.001 |
| ZHANG_INTERFERON_RESPONSE | −0.82431 | 2.363861 | 2.330738 | 0.893 | <0.001 | <0.001 |
| EINAV_INTERFERON_SIGNATURE_IN_CANCER | 0.452984 | 2.327512 | 2.303409 | 1.000 | <0.001 | <0.001 |
| TSENG_IRS1_TARGETS_DN | 1.086487 | 1.9622 | 2.110712 | 0.595 | 0.002 | <0.001 |
| DAUER_STAT3_TARGETS_DN | 0.49214 | 2.332711 | 2.044684 | 1.000 | <0.001 | 0.001 |
| KEGG_CYTOKINE_CYTOKINE_RECEPTOR_INTERACTION | −1.05504 | 2.092004 | 1.973682 | 0.575 | <0.001 | 0.001 |
| DER_IFN_ALPHA_RESPONSE_UP | −0.61103 | 2.152106 | 1.965927 | 0.997 | <0.001 | 0.002 |
| KEGG_JAK_STAT_SIGNALING_PATHWAY | 1.323564 | 1.822952 | 1.868087 | 0.360 | 0.012 | 0.004 |
| BIOCARTA_INFLAM_PATHWAY | −1.03839 | 1.934039 | 1.792361 | 0.600 | 0.004 | 0.012 |
| DASU_IL6_SIGNALING_SCAR_UP | 1.249695 | 2.034135 | 1.658946 | 0.426 | 0.001 | 0.033 |
| REACTOME_PI3K_AKT_SIGNALING | 1.451631 | 0.752397 | 1.376669 | 0.238 | 0.922 | 0.139 |
| CROONQUIST_NRAS_SIGNALING_DN | −2.47053 | −1.06302 | −1.86165 | <0.001 | 0.667 | 0.020 |
| CROONQUIST_IL6_DEPRIVATION_DN | −2.64986 | −1.10654 | −2.33675 | <0.001 | 0.586 | <0.001 |
| Protein synthesis | ||||||
| KEGG_RIBOSOME | 2.226323 | 1.02579 | 1.438408 | 0.005 | 0.552 | 0.106 |
| KRIGE_AMINO_ACID_DEPRIVATION | 2.124014 | 1.422338 | 1.829972 | 0.007 | 0.129 | 0.008 |
| Adhesion | ||||||
| REACTOME_ADHERENS_JUNCTIONS_INTERACTIONS | −0.6271 | 1.994029 | 1.614459 | 0.995 | 0.002 | 0.044 |
| CROONQUIST_STROMAL_STIMULATION_UP | −1.17825 | 1.87128 | 2.138987 | 0.400 | 0.007 | <0.001 |
| KEGG_CELL_ADHESION_MOLECULES_CAMS | −1.02396 | 2.055671 | 2.157459 | 0.621 | 0.001 | <0.001 |
| Metabolism | ||||||
| KEGG_VALINE_LEUCINE_AND_ISOLEUCINE_DEGRADATION | −2.35516 | 0.562928 | −1.52169 | <0.001 | 0.996 | 0.121 |
| KEGG_GLYCOLYSIS_GLUCONEOGENESIS | −2.53665 | 1.257924 | −1.82849 | <0.001 | 0.252 | 0.025 |
| KEGG_N_GLYCAN_BIOSYNTHESIS | 1.491404 | −1.90203 | −1.87135 | 0.206 | 0.042 | 0.019 |
| KEGG_GLYCOSAMINOGLYCAN_DEGRADATION | 0.943999 | −1.89651 | −2.00295 | 0.801 | 0.040 | 0.007 |
| REACTOME_METABOLISM_OF_CARBOHYDRATES | −2.04296 | 0.833918 | −2.01312 | 0.001 | 0.835 | 0.007 |
| REACTOME_GLUCOSE_METABOLISM | −2.31217 | 0.88233 | −2.23553 | <0.001 | 0.771 | 0.002 |
| KEGG_STEROID_BIOSYNTHESIS | −2.28708 | −1.82998 | −2.25745 | <0.001 | 0.052 | 0.003 |
| REACTOME_STEROID_METABOLISM | −2.27821 | −1.58039 | −2.67395 | <0.001 | 0.156 | <0.001 |
| REACTOME_CHOLESTEROL_BIOSYNTHESIS | −2.44969 | −1.7996 | −2.7018 | <0.001 | 0.063 | <0.001 |
| Cell cycle regulation | ||||||
| WU_APOPTOSIS_BY_CDKN1A_VIA_TP53 | −2.30721 | 1.345716 | −1.0242 | <0.001 | 0.176 | 0.607 |
| TANG_SENESCENCE_TP53_TARGETS_DN | −2.26777 | 0.669011 | −1.25232 | <0.001 | 0.973 | 0.322 |
| CHANG_CYCLING_GENES | −2.21082 | 0.580398 | −1.90182 | <0.001 | 0.993 | 0.014 |
| EGUCHI_CELL_CYCLE_RB1_TARGETS | −2.00287 | −1.28341 | −2.06781 | 0.001 | 0.375 | 0.005 |
| MOLENAAR_TARGETS_OF_CCND1_AND_CDK4_DN | −2.2664 | −0.99657 | −2.17995 | 0.000 | 0.764 | 0.002 |
NES, Normalized enrichment score.
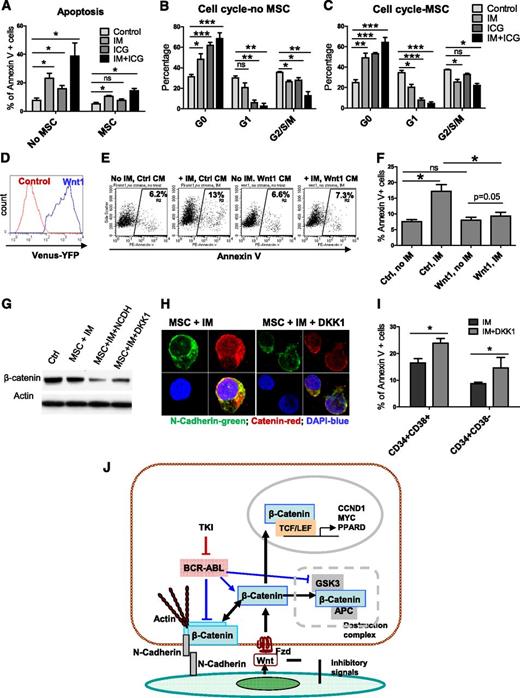
MSC-mediated Wnt–β-catenin signaling protects CML stem/progenitor cells from IM treatment
We next studied the functional significance of β-catenin signaling in the protection of CML CD34+ cells from IM treatment. The small molecule β-catenin inhibitor ICG001 increased apoptosis and reduced cell cycling in CML CD34+ cells cultured with or without MSCs (Figure 6A-C), supporting an important role for Wnt–β-catenin signaling in maintaining proliferation and survival of CML stem/progenitor cells. ICG001 treatment resulted in reduced cyclin D1 and c-Myc expression (supplemental Figure 4A), showing an important role for β-catenin signaling in increased expression of these genes. ICG001 treatment enhanced adhesion of CML CD34+ cells to MSCs (supplemental Figure 4B), indicating that altered cyclin D1 and c-Myc levels are not simply related to adherence or detachment to MSCs. Culture with conditioned medium (CM) from cells expressing Wnt1 resulted in significantly enhanced Wnt reporter activity in CML CD34+ cells compared with control CM (Figure 6D). Wnt1 CM significantly reduced apoptosis of CML CD34+ cells following IM treatment compared with control CM, mimicking the effect of coculture with MSCs (Figure 6E-F) and indicating that exogenous Wnt can protect CML CD34+ cells from TKI treatment. Treatment with purified Wnt3a (100 ng/mL) reduced N-cadherin mRNA expression in CML CD34+ cells after 24 hours but did not change N-cadherin mRNA and protein expression at 72 hours (supplemental Figure 4C-D), suggesting that increased N-cadherin expression in cells cultured with IM and MSCs reflects mechanisms other than Wnt–β-catenin signaling. Finally, we used the Wnt receptor inhibitor DKK1 (1 µg/mL), which prevents Wnt binding to the LRP6 receptor, to study the role of exogenous Wnt signaling in MSC-mediated protection of CML stem/progenitor cells from TKIs. DKK1 treatment prevented MSC-mediated increase in β-catenin expression (Figure 6G), nuclear localization in CML CD34+ cells (Figure 6H), and increased IM-induced apoptosis in CML CD34+CD38− and CD34+CD38+ cells cocultured with MSCs (Figure 6I). These results indicate an important role for MSC-mediated exogenous Wnt signaling in the activation of β-catenin and protection of CML progenitors from TKI treatment.
Role of Wnt–β-catenin signaling in MSC-mediated protection of CML stem/progenitor cells from TKI treatment. (A) Apoptosis of CML CD34+ cells treated with IM (5 µM), ICG001 (5 µM), or IM plus ICG001 in the absence and presence of MSCs. Cell cycle of CML CD34+ cells treated with IM, ICG001, or IM plus ICG001 in the absence (B) and presence (C) of MSCs. (D) CML CD34+ cells were exposed to CM from Wnt1-transfected cells and Wnt reporter activity measured after 2 days. (E) Flow cytometry plots and (F) graph showing apoptosis of CML CD34+ cells cocultured with Wnt1-CM with or without IM treatment. (G) β-catenin protein expression and (H) nuclear localization of β-catenin in IM-treated CML CD34+ cells after addition of Wnt receptor antagonist DKK1(1 µg/mL). Results shown are representative of 100 cells analyzed per slide. (I) Apoptosis of IM-treated CML CD34+ cells cocultured with MSCs in the presence and absence of DKK1. (J) Proposed Wnt–β-catenin and N-cadherin interactions in CML CD34+ cells treated with TKI in the presence of MSCs. TKI treatment stabilizes β-catenin by reducing β-catenin phosphorylation, increasing N-cadherin–mediated adhesion to MSCs, and enhancing N-cadherin–β-catenin interaction. Wnt proteins secreted by MSCs activate β-catenin signaling in MSC-adherent CML stem/progenitor cells, leading to enhanced nuclear translocation of β-catenin and transcription of target genes. Complex formation with N-cadherin in MSC-adherent CML stem/progenitor cells may protect β-catenin from degradation and provide a β-catenin pool that can be activated by exogenous Wnt ligands. MSC-induced N-cadherin and Wnt–β-catenin signaling protects and preserves CML stem/progenitor cells from TKI treatment. ns, not significant. n = 3. *P < .05.
Role of Wnt–β-catenin signaling in MSC-mediated protection of CML stem/progenitor cells from TKI treatment. (A) Apoptosis of CML CD34+ cells treated with IM (5 µM), ICG001 (5 µM), or IM plus ICG001 in the absence and presence of MSCs. Cell cycle of CML CD34+ cells treated with IM, ICG001, or IM plus ICG001 in the absence (B) and presence (C) of MSCs. (D) CML CD34+ cells were exposed to CM from Wnt1-transfected cells and Wnt reporter activity measured after 2 days. (E) Flow cytometry plots and (F) graph showing apoptosis of CML CD34+ cells cocultured with Wnt1-CM with or without IM treatment. (G) β-catenin protein expression and (H) nuclear localization of β-catenin in IM-treated CML CD34+ cells after addition of Wnt receptor antagonist DKK1(1 µg/mL). Results shown are representative of 100 cells analyzed per slide. (I) Apoptosis of IM-treated CML CD34+ cells cocultured with MSCs in the presence and absence of DKK1. (J) Proposed Wnt–β-catenin and N-cadherin interactions in CML CD34+ cells treated with TKI in the presence of MSCs. TKI treatment stabilizes β-catenin by reducing β-catenin phosphorylation, increasing N-cadherin–mediated adhesion to MSCs, and enhancing N-cadherin–β-catenin interaction. Wnt proteins secreted by MSCs activate β-catenin signaling in MSC-adherent CML stem/progenitor cells, leading to enhanced nuclear translocation of β-catenin and transcription of target genes. Complex formation with N-cadherin in MSC-adherent CML stem/progenitor cells may protect β-catenin from degradation and provide a β-catenin pool that can be activated by exogenous Wnt ligands. MSC-induced N-cadherin and Wnt–β-catenin signaling protects and preserves CML stem/progenitor cells from TKI treatment. ns, not significant. n = 3. *P < .05.
Discussion
BCR-ABL expressing LSCs persist in CML patients receiving TKI treatment, despite effective inhibition of BCR-ABL tyrosine kinase activity.6,7,9 LSCs are exposed to TKI in the context of the BM microenvironment, and our results show that BM-derived MSCs protect CML stem and progenitor cells from TKI-mediated cell death and depletion. The supportive effects of MSCs extend to both dividing and nondividing cells. MSC enhanced the proportion of nondividing CML cells, maintained CD34+ expression, and enhanced retention of CML stem cells with NSG mouse repopulating capacity in the presence and the absence of TKI. Gene expression analysis confirmed that MSC coculture induced similar patterns of gene expression in the presence or absence of IM.
Our observation that N-cadherin–mediated adhesion plays an important role in protection of CML stem/progenitor cells from TKI treatment by BM MSCs reveals a novel and previously unrecognized mechanism of resistance of CML stem/progenitor cells to TKI treatment. The role of N-cadherin in HSC regulation has been controversial. Although initially reported to play an important role in murine HSC adhesion to osteoblasts and maintenance of HSC quiescence, subsequent studies using knockout mice have not supported an important role for N-cadherin in murine HSC function.23,25,26,47 Indeed, we could not detect N-cadherin mRNA expression in purified murine HSCs (data not shown). In contrast, but consistent with previous reports,27 we found that N-cadherin was expressed on human stem/progenitor cell populations, indicating species-dependent differences in expression and function of N-cadherin between human and murine HSCs. CML stem/progenitor cells showed reduced N-cadherin expression compared with normal progenitors. An increase in N-cadherin expression was seen in MSC-adherent CML cells. CML stem/progenitor cells cultured on MSCs demonstrated increased association of N-cadherin and cytoplasmic β-catenin, which was the highest in IM-treated cells. Increased N-cadherin and β-catenin interactions may contribute to enhanced MSC adhesion and, ultimately, to resistance to TKIs. The mechanisms underlying increased N-cadherin expression are not clear but could reflect increased retention of N-cadherin–expressing MSC-adherent cells, which are protected from TKI treatment.
We show that coculture with MSCs activates Wnt–β-catenin signaling in CML CD34+ cells, as evidenced by increase in β-catenin levels, translocation to the nucleus, increased transcriptional activity, and enhanced expression of β-catenin target genes. Increased β-catenin activity is restricted to MSC-adherent CML cells. IM treatment reduced β-catenin levels in CML stem/progenitor cells but did not affect Wnt reporter activity or expression of Wnt target genes, indicating that additional kinase-independent mechanisms contribute to increased β-catenin signaling in CML cells. Importantly, coculture with MSCs also activated Wnt–β-catenin signaling in IM-treated CML stem/progenitor cells. The β-catenin inhibitor ICG001 reduced survival and proliferation of CML stem/progenitor cells with and without IM, whereas addition of purified Wnt1 activated β-catenin and protected CML cells from IM-induced apoptosis, confirming an important role for Wnt–β-catenin signaling in maintaining CML stem/progenitor cells. Our results suggest that engagement of Wnt receptors by microenvironmental Wnt proteins can activate β-catenin signaling and enhance survival of TKI-treated CML stem/progenitor cells, since the LRP6 antagonist DKK1 inhibited Wnt signaling and enhanced apoptosis of IM-treated CML stem/progenitor cells cocultured with MSCs. These results extend previous observations of increased Wnt–β-catenin signaling in LSCs33-35,37 and show that physiological activation of this pathway via microenvironmental Wnts is a novel BCR-ABL kinase-independent mechanism of resistance to TKI treatment in human CP CML stem/progenitor cells.
Cadherin cytoplasmic domains bind to submembranous β-catenin, which in turn is linked to the actin cytoskeleton. Cadherin receptor association with the cytoskeleton is necessary for stabilization of cell–cell adhesion. However, the role of N-cadherin and β-catenin interactions in regulating HSC growth is not well studied.29,45,48,49 We observed increased cytoplasmic N-cadherin–β-catenin complex formation in IM-treated CML CD34+ cells cultured with MSCs, which could contribute to increased N-cadherin–mediated adhesion to MSCs. Although cadherins are reported to modulate Wnt–β-catenin signaling by sequestering β-catenin at the cell surface, which prevents translocation to the nucleus,49 we found that N-cadherin–mediated adhesion to MSCs was associated with increased expression of β-catenin target genes in CML progenitors. Since secreted Wnts are bound to the cell surface or extracellular matrix, physiological Wnt signaling requires close adhesion of stem/progenitor cells to MSCs. The critical role for N-cadherin in MSC-mediated Wnt signaling may be related to its role in adhesion of CML progenitors to MSCs. Another possibility is that β-catenin in complex with N-cadherin in adherent cells may be protected from degradation and may remain available for activation by exogenous Wnt ligands.49
Previous studies have suggested a potential role for the tumor microenvironment in protecting leukemia cells from the effects of drug therapy.14,18,19,50 However, it has been unclear whether such interactions could determine resistance of human CML LSCs to TKI treatment. With an increasing number of CML patients receiving continuous TKI treatment, concerns regarding high financial burden, long-term side effects, and loss of compliance have generated interest in developing approaches to target residual LSCs to facilitate cessation of treatment and achieve cures. Our results reveal a close interplay between microenvironmental signals mediated by N-cadherin and Wnt–β-catenin in protecting CML stem/progenitor cells during TKI treatment. These results elucidate a novel, previously unrecognized mechanism of resistance of CML stem/progenitor cells to TKI treatment and support further testing of measures aimed at interfering with N-cadherin–mediated interactions or Wnt secretion or signaling from the microenvironment to promote eradication of residual malignant cells.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by National Institutes of Health (NIH) grants R01 HL77847 and R01 CA95684 (R.B.), NIH Cancer Center Core grant CA34196 (The Jackson Laboratory), U01 HL100395 (R.T.M), and Cancer Research-UK Programme grant C11074/A11008 (T.L.H.). We acknowledge the excellent technical support of the City of Hope National Medical Center Analytical Cytometry and Functional Genomics cores, and the Animal Resources Center. We thank StemCyte for their generous gift of cord blood samples, and Allen Lin, Jennifer Arceo, Alan Hair, and Linda Seymour for collecting and processing patient samples.
National Institutes of Health
Authorship
B.Z. designed and performed research, analyzed data, and wrote the manuscript. M.L. performed data analysis and wrote and reviewed the manuscript. T.M. performed experiments and reviewed the manuscript. T.L.H., R.T.M., D.C., and L.S. provided material, interpreted data, and reviewed the manuscript. R.B. designed the study, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ravi Bhatia, MD, Division of Hematopoietic Stem Cell and Leukemia Research, City of Hope National Medical Center, Duarte, CA 91010; e-mail: rbhatia@coh.org

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal