Abstract
Birt-Hogg-Dubé (BHD) syndrome is an autosomal dominant disorder characterized by cutaneous fibrofolliculomas, pulmonary cysts, and kidney malignancies. Affected individuals carry germ line mutations in folliculin (FLCN), a tumor suppressor gene that becomes biallelically inactivated in kidney tumors by second-hit mutations. Similar to other factors implicated in kidney cancer, FLCN has been shown to modulate activation of mammalian target of rapamycin (mTOR). However, its precise in vivo function is largely unknown because germ line deletion of Flcn results in early embryonic lethality in animal models. Here, we describe mice deficient in the newly characterized folliculin-interacting protein 1 (Fnip1). In contrast to Flcn, Fnip1−/− mice develop normally, are not susceptible to kidney neoplasia, but display a striking pro-B cell block that is entirely independent of mTOR activity. We show that this developmental arrest results from rapid caspase-induced pre-B cell death, and that a Bcl2 transgene reconstitutes mature B-cell populations, respectively. We also demonstrate that conditional deletion of Flcn recapitulates the pro-B cell arrest of Fnip1−/− mice. Our studies thus demonstrate that the FLCN-FNIP complex deregulated in BHD syndrome is absolutely required for B-cell differentiation, and that it functions through both mTOR-dependent and independent pathways.
Introduction
Human Birt-Hogg-Dubé (BHD) syndrome is an autosomal dominant hereditary cancer syndrome characterized by the development of cutaneous fibrofolliculoma, multiple pulmonary cysts, spontaneous pneumothorax, and increased risk of renal cell neoplasia.1,2 Affected individuals carry germ line mutations in FLCN, a recently characterized gene that encodes a 64-kDa protein, folliculin.2 In kidney tumors, the FLCN wild-type (WT) allele is deleted by somatic mutation or loss of heterozygosity,3-5 supporting the idea that FLCN is a classic tumor suppressor gene that follows the Knudson 2-hit hypothesis.6 FLCN lacks predicted functional domains or homology to known proteins; however, it is also highly conserved across species indicating that it might play an essential role in development.2 In agreement with this notion, homozygous loss of Flcn in mice causes visceral endoderm defects and embryonic lethality at E5.5-E6.5.5
Although its precise in vivo function is unknown, folliculin may regulate cellular energy sensing by controlling mTOR signaling.7-10 For instance, loss of Flcn in mouse kidneys leads to cellular hyperproliferation and overactivation of mTORC1, mTORC2, and MAPK pathways, whereas rapamycin treatment partially rescues this phenotype.5,11-14 In this context, FLCN is not unlike the 6 additional genes that have so far been implicated in kidney cancer (VHL, MET, TSC1, TSC2, FH, and SDH), in that they all interact with metabolic pathways that respond to stress or nutrient stimulation (reviewed by Linehan et al15 ). However, mounting evidence indicates that folliculin may also regulate additional signal transduction pathways independent of mTOR activity. Flcn−/− embryonic stem (ES) cells for instance are resistant to apoptosis as a result of aberrant transforming growth factor (TGF)β signaling.14 In kidney cell lines, FLCN deficiency results in increased activity of TFE3, a member of the MiTF/TFE transcription factor family.16 Furthermore, deletion of the FLCN homolog in fruit flies disrupts male germ line stem cell maintenance, a defect that appears to be linked to the JAK/STAT (Janus kinase/signal transducer and activator of transcription) pathway.17
Two folliculin interacting proteins, FNIP1 and FNIP2, were recently isolated.7-9 These factors are broadly expressed in human tissues and specifically associate with folliculin C-terminal domain in ex vivo studies.7-9 The significance of this interaction is implicit by the fact that most BHD-causing mutations delete the folliculin C-terminal domain.1 Further supporting the link between BHD syndrome and mTOR signaling, the FLCN-FNIP complex has been shown to interact with and be phosphorylated by 5′-adenosine monophosphate (AMP)–activated protein kinase (AMPK), a key enzyme in cellular energy sensing that negatively regulates mTOR activity.11 However, whether FNIP proteins are true functional partners of FLCN in vivo and are thus implicated in BHD syndrome has not been directly established.
Here, we report the analysis of Fnip1 knockout (KO) mice. In contrast to Flcn−/−, we show that Fnip1 germ line targeting does not lead to embryonic lethality or kidney hyperplasia. Unexpectedly, the mice display a marked pro-B cell arrest (the Hardy fraction C-C′), which unlike tumor development in BHD mice, cannot be rescued by rapamycin treatment and is thus mTOR independent. Transcriptome analyses of Fnip1−/− pro-B cells reveal compromised expression of key genes implicated in B-cell surface expression, including VpreB1/VpreB2, λ5, and Rag1/Rag2. We also show that expression of prerecombined heavy and light-chain genes in Fnip1−/− mice helps bypass the pro-B cell arrest to the transitional bone marrow (BM) IgM+ compartment. Strikingly, the reconstituted immature B cells fail to migrate to the periphery. We show that this block is driven by caspase activation and intrinsic cell death, and that expression of a Bcl2 antiapoptotic transgene promotes normal numbers of peripheral B cells in the knockout. Finally, in support of an in vivo functional link between Flcn and Fnip1, we show that tamoxifen-inducible deletion of folliculin in mice recapitulates the Fnip1−/− pro-B cell arrest.
We conclude that (1) the folliculin/Fnip pathway mutated in BHD syndrome is strictly required for BM B-cell survival, (2) folliculin works through both mTOR-dependent (ie, kidney tumor development) and independent (ie, B-cell developmental arrest) mechanisms, and (3) Fnip1−/− mice phenocopy a subset of abnormalities associated with Flcn deletion.
Methods
Gene targeting and drug treatments
Fnip1 gene-trap ES cells (RRM154) were purchased from BayGenomics. In this clone, the gene-trap vector (pGT0Lxf) carrying a splice-acceptor sequence was inserted within intron 2. ES cells were injected into C57BL/6J blastocysts. After germ line transmission mice were backcrossed to the C57BL/6J background for 3 generations. The Fnip1lox allele was introduced by targeting a neomycin resistance cassette (Neor), flanked by Frt and loxP sequences, into Fnip1 intron 5. A second loxP sequence was inserted within intron 6. The targeting vector also carried a thymidine kinase gene for negative selection. ES cells were selected for G418 resistance and gancyclovir sensitivity. Proper targeting was assessed by Southern blotting. Backcrossing of chimeras to C57BL/6J mice produced heterozygous F1 offspring with germ line transmission of the Fnip1 floxed (f) allele. The Neo cassette flanked by Frt sites was excised in vivo by a Flp recombinase transgene expressed under the β-actin promoter.18 To generate a Fnip1 deleted allele (Fnip1+/−), a β-actin-Cre recombinase transgene19 was introduced into FNIP1f/+ mice, followed by backcrossing to the C57BL/6J for Cre transgene removal. The Flcn conditional knockout allele was previously described.11 To conditionally delete the Flcnf allele, a CreER transgene20 was introduced into Flcnf/− mice, and at 6 weeks of age Flcnf/−CreER mice were intraperitoneally injected with tamoxifen dissolved in corn oil at the dosage of 130 mg/kg. For rapamycin treatment, rapamycin (LC Laboratories) was dissolved in 100% ethanol at a stock concentration of 10 mg/mL and kept at −20°C. Rapamycin stock solution was diluted to 200 μg/mL in buffer (5% Tween 80, 5% PEG400) and injected intraperitoneally at a dose of 2 mg/kg daily. Animal care procedures followed National Cancer Institute (NCI)–Frederick Animal Care and Use Committee guidelines.
Flow cytometry
With the exception of IgM-Cy5 (Jackson ImmunoResearch Laboratories), antibodies were from BD Bioscience: B220-PercP-Cy5.5, B220-APC, CD4-PE, CD8-FITC, CD43-FITC, CD43-PE, CD19-APC, CD19-PercP-Cy5.5, CD24-FITC, BP-1-biotin, CD25-APC, CD25-PE, Igκ-PE, CCR9-PE, streptavidin-APC, and FcR. Dead cells were gated using DAPI (Sigma-Aldrich) or TO-PRO-3. Flow cytometers were FACSCalibur (BD Bioscience) and CyanADP (Beckman Coulter), and as cell sorter we used BD Bioscience FACSAriaII. Flow cytometry analysis was done with FlowJo Version 9.4.10 software (TreeStar). Intracellular staining was performed using BD Cytofix/Cytoperm fixation/permeabilization kit. For apoptotic cell detection, Vybrant FAM poly caspase assay kit (Molecular Probes) was used with DAPI according to the manufacturer's protocol.
qRT-PCR
Total RNA was isolated with TRIzol (Invitrogen) and reverse transcribed using Superscript III reverse transcriptase kit (Invitrogen). Quantitative PCR (qPCR) was performed via 7300 Realtime PCR System (Applied Biosystems) using SYBR Green PCR Master Mix (Fermentas). Relative gene expression was measured by normalizing Ct values based on expression of Rplp0 (36B4) gene control. PCR primer sequences are provided as supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
mRNA-Seq
BM cells from FNIP1+/+ and FNIP1−/− mice were stained with CD24(heat shock antigen)–FITC, CD43−PE, B220-PerCP-Cy5.5, and BP-1-APC. Fraction B (B220lowCD43+CD24+BP-1−) and Fraction C-C′ (B220lowCD43+CD24+BP-1+) pro-B cells were sorted and mRNA libraries were generated according to the mRNA TrueSeq sample prep kit (Illumina), sequenced on a HiSeq 2000 (Illumina) and analyzed as previously described.21
Western blotting
Kidney tissues were flash-frozen in liquid nitrogen immediately after dissection. Frozen tissues were homogenized by polytron homogenizer on ice in radio immunoprecipitation assay (RIPA) buffer (20mM tris-HCl, pH 7.5, 150mM NaCl, 1mM ethylenediaminetetraacetic acid, 1.0% Triton X-100, 0.5% deoxycholate, 0.1% sodium dodecylsufate, PhosSTOP [Roche], phosphatase inhibitor cocktail [Roche], and complete protease inhibitor cocktail [Roche]), followed by centrifugation at 13 200g for 30 minutes. Protein concentrations of cleared supernatants were measured with BCA protein assay kit (Pierce) and adjusted to 1.33 mg/mL. Four times SDS sample buffer was then added and samples were boiled for 5′ to produce 1 mg/mL sample lysates. A total of 20 μg of protein was loaded onto 4% to 20% tris-glycin SDS-PAGE gels. Immunoblotting was performed as previously described 11 with minor modifications optimized for Odyssey infrared imaging system (LI-COR). In brief, separated proteins were transferred to Immobilon-FL polyvinylidene difluoride (PVDF) membrane (Millipore), blocked with Odysey blocking buffer (LI-COR) at room temperature for 1 hour. Blocked membranes were incubated overnight at 4°C with primary antibodies, diluted with 0.1% bovine serum albuminTris-buffered saline with Tween 20 (TBST; 20mM Tris-HCl pH8.0, 150mM NaCl, 0.05% Tween 20) as follows: p-mTOR (Ser2448) 1:1000, p-mTOR (Ser2448) 1:1000, mTOR 1:1000, p-S6 ribosomal protein (Ser240/244) 1:1000, p-S6 ribosomal protein (Ser235/236) 1:1000, S6 ribosomal protein 1:1000, and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) 1:1000. All antibodies were purchased from Cell Signaling. After 3 washes with TBST, membranes were incubated at room temperature for 1 hour with infrared dyes conjugated secondary antibody IRDye 800CW goat anti–rabbit IgG (LI-COR), diluted 1:15 000 with 0.2% Tween 20/Odysey blocking Buffer. After 3 additional washes with TBST and brief soaking in phosphate-buffered saline, images were acquired with an Odyssey infrared imaging system (LI-COR).
V(D)J recombination
Sorted C/C′ fraction cells were digested with proteinase K. Digested solution was directly used for PCR after heat inactivation of proteinase K. D-J and V-DJ recombination events were amplified by PCR using primers and conditions as previously described.22 PCR products were separated on agarose gels, transferred to a membrane and analyzed by Southern blot using published oligonucleotide probes.22
Statistical analysis
All the statistical analysis were performed with the Mann Whitney U test or Student t test as appropriate, and represented as mean ± SD. P values were defined as: P < .05; P < .01; NS (nonsignificant) P > .05.
Results
Intrinsic pro-B to pre-B cell block in Fnip1−/− mice
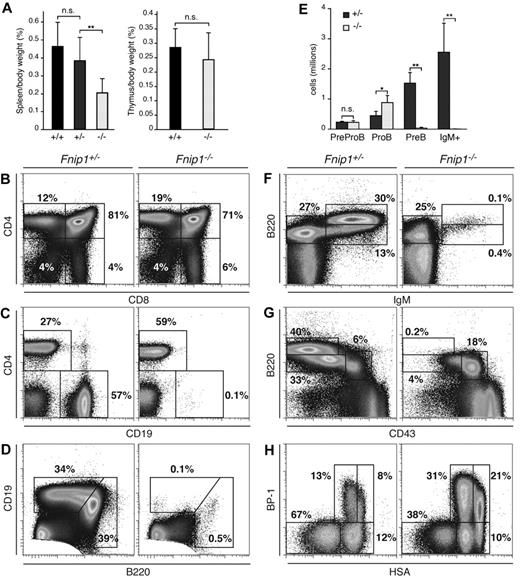
To determine the in vivo function of Fnip1, we targeted the Fnip1 gene in the mouse germ line (Figure 1, Fnip1lox). In parallel, we generated a second Fnip1 knockout line using GeneTrap ES cells (IGTC clone RRM154, Figure 1, Fnip1β-Geo). As no phenotypic differences were observed between the 2 lines, we used them interchangeably throughout the studies and they will be referred to hereafter as Fnip1−/− mice. In contrast to Flcn,5 deletion of Fnip1 did not compromise viability or fertility, as progeny from heterozygous crosses were born in normal Mendelian ratios (not shown). Unexpectedly, gross anatomical inspection showed a significant reduction in the spleen size of Fnip1 KO mice compared with heterozygous or WT controls (P < .01, Figure 2A left graph). To examine whether this feature was the result of defective lymphopoiesis we analyzed BM, thymus, and spleen from all 3 strains by flow cytometry. We found that total thymocyte numbers and peripheral T cells were normal in the absence of Fnip1 (Figure 2B). In agreement with this observation, thymus size was comparable between knockout and littermate controls (Figure 2A right graph). In contrast, Fnip1−/− mice were severely B-cell lymphopenic, in that they displayed a nearly complete lack of conventional splenic CD19+ cells (Figure 2C). Similarly, B220lowCD19high peritoneal B1 cells were depleted in the knockout (Figure 2D). In the BM, B220high mature recirculating, B220lowIgMhigh transitional, and B220lowCD43−CD25+ pre-B cells were also markedly reduced in Fnip1−/− mice (Figure 2E-G and not shown). Conversely, there was a consistent accumulation of B220lowCD43+CD25−pro-B cells in mutant mice, as their absolute number was 2- to 3-fold increased relative to control (P < .05, Figure 2E-G).
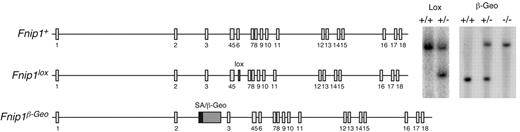
Generation of Fnip1 KO mice. Left schematics show Fnip1 WT (Fnip1+), targeted (Fnip1lox), and gene-trapped (Fnip1β-Geo) alleles. The Fnip1lox mutation was introduced in the germ line by targeting a neomycin resistance cassette (flanked by LoxP sites) within intron 5, followed by a second LoxP site within intron 6. The Neo-exon 6 sequence was removed by crossing targeted mice with β-actin-Cre transgenic mice.19 Exon 6 deletion generates a premature termination codon in exon 7. Fnip1β-Geo mice were derived from GeneTrap ES cell clone RRM154, which carries a splice acceptor (SA) β-Geo cassette inserted within intron 2. Proper gene targeting in both cases was confirmed by Southern blotting (right gel pictures).
Generation of Fnip1 KO mice. Left schematics show Fnip1 WT (Fnip1+), targeted (Fnip1lox), and gene-trapped (Fnip1β-Geo) alleles. The Fnip1lox mutation was introduced in the germ line by targeting a neomycin resistance cassette (flanked by LoxP sites) within intron 5, followed by a second LoxP site within intron 6. The Neo-exon 6 sequence was removed by crossing targeted mice with β-actin-Cre transgenic mice.19 Exon 6 deletion generates a premature termination codon in exon 7. Fnip1β-Geo mice were derived from GeneTrap ES cell clone RRM154, which carries a splice acceptor (SA) β-Geo cassette inserted within intron 2. Proper gene targeting in both cases was confirmed by Southern blotting (right gel pictures).
Severe pro-B cell arrest in Fnip1−/− mice. (A) Left bar graph: spleen weight (given as percentage of body weight) in Fnip1−/− mice relative to littermate controls graphed as the mean ± SD; NS = no statistical significance, **P < .01 (Student t test), n = 9 (Fnip1+/+), 17 (Fnip1+/−), and 13 (Fnip1−/−). Right graph shows the same for thymuses. n = 6 (Fnip1+/+), and 8 (Fnip1−/−). (B-D) Fluorocytometric analysis of CD4+ and CD8+ thymocytes (B), CD4+ and CD19+ splenocytes (C), and B1 (B220highCD19low) and B2 (B220lowCD19high) peritoneal cavity B cells (D). (E) Absolute number of pre–pro-B (B220+IgM−CD19−), pro-B (B220+IgM−CD19+CD43+CD25−), pre-B (B220+IgM−CD19+CD43−CD25+), and immature (B220+IgM+) B cells in BM of Fnip1+/− and Fnip1−/−mice. *P < .05, **P < .01 (Student t test), n = 4 for both strains. (F-H) Fluorocytometric analysis of B220 and IgM (F), B220 and CD43 (G), and BP-1 and HSA (H) expression in BM cells from Fnip1+/−and Fnip1−/−mice. Each analysis is representative of at least 3 independent experiments.
Severe pro-B cell arrest in Fnip1−/− mice. (A) Left bar graph: spleen weight (given as percentage of body weight) in Fnip1−/− mice relative to littermate controls graphed as the mean ± SD; NS = no statistical significance, **P < .01 (Student t test), n = 9 (Fnip1+/+), 17 (Fnip1+/−), and 13 (Fnip1−/−). Right graph shows the same for thymuses. n = 6 (Fnip1+/+), and 8 (Fnip1−/−). (B-D) Fluorocytometric analysis of CD4+ and CD8+ thymocytes (B), CD4+ and CD19+ splenocytes (C), and B1 (B220highCD19low) and B2 (B220lowCD19high) peritoneal cavity B cells (D). (E) Absolute number of pre–pro-B (B220+IgM−CD19−), pro-B (B220+IgM−CD19+CD43+CD25−), pre-B (B220+IgM−CD19+CD43−CD25+), and immature (B220+IgM+) B cells in BM of Fnip1+/− and Fnip1−/−mice. *P < .05, **P < .01 (Student t test), n = 4 for both strains. (F-H) Fluorocytometric analysis of B220 and IgM (F), B220 and CD43 (G), and BP-1 and HSA (H) expression in BM cells from Fnip1+/−and Fnip1−/−mice. Each analysis is representative of at least 3 independent experiments.
To precisely define the B-cell population(s) affected by the absence of Fnip1, we fractionated B220lowCD43+ pro-B cells into the Hardy A, B, C, and C′ subsets based on differential cell-surface expression of HSA and BP-1.23 We found that relative to control, there was a clear accumulation of pro-B cell fractions C and C′ in Fnip1-deficient mice (Figure 2H). Outside the B-cell compartment, BM monocyte (Mac-1+Gr-1−), granulocyte (Mac-1+Gr-1+), and erythrocyte (CD71+Ter119+) lineages appeared normal (not shown). On the basis of these findings we conclude that germ line deletion of Fnip1 leads to a specific developmental block at pro-B cell fractions C-C′.
To explore whether the Fnip1−/− B-cell lymphopenia was the result of cell autonomous or environmental abnormalities we performed BM transplantation experiments. We found that whereas Fnip1+/− donor BM cells reconstituted the B-cell compartment of irradiated Ly5.2 congenic mice, transplanted Fnip1−/− cells recapitulated the same pro-B cell developmental arrest observed in Fnip1−/− mice. Conversely, T cells and other hematopoietic lineages were equally reconstituted in the 2 strains (not shown). We conclude that the B lymphocyte defect of Fnip1 deficient mice is cell intrinsic.
Fnip1−/− mice recapitulate some but not all Flcn−/− defects
We sought to ascertain whether folliculin deficiency was also associated with a pro-B cell defect. Because Flcn germ line deletion is embryonic lethal,5 we generated Flcnflox/− mice expressing Cre-ER fusion recombinase under the pCAGGS promoter (CreER).20 Six-week-old mice were injected with 1 dose of tamoxifen (130 mg/kg) and BM, spleen, and thymuses were analyzed by flow cytometry 4 or 8 weeks after treatment. Consistent with the Fnip1−/− phenotype, Flcnflox/−CreER mice showed few CD19+ B cells in the spleen and displayed a pro-B cell block in the BM (Figure 3A and not shown). Thus, conditional Flcn deletion phenocopies the B-cell developmental defect observed in Fnip1−/− mice.
Comparison of Fnip1−/− and tamoxifen-induced Flcn deficient mice. (A) BM B-cell analysis in Flcnf/+ERCre+ and Flcnf/−ERCre+ mice 4 weeks posttamoxifen (TM) injection. Cell samples were stained with CD43 (FITC) and B220 (PE). (B) Flcnf/−ERCre+ mice treated with tamoxifen (right micrograph) show hyperplastic cystic lesions (denoted with arrowheads) in kidney. Conversely, Fnip−/− mice rarely display cysts (left micrograph). Scale bar = 200 μm. (C) Western blot analysis of kidney cell lysates from Fnip1+/−, Fnip1−/−, and Flcn−/− mice. mTOR activation was assessed via phosphorylation of mTOR serines 2481 and 2448, as well as phosphorylation of ribosomal protein S6, serines 240/244 and 235/236. GAPDH expression was used as a loading control. (D) Failure to rescue peripheral CD19+ B lymphocytes in Fnip1−/− mice treated with buffer or rapamycin (2 mg/kg) for 3 weeks. Each fluorescence-activated cell sorter (FACS) analysis is representative of at least 3 independent experiments.
Comparison of Fnip1−/− and tamoxifen-induced Flcn deficient mice. (A) BM B-cell analysis in Flcnf/+ERCre+ and Flcnf/−ERCre+ mice 4 weeks posttamoxifen (TM) injection. Cell samples were stained with CD43 (FITC) and B220 (PE). (B) Flcnf/−ERCre+ mice treated with tamoxifen (right micrograph) show hyperplastic cystic lesions (denoted with arrowheads) in kidney. Conversely, Fnip−/− mice rarely display cysts (left micrograph). Scale bar = 200 μm. (C) Western blot analysis of kidney cell lysates from Fnip1+/−, Fnip1−/−, and Flcn−/− mice. mTOR activation was assessed via phosphorylation of mTOR serines 2481 and 2448, as well as phosphorylation of ribosomal protein S6, serines 240/244 and 235/236. GAPDH expression was used as a loading control. (D) Failure to rescue peripheral CD19+ B lymphocytes in Fnip1−/− mice treated with buffer or rapamycin (2 mg/kg) for 3 weeks. Each fluorescence-activated cell sorter (FACS) analysis is representative of at least 3 independent experiments.
Analogous to other factors that suppress kidney cancer, folliculin interacts with the energy and nutrient sensing AMPK-mTOR signaling pathway,5 the target of rapamycin. In support of this, Flcn deletion in kidneys results in mTOR overactivation (hyperphosphorylation), uncontrolled kidney cell proliferation, polycystic kidneys, and death from renal failure by 3 weeks of age.11 Rapamycin treatment on the other hand suppresses uncontrolled kidney cell proliferation and significantly extends median survival.11 In contrast to Flcn−/−, we detected few or no cysts in Fnip1−/− kidneys (Figure 3B), and there was no renal failure in the absence of Fnip1 (not shown), implying that mTOR activity was unaffected in the absence of Fnip1. To directly assess mTOR activation in Fnip1−/− mice we measured phosphorylation of both mTOR (Ser2448 and Ser2481) and one of its downstream targets, the S6 subunit of the 40S ribosomal complex (Ser235/236 and Ser240/244). As previously shown,5,7 both proteins were hyperphosphorylated in Flcn deleted kidney cells (Figure 3C). Conversely, the levels of mTOR and S6 phosphorylation in kidney or pro-B cells were comparable between Fnip1−/− and littermate controls (Figure 3C and not shown), demonstrating that mTOR signaling is not compromised by Fnip1 depletion. To confirm this result, Fnip1−/− newborn mice were injected intraperitoneally with rapamycin (2 mg/kg) or buffer control on a daily basis for a total of 3 weeks. Although rapamycin treatment clearly reverted the kidney phenotype of Flcn−/− mice11 and decreased mTOR phosphorylation in Fnip1−/− cells (supplemental Figure 1), it could not bypass the B-cell lymphopenia of Fnip1 deficient mice (Figure 3D). Thus the Fnip1−/− pro-B cell block is independent of mTOR signaling. Taken together the observations demonstrate that Fnip1−/− mice recapitulate some but not all developmental defects associated with Flcn deletion.
A pre-recombined B-cell receptor does not reconstitute Fnip1−/− B cells
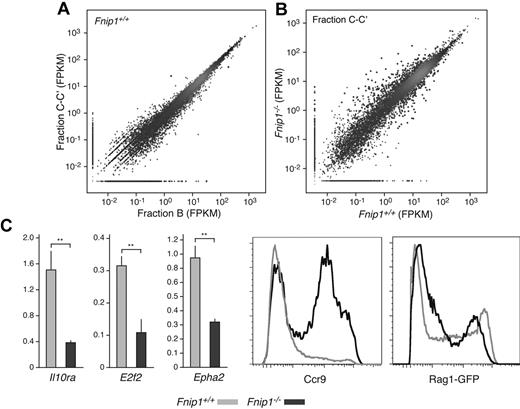
To clarify the nature of the Fnip1−/− B-cell lymphopenia, we sought to compare transcription profiles of WT and mutant pro-B cells. To this end pro-B cell fractions B and C-C′ were sorted to ∼ 95% purity and from each fraction, 2 biologic replicate transcriptomes were obtained by deep-sequencing.21 Interestingly, with few exceptions the transcriptomes of B and C-C′ fractions were nearly identical both in WT and KO mice (Figure 4A and not shown). In contrast, Fnip1+/+ and mutant cells showed significant differences in both fractions, with at least 400 transcripts displaying greater than 2 fold change between the 2 cell types (false discovery rate = 1%, Figure 4B and supplemental Table 1). Among these we found key genes implicated in B-lymphocyte development, including Cd22, Cd5, Prkcq, Il10rα, E2f2, Epha2, Ccr9, Vpreb1, Vpreb2, λ5, and Rag1/Rag2. Figure 4C shows qPCR and flow cytometric results that validate defective expression of a subset of these genes in Fnip1−/− pro-B cells.
Transcriptome analysis of Fnip1−/− pro-B cells. (A) Scatter plot showing a comparative analysis between Fnip1+/+ transcriptomes (mRNA-Seq FPKM values) obtained from the Hardy pro-B cell fractions B and C-C′. Red dots represent genes whose transcription is statistically different between the 2 fractions; n = 1 (using pooled estimates of dispersion). (B) Same analysis as in panel A but comparing fractions C-C′ between Fnip1+/+ and Fnip1−/− mice; n = 2 for each genotype. (C) Bar graphs showing RT-qPCR measurements of transcription in Fnip1+/+ and Fnip1−/− pro-B cells from selected genes (n = 3). Histograms showing ccr9 and Rag1-GFP expression, as determined by flow cytometry, on B220+IgM−CD25− pro-B cells from KO and control mice. Each FACS analysis is representative of at least 3 independent experiments.
Transcriptome analysis of Fnip1−/− pro-B cells. (A) Scatter plot showing a comparative analysis between Fnip1+/+ transcriptomes (mRNA-Seq FPKM values) obtained from the Hardy pro-B cell fractions B and C-C′. Red dots represent genes whose transcription is statistically different between the 2 fractions; n = 1 (using pooled estimates of dispersion). (B) Same analysis as in panel A but comparing fractions C-C′ between Fnip1+/+ and Fnip1−/− mice; n = 2 for each genotype. (C) Bar graphs showing RT-qPCR measurements of transcription in Fnip1+/+ and Fnip1−/− pro-B cells from selected genes (n = 3). Histograms showing ccr9 and Rag1-GFP expression, as determined by flow cytometry, on B220+IgM−CD25− pro-B cells from KO and control mice. Each FACS analysis is representative of at least 3 independent experiments.
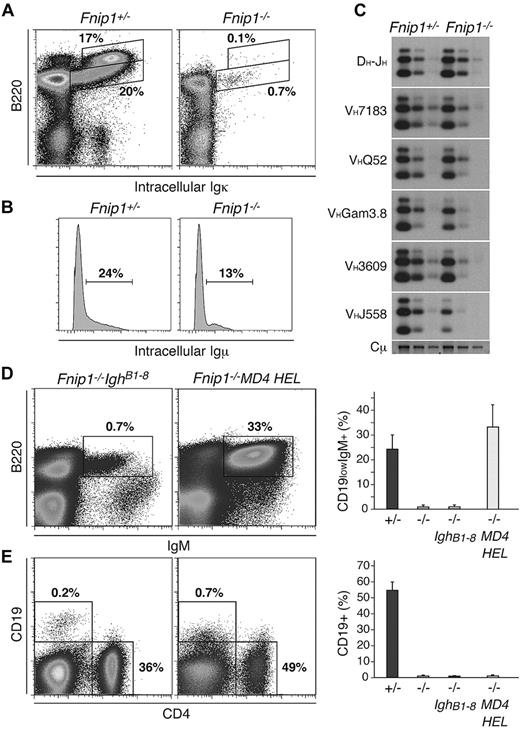
Because Vpreb, λ5 and Rag proteins control recombination and expression of the B-cell surface receptor we assessed intracellular Igκ and Igμ expression by flow cytometry. As expected based on the reduced number of pre-B cells, there were but background levels (< 1%) of intracellular Igκ in Fnip1−/− BM B220low cells (Figure 5A). However, in spite of the pro-B cell accumulation, we also detected lower levels of intracellular Igμ within B220lowCD43+ cells in KO mice compared with controls (13% vs 24%, Figure 5B). To investigate whether V(D)J recombination is compromised in Fnip1−/− progenitor cells we cell sorted fraction C/C′ lymphocytes from KO and control mice and amplified D-J and V-DJ recombination products by PCR as described.22 We found no significant differences in the levels of D-J recombination between the 2 strains (Figure 5C). V-DJ recombination products were also amplified from Fnip1−/− pro-B cells, although less efficiently for distal recombination events, such as VH3609 and VHJ558 (Figure 5C), a result perhaps explained by the reduced Rag expression in knockout cells. The data therefore show no overt defects in Rag-mediated recombination in Fnip1−/− BM B cells. Likewise, arguing against the idea that KO mice lack a pre-B cell surface receptor, flow cytometry showed λ5 staining in Fnip1−/− pro-B cells (supplemental Figure 2). Consistent with the transcriptome results, λ5+ pro-B cell numbers were modestly decreased in the knockout (supplemental Figure 2).
Prerecombined antibody genes do not rescue Fnip1−/− B cells. (A) Igκ staining in Fnip1+/− and Fnip1−/− BM B220+ B cells. (B) Histogram of intracellular Igμ expression in B220+CD43+ pro-B cells from Fnip1+/− and Fnip1−/− mice. (C) D-J and V-DJ recombination analysis in WT and KO fraction C/C′ B cells as determined by PCR-Southern blot. (D) B220/IgM expression profiles in Fnip1−/− mice expressing B1-8 heavy chain (IghB18)24 or the αHEL heavy and light chain transgene (MD4 HEL).25 Right bar graph represents the mean ± SD of IgM+B220+ B cells from each strain; n values were as follows: 6 for Fnip1+/− and Fnip1−/−, and 3 for Fnip1−/−IghB1-8 and Fnip1−/−MD4 HEL. (E) Same analysis as in panel D but using CD19 and CD4 staining of splenocytes; n values: 5 mice for Fnip1+/− and Fnip1−/−, and 4 mice for Fnip1−/−IghB1-8 and Fnip1−/−MD4 HEL. Each FACS analysis is representative of at least 3 independent experiments.
Prerecombined antibody genes do not rescue Fnip1−/− B cells. (A) Igκ staining in Fnip1+/− and Fnip1−/− BM B220+ B cells. (B) Histogram of intracellular Igμ expression in B220+CD43+ pro-B cells from Fnip1+/− and Fnip1−/− mice. (C) D-J and V-DJ recombination analysis in WT and KO fraction C/C′ B cells as determined by PCR-Southern blot. (D) B220/IgM expression profiles in Fnip1−/− mice expressing B1-8 heavy chain (IghB18)24 or the αHEL heavy and light chain transgene (MD4 HEL).25 Right bar graph represents the mean ± SD of IgM+B220+ B cells from each strain; n values were as follows: 6 for Fnip1+/− and Fnip1−/−, and 3 for Fnip1−/−IghB1-8 and Fnip1−/−MD4 HEL. (E) Same analysis as in panel D but using CD19 and CD4 staining of splenocytes; n values: 5 mice for Fnip1+/− and Fnip1−/−, and 4 mice for Fnip1−/−IghB1-8 and Fnip1−/−MD4 HEL. Each FACS analysis is representative of at least 3 independent experiments.
To formally exclude the possibility that defects in B-cell receptor assembly or expression might explain the Fnip1−/− B-cell phenotype, we expressed a pre-recombined heavy chain knock-in (IghB1-8,24 ), or heavy and light-chain transgenes (MD4 HEL,25 ) in Fnip1−/− mice. We found that preassembled heavy and light-chain genes (but not heavy-chain only) resulted in a substantial number of BM immature B220lowIgM+ cells in the knockout (Figure 5C middle panel and bar graph). Strikingly, in neither case did we observe peripheral splenic B2 or peritoneal B1 cells after reconstitution (Figure 5D and not shown). The findings are thus consistent with the notion that the Fnip1−/− BM B-cell block is not caused by defects in V(D)J recombination or cell-surface expression of a functional B-cell receptor.
Fnip1−/− pre-B cells undergo rapid apoptosis
We entertained the possibility that lack of peripheral B cells in Fnip1−/− mice might result from BM precursor cell death. Therefore we measured cell death in pro-B and pre-B cells from KO and WT controls with the fluorescent DNA stain DAPI. We found increased numbers of DAPI+ pro-B cells in the KO but this was not statistically significant (P = .1, Figure 6A). Conversely, there were on average 10 times more DAPI+ pre-B cells in Fnip1−/− relative to littermates (P < .02, Figure 6A). In addition, as determined by affinity labeling assays, deficient pre-B cells displayed enhanced caspase activity relative to Fnip1 proficient cells (Figure 6B). The results suggest that Fnip1 modulates BM B-cell survival. To confirm this idea we explored whether ectopic overexpression of antiapoptotic Bcl2 in the B-cell compartment (Eμ-Bcl2)26 was sufficient to rescue the developmental defect associated with Fnip1 deficiency. We found that Bcl-2 expression led to increased numbers of B220+IgM+ B cells in the BM (Figure 6C). Importantly, both splenic CD19+ and peritoneal B1 cell populations were reconstituted in Eμ-Bcl2 Fnip1−/− mice (Figure 6D and not shown). This was in direct contrast to the lack of peripheral B cells observed in MD4 Fnip1−/− mice (Figure 5E). We conclude that deletion of Fnip1 leads to caspase activation and apoptosis of precursor BM B cells, a feature that can be rescued by antiapoptotic Bcl2 expression.
Increased caspase activation and apoptosis in Fnip1−/− B cells. (A) Bar graph showing the frequency of dead cells within pro-B (B220lowIgM−CD25−) and pre-B (B220lowIgM−CD25+) populations from Fnip1+/+ and Fnip1−/− mice as measured by DAPI staining. Mean ± SD from 4 independent experiments, (pro-B) P = .1, (pre-B) P < .02, Student t test. (B) Flow cytometric measurement of caspase activity in Fnip1+/+ and Fnip1−/− pre-B cells from interleukin 7 ex vivo cultures. Twenty-four hours after interleukin 7 removal cells were stained with DAPI and FAM, a carboxyfluorescein moiety that becomes covalently linked to the active site of caspases. (C) Flow cytometric analysis of BM and (D) spleen cells from Fnip1−/− mice reconstituted with an Eμ-Bcl2 transgene. BM cells were stained with anti-B220 and anti-IgM antibodies; staining for splenocytes was with anti-CD4 and anti-CD19. Each FACS analysis is representative of 3 independent experiments.
Increased caspase activation and apoptosis in Fnip1−/− B cells. (A) Bar graph showing the frequency of dead cells within pro-B (B220lowIgM−CD25−) and pre-B (B220lowIgM−CD25+) populations from Fnip1+/+ and Fnip1−/− mice as measured by DAPI staining. Mean ± SD from 4 independent experiments, (pro-B) P = .1, (pre-B) P < .02, Student t test. (B) Flow cytometric measurement of caspase activity in Fnip1+/+ and Fnip1−/− pre-B cells from interleukin 7 ex vivo cultures. Twenty-four hours after interleukin 7 removal cells were stained with DAPI and FAM, a carboxyfluorescein moiety that becomes covalently linked to the active site of caspases. (C) Flow cytometric analysis of BM and (D) spleen cells from Fnip1−/− mice reconstituted with an Eμ-Bcl2 transgene. BM cells were stained with anti-B220 and anti-IgM antibodies; staining for splenocytes was with anti-CD4 and anti-CD19. Each FACS analysis is representative of 3 independent experiments.
Discussion
FNIP1 and FNIP2 were originally isolated as folliculin interacting proteins in human cells.7-9 Although lacking known functional domains, FNIP proteins were proposed to play a role in cellular energy sensing based on the observation that they associate with AMPK, a negative regulator of the mTOR pathway. Consistent with this view, homozygous loss of FLCN in human or mouse kidneys promotes overactivation of mTORC1 and mTORC2 leading to cell hyperproliferation and tumor development.15 Furthermore, rapamycin treatment of kidney-targeted Flcn deficient mice partially rescues this phenotype.11 However, whether folliculin works primarily through one or both FNIP proteins has been unclear. Our findings address this issue by comparing the phenotype of conditional Flcn to germ line deleted Fnip1 mice. In both models we find that B-cell development past the pro-B cell stage is defective, indicating that Flcn and Fnip1 function cooperatively at least within the context of B-cell ontogeny. On the other hand, Fnip1−/− mice fail to phenocopy essential features of BHD syndrome, most particularly mTOR overactivation and uncontrolled kidney cell proliferation. Also contrary to results with Flcn−/− mice, Fnip1 deletion does not lead to early embryonic lethality. These data suggest that, at least for activities related to kidney and embryonic development, Fnip2 plays an important role. A second possibility is that Fnip proteins are functionally redundant in this regard. In favor of the latter hypothesis, FNIP1 and FNIP2 are highly homologous, displaying ∼ 75% sequence similarity.8,9 In addition, none of the BHD-associated mutations described so far targets FNIP1 or FNIP2 genes. Instead, most mutations disrupt folliculin's capacity to associate both with FNIP1 and FNIP2.7-9 A formal test of these 2 scenarios awaits germ line deletion of Fnip2 as well as a combined deletion of Fnip1 and Fnip2 in the mouse.
Another novel aspect of our studies concerns the observation that, independent of mTOR signaling, Fnip1 and folliculin control BM B-cell survival and pro-B-cell differentiation. This result was unexpected because no B cell defects have been reported so far in conjunction with BHD syndrome. At the same time, FLCN mutations are monoallelic in nature, and thus, analogous to Flcn+/− mice, BHD patients display normal B-cell ontogeny. In light of this discussion on Fnip1-Fnip2 functional redundancy, our findings reveal for the first time Folliculin-Fnip1 activities within the hematopoietic system that are independent of Fnip2. Whether Fnip2 can also function independently of Fnip1 remains to be determined, but based on these findings we propose that different follicullin-Fnip complexes function at multiple levels during mammalian development and kidney cancer, and that these complexes may or may not regulate the mTOR pathway.
We have shown that the Fnip1−/− pro-B cell block is not caused by defects in V(D)J recombination or failure to express a functional B-cell surface receptor in knockout cells. Particularly, enforced expression of a pre-rearranged antibody receptor only rescued development to the transitional BM B-cell stage. On the other hand, prompted by the overactivation of caspases and enhanced apoptosis observed in Fnip1−/− pre-B cells, we achieved full peripheral reconstitution on ectopic expression of Bcl2 in the B compartment, demonstrating that absence of Flcn or Fnip primarily compromises B-cell viability at a very specific stage. Future experiments will address the precise nature of this defect, which in turn might provide us with a better understanding of the tumor suppressor function of FLCN.
In conclusion, our studies demonstrate that folliculin works through both mTOR-dependent and independent pathways, and that it controls B lymphopoiesis together with its interacting partner Fnip1.
All sequence data are available at the NCBI GEO database under accession no. GSE38741.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank members of the Casellas and Linehan laboratories for helpful discussions; G. Gutierrez from National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) genomics facility for technical assistance; James M. Simone and Jeffrey Lay for cell sorting; and Louise Cromwell for technical support with the mouse studies.
This work was supported in part by the Intramural Research Program of NIAMS and NCI, Center for Cancer Research, National Institutes of Health (NIH). This project was funded in part with federal funds from the NCI, NIH, under contract no. HHSN261200800001E.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. NCI-Frederick is accredited by AAALAC International and follows the Public Health Service Policy for the Care and Use of Laboratory Animals. Animal care was provided in accordance with the procedures outlined in the “Guide for Care and Use of Laboratory Animals” (National Research Council; 1996; National Academy Press; Washington, DC). None of the authors of this paper have a financial interest related to this work. This study made use of the high-performance computational capabilities of the Biowulf Linux cluster at the NIH (http://biowulf.nih.gov).
National Institutes of Health
Authorship
Contribution: M.B., J.R.K., L.S.S., W.M.L., and R.C. designed the research; M.B., S.K., H.C.S., R.M.H., M.E.K., H.F.O., and L.S.S. performed research; M.B., H.H., Y.H., C.G.G., and P.B. contributed vital new reagents or analytical tools; M.B., J.R.K., H.-W.S., W.R., K.-R.K.-K., W.M.L., and R.C. analyzed and interpreted data; M.B., E.S., and L.T. contributed Fnip1−/− mice generation; and M.B., W.M.L., and R.C. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Rafael Casellas, NIAMS, NIH, 10 Center Dr, MSC 1930, 10/13C103-D, Bethesda, MD 20892; e-mail: rafael.casellas@nih.gov; or W. Marston Linehan, NCI, NIH, 10 Center Dr, MSC 1107, 10 CRC 1-5940W, Bethesda, MD 20892; e-mail: wml@nih.gov.
References
Author notes
W.M.L. and R.C. contributed equally to this work.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal