In this issue of Blood, Gilbert and colleagues present an elegant study on the membrane-interactive domains of factors VIII (FVIII) and V (FV).1 The 3-dimensional structure of these domains exposes a few characteristic loops, referred to as “fatty feet,” which mediate the interaction with lipid membranes. These are similar, but not identical in FVIII and FV. Swapping feet between FV and FVIII resulted in remarkable changes in lipid binding and biologic activity. These data help in understanding how these cofactors assemble with their catalytic partners in the coagulation cascade.
During evolution, nature has duplicated 2 homologous biologic amplifiers, the intrinsic factor Xase and the prothrombinase complex. The heart of these complexes is formed by FVIII and FV, which already occur as 2 separate cofactors in the Fugu (pufferfish) genome.2 In the past few decades, we learned that these cofactors assemble with their enzyme partners and substrates on biologic membranes containing the negatively-charged phosphatidyl serine (PS).3 More recently, the crystal structure of FVIII and a large part of FV have been resolved.4,5 Knowing the 3-dimensional structure has long been regarded as the Holy Grail in structure/function analysis of the coagulation factors. Now that we have these structures at our disposal, we can target the numerous issues that remain unexplained. For instance, while the structures of factors VIII and V are remarkably similar, these proteins serve different functions. FVIII accelerates the activation of factor X by activated FIX, while FV amplifies the activation of prothrombin by activated FX. Both cofactors promote coagulation by assembling with their enzymatic partners and substrates on membranes that expose PS, such as activated platelets. On the one hand, it seems reasonable to assume that these mechanisms have just been duplicated by nature and thus are very similar. On the other hand, the plasma concentration of FV is 100-fold higher than that of FVIII, which would make FV a very effective competitor in FX activation. This conceptual problem might be resolved by the existence of specific membrane receptors on activated platelets that distinguish between FVIII and FV.6,7 However, such receptors have not yet been identified. It therefore remains of particular interest to address potential differences between FVIII and FV with respect to their requirements for binding to PS-containing membranes. This is what Gilbert and colleagues report in their present study.1
Their approach is a very elegant one. They focus on the C-terminal part of FVIII and FV, the C2 domain, which comprises a major lipid-binding part of these proteins. Since the crystallization of the isolated C2 domain of FVIII and FV, it has been recognized that these domains contain solvent-exposed hydrophobic amino acids combined with positively charged amino acids (see figure). These “hydrophobic spikes” or fatty feet seem ideally designed to interact with lipid membranes containing the negatively charged lipid PS. Gilbert and colleagues have addressed 2 feet in the C2 domain, and swapped these between FVIII and FV. For each foot, this swap comprised no more than 2 amino acids. The hybrid proteins were expressed, purified, and analyzed for lipid binding and cofactor function. Surprisingly, these minor changes did have significant effects on cofactor function. FVIII activity increased by introducing the Trp2063/Trp2064 motif from FV instead of its own Met2199/Phe2200 foot (see figure). On the other hand, FV activity did benefit from the introduction of the Leu2251/Leu2252 motif from FVIII instead of its own Leu2116/Ser2117 foot. Why would swapping 1 foot from FVIII improve FV and vice versa? And why are these effects gain-of-function and not loss-of-function? Don't these cofactors stand on the “wrong” feet, in particular when the 2 feet are swapped together? These intriguing questions remain difficult to answer at this point.
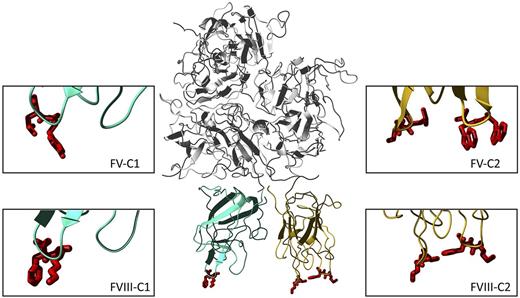
The factor VIII molecule and its fatty feet. The complete FVIII molecule in the center is derived from the PDB entry 2R7E. The top part represents the 3 A domains (gray). The C1 and C2 domains are depicted in cyan blue and orange, respectively, and the lipid binding amino acids are depicted in red. The right panels show close-ups of the bottom of the C2 domain of factors VIII and V. The swapped residues are Met2199/Phe2200 (right) and Leu2251/Leu2252 (left) in FVIII and Trp2063/Trp2064 (right) and Leu2116/Ser2117 (left) in FV. The left panels display the bottom of the C1 domain, with the Lys2092/Phe2093 foot of FVIII and its homologue Tyr1956/Leu1957 in FV. FV loops in the C2 domain are derived from PDB entry 1CZT, and those of the C1 domain by modeling the human sequence on the bovine template (entry 1SDD). These structures suggest that the 2 C domains stand side-by-side on the membrane and contribute in a cooperative manner to membrane binding. Because the C2 domain is only loosely attached in the structure by only few interdomain contacts, it may be capable of some reorientation in response to changes in the fatty feet or in the composition of the procoagulant membrane.
The factor VIII molecule and its fatty feet. The complete FVIII molecule in the center is derived from the PDB entry 2R7E. The top part represents the 3 A domains (gray). The C1 and C2 domains are depicted in cyan blue and orange, respectively, and the lipid binding amino acids are depicted in red. The right panels show close-ups of the bottom of the C2 domain of factors VIII and V. The swapped residues are Met2199/Phe2200 (right) and Leu2251/Leu2252 (left) in FVIII and Trp2063/Trp2064 (right) and Leu2116/Ser2117 (left) in FV. The left panels display the bottom of the C1 domain, with the Lys2092/Phe2093 foot of FVIII and its homologue Tyr1956/Leu1957 in FV. FV loops in the C2 domain are derived from PDB entry 1CZT, and those of the C1 domain by modeling the human sequence on the bovine template (entry 1SDD). These structures suggest that the 2 C domains stand side-by-side on the membrane and contribute in a cooperative manner to membrane binding. Because the C2 domain is only loosely attached in the structure by only few interdomain contacts, it may be capable of some reorientation in response to changes in the fatty feet or in the composition of the procoagulant membrane.
There are several factors that complicate the full appreciation of these findings. First, the C2 domain mutants still have their C1 domain to mediate lipid binding. In collaboration with our group, Gilbert and colleagues recently identified 1 foot in the C1 domain of FVIII (Lys2902/Phe2093, see figure) that also contributes to lipid binding, in particular to membranes of low (4%), but not higher (> 15%) PS content.8 The swap variants in the present study have been analyzed on membranes containing 4% PS only. Thus, lipid binding of some C2 domain hybrids at 4% PS might be mediated by the C1 domain as well. Perhaps this explains why major loss-of-function effects have not been observed. In this regard it is interesting that the Lys2902/Phe2093 foot in FVIII has a counterpart in FV (Tyr1956/Leu1957) that is known to contribute to membrane-dependent prothrombin activation.9 FV with Alanine in these 2 positions was defective at low PS content only. However, this defect became prominent at high PS content as well when these substitutions were combined with mutation of the Trp2063/Trp2064 foot in the C2 domain.9 This suggests that in FV the C1 and C2 domain feet contribute to membrane assembly in a cooperative manner. It seems very likely that this also holds true for FVIII. The importance of the FVIII C1 domain is further underlined by the finding that deletion of the entire C2 domain from FVIII still retains substantial lipid-dependent FX activation.10 Apparently, the 2 C domains of FVIII and FV cooperate together by an intricate mechanism that is largely dependent on the membrane composition. Although the details thereof remain to be established, the current work is inspiring, and facilitates further unraveling these complex interactions in the near future.
Conflict-of-interest disclosure: The authors declare no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal