Abstract
Dysregulation of cyclin-dependent kinase 4 (CDK4) and CDK6 by gain of function or loss of inhibition is common in human cancer, including multiple myeloma, but success in targeting CDK with broad-spectrum inhibitors has been modest. By selective and reversible inhibition of CDK4/CDK6, we have developed a strategy to both inhibit proliferation and enhance cytotoxic killing of cancer cells. We show that induction of prolonged early-G1 arrest (pG1) by CDK4/CDK6 inhibition halts gene expression in early-G1 and prevents expression of genes programmed for other cell-cycle phases. Removal of the early-G1 block leads to S-phase synchronization (pG1-S) but fails to completely restore scheduled gene expression. Consequently, the IRF4 protein required to protect myeloma cells from apoptosis is markedly reduced in pG1 and further in pG1-S in response to cytotoxic agents, such as the proteasome inhibitor bortezomib. The coordinated loss of IRF4 and gain of Bim sensitize myeloma tumor cells to bortezomib-induced apoptosis in pG1 in the absence of Noxa and more profoundly in pG1-S in cooperation with Noxa in vitro. Induction of pG1 and pG1-S by reversible CDK4/CDK6 inhibition further augments tumor-specific bortezomib killing in myeloma xenografts. Reversible inhibition of CDK4/CDK6 in sequential combination therapy thus represents a novel mechanism-based cancer therapy.
Introduction
Cyclin-dependent kinase 4 (CDK4) and CDK6 associate with D-type cyclins to promote cell-cycle entry and progression through G1 by inactivating the retinoblastoma protein Rb in opposition to the INK4 family of CDK inhibitors (CKIs). They also facilitate progression to S phase by titrating the Cip/Kip CKIs that inhibit CDK2/cyclin E and CDK2/cyclin A.1 Although germline mutations of CDK4 or CDK6 in human cancer are rare, dysregulation of CDK4 and CDK6 by gain of function or loss of inhibition is common.2 For example, amplification of CDK4 and deletion of CDKN2A and CDKN2B of the INK4 family are among the most frequent genomic aberrations in human metastatic lung adenocarcinoma.3 CDKN2C (p18INK4c),4,5 which inhibits pituitary adenoma development in mice,6 also suppresses glioblastoma growth in humans.7 In preclinical models, p18INK4c is essential for restraining proliferation of luminal mammary progenitor cells and tumorigenesis,8 consistent with the requirement for Cdk4 in inducing mammary and breast tumors.9,10 Collectively, these findings suggest that halting unscheduled cell-cycle progression by inhibition of CDK4/CDK6 may significantly improve cancer therapy.
Multiple lines of evidence further suggest that dysregulation of CDK4/CDK6 is pivotal for the loss of cell-cycle control in multiple myeloma (MM), which remains incurable. In MM, malignant plasmacytoid cells retain the self-renewing potential as opposed to normal plasma cells, which are permanently arrested in early G1 because of inhibition of CDK4 and CDK6 by p18INK4c.11,12 During the stable phase of the disease, myeloma cells accumulate in the BM mainly because of impaired apoptosis. However, they inevitably reenter the cell cycle and proliferate without restraint in relapse.13 Although the genetic basis for cell-cycle dysregulation in MM is unknown, deletion and inactivation of CDKN2C and other INK4 CKIs have been noted.14-16 Cyclin D1 is aberrantly expressed at a significant frequency in MM because of t (11:14) chromosomal translocation,17,18 but this alone is insufficient to drive the cell cycle.19 Instead, proliferation of BM myeloma cells is preceded by coordinated overexpression of CDK4/cyclin D1, or CDK6/CDK4 together with cyclin D2, that is specific for each case of MM.19 Deletion of CDKN2C or overexpression of CDK6 in MM further correlates with unfavorable overall survival.15 CDK4 and CDK6 thus appear to be promising targets for cell-cycle control of MM.
Success in targeting the cell cycle in cancer with broad-spectrum CDK inhibitors has been modest, mainly because of a lack of selectivity and high toxicity.20 However, PD 0332991, a cell-permeable pyridopyrimidine with oral bioavailability,21 has shown significant promise. PD 0332991 is the only known selective and potent inhibitor for CDK4 and CDK6. Unlike other CDK inhibitors, at concentrations specific for inhibition of CDK4/CDK6 (< 5μM), PD 0332991 has little or no activity against at least 38 additional kinases,21 including CDK2 because of induction of sterical clash in the hinge region.22 Providing the first evidence for its bioactivity in primary cancer cells, PD 0332991 rapidly inhibits CDK4 and CDK6 (IC50, 60nM) and induces early G1 arrest in primary human myeloma cells in the presence of BM stromal cells (BMSCs) ex vivo.23 It is similarly effective in other cancers in vitro, including mantle cell lymphoma (MCL), acute myeloid leukemia cells, and breast cancer cells.24-26 In vivo, it suppresses tumor development in xenografts.21,23,25
However, PD 0332991 acts reversibly.21 Tumor growth resumed on discontinuation of PD 0332991.23 These findings highlight the promise of PD 0332991 as a selective and effective CDK inhibitor and the importance of targeting CDK4/CDK6 in combination therapy. Consistent with this possibility, in a pilot study PD 0332991 cooperated with bortezomib in prolonging the survival of mice developing tumors in the immune-competent 5T myeloma model.27
PD 0332991 has now shown promise in the first single-agent phase 1 clinical study of MCL28 and is being actively investigated in MM as well as many other human cancers (www.clinicaltrials.gov). Elucidating the mechanism by which CDK4/CDK6 inhibition sensitizes tumor cells to cytotoxic killing is thus timely and critical for targeting the cell cycle in human cancer.
Taking advantage of the exceptional specificity and reversibility of PD 0332991, we have developed a novel strategy to both inhibit proliferation and enhance cytotoxic killing of myeloma cells in vitro and in vivo. Targeting CDK4/CDK6 in sequential combination with cytotoxic agents thus represents a novel mechanism-based cancer therapy.
Methods
Isolation of primary BM myeloma cells and cell culture
BM specimens were obtained from multiple myeloma patients at the New York–Presbyterian Hospital under informed consent as part of an Institutional Review Board-approved study in accordance with the Declaration of Helsinki. Primary CD138+ human BM myeloma cells were isolated and cultured as previously described.23 Human myeloma cell lines (HMCLs) MM1.S and MM1.R were obtained from Dr N. Krett (Northwestern University, Chicago, IL) and CAG from Dr J. Epstein (University of Arkansas, Little Rock, AK). HMCLs U266, RPMI8226, and 2132, and Rb-negative MCL cell line REC-1 were purchased from ATCC, and LP-1 from DSMZ. KMS12PE has been described previously.29 Cells were treated with PD 0332991 (Pfizer) at 0.25μM unless otherwise indicated, bortezomib (ChemieTek) or dexamethasone (Sigma-Aldrich) at concentrations and for time indicated. The pan-caspase inhibitor Q-VD-OPh (R&D Systems) was used at 20μM.
BrdU uptake and DNA content analysis
5-Bromo-2-deoxyuridine (BrdU, 5 μg/mL; Sigma-Aldrich) was added to HMCL cells and CD138+ BM myeloma cells for the time indicated. BrdU uptake and DNA content per cell were determined as described.30
Cell death assays
Apoptotic and dead cells were analyzed by staining the cells with MitoProbe JC-1 (66nM) or MitoTracker Red CMXRos (33nM; Invitrogen) to detect mitochondrial outer membrane permeabilization, or using a To-Pro-3 (Invitrogen) or annexin V-FITC Apoptosis Detection Kit (Calbiochem) according to the manufacturer's instructions. Live cells were determined by trypan blue exclusion staining in triplicate.
RNA microarray
Total RNA was isolated using an RNeasy Plus Mini Kit (QIAGEN). RNA (1 μg) was amplified, labeled with Cy3, and hybridized to Agilent 4 × 44K Whole Human Genome Microarray (Agilent Technologies) according to the manufacturer's protocols. The microarray was scanned to obtain fluorescent signal using Agilent Feature Extraction Version 9.1 software (Agilent Technologies). Data were further analyzed with GeneSpring GX11.0 (Agilent Technologies). Microarray data were deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under the accession number GSE35728.
shRNA knockdown
To knockdown CDK4 or CDK6, cells were infected with CDK4 TRCN0000000364(364), 365 or 520, or CDK6 TRCN0000039743(743), 744, or 746 MISSION shRNA lentiviral transduction particles (Sigma-Aldrich), using the MISSION nontarget (n-t) shRNA lentiviral transduction particles as a control. To knockdown IRF4, Bim, or Noxa, the pKLO.1 vector carrying IRF4, BCL2L11, PMAIP1, or the GFP437 control shRNA (RNAi Consortium at Broad Institute, Cambridge, MA) was cotransfected with pCMV-dR8.74psPAX2 and pMD2.G plasmids (Addgene) into 293T cells to generate the desired shRNA lentivirus. Of the effective shRNA lentiviruses generated, IRF4 [TRCN0000014766 (766) and 767)], BCL2L11 (TRCN0000001053), PMAIP1 shRNA [TRCN0000150555 (555) and TRCN0000151311 (311)], and GFP-437 control shRNA lentiviruses were chosen for this study. Knockdown of each target was validated by quantitative RT-PCR and immunoblotting at 60 to 72 hours after transduction.
Ectopic expression of human IRF4
To express human IRF4, the IRF4 inducible-retroviruses were produced by cotransfecting 293T cells with the ToIRF4 plasmid and the ecotropic envelope and gag-pol plasmids. The control retrovirus was generated using the Vxy vector. Infection of KMS12PE cells with viral supernatants and doxycycline induction were performed as described.29
Quantitative RT-PCR
Total RNA was isolated using the TRIzol reagent (Invitrogen). The first-strand cDNA was synthesized using SuperScript III (Invitrogen) and subjected to real-time RT-PCR using the Assays-on-Demand gene expression mixes specific for each indicated human gene or ACTB and the TaqMan Universal PCR Master Mix (Applied Biosystems). Reactions were carried out in triplicate in the ABI PRISM 7900 HT Sequence Detection System. The relative amount of products was determined by the comparative Ct method according to the manufacturer's instructions.
Immunoblotting
Live cells were enriched by Ficoll (GE Healthcare) density gradient centrifugation before preparation of whole-cell lysates for immunoblotting of cell-cycle proteins as described,30 or lysates for analysis of Bcl-2 family proteins as described.31 The antibodies used are summarized in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Signals were developed with the SuperSignal West Femto Maximum Sensitivity Substrate (Pierce Biotechnology). The protein level was quantitated using the ImageJ program developed by Wayne Rasband (National Institute of Mental Health, Bethesda, MD).
Myeloma xenograft model and therapy
MM1.S cells (1 × 107) stably expressing the HSV-TK-eGFP-luciferase fusion protein were injected intravenously into NOD/SCID (NOD/LTSZPrko/J) mice as described.32 PD 0332991 was dissolved in vehicle (50mM sodium lactate, pH 4.0) and administered daily by gavage at 150 mg/kg or 80 mg/kg for the days indicated. Bortezomib (0.25 mg/kg) was administered intravenously as indicated. The control mice received the vehicle through the same route and on the same schedule.
Immunohistochemistry
Sections (4-μm) of paraffin-embedded BM tissue from femurs of the MM1.S xenograft NOD/SCID mice were analyzed using a TechMate500 BioTek automated immunostainer (Ventana Medical Systems) according to the manufacturer's specifications. Simultaneous expression of IRF4 and phosphorylated Rb or Ki67 was detected with mAb to human MUM1 (IRF4) protein (Dako North America) and polyclonal rabbit antibody to phospho-Ser807/811 of human Rb (Cell Signaling Technology) or mAb to Ki67 (Zymed).
Statistical analysis
All statistical analyses were performed by the use of Student t test, assuming unequal variances, where the significance level was set at P less than .05. Unless otherwise noted, error bars in figures are displayed as plus or minus SEM, and data presented represent at least 3 independent experiments.
Results
Selective and reversible inhibition of CDK4/CDK6 induces prolonged early G1 arrest followed by cell-cycle synchronization
The exceptional selectivity of PD 0332991 suggests that induction of prolonged early G1 arrest (pG1) by inhibition of CDK4/CDK6 may halt gene expression in early G1 and prevent the expression of genes programmed for other phases of the cell cycle, thereby sensitizing tumor cells for cytotoxic killing. As PD 0332991 acts reversibly, release of pG1 may lead to synchronous progression to S phase (pG1-S) with incomplete restoration of scheduled gene expression, thus further heightening the susceptibility to cytotoxic killing.
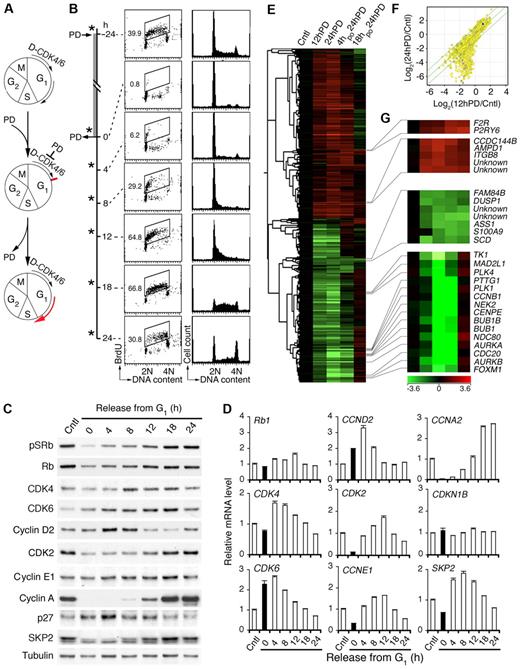
To test this hypothesis, we first established that release from pG1 was followed by synchronous progression to S phase (Figure 1A). Inhibition of CDK4/CDK6 with PD 0332991 (0.25μM) induced a complete early G1 arrest within 12 hours independent of p53 in all Rb-positive HMCLs tested, including MM1.S.23 By 4 hours after release from prolonged PD 0332991 treatment (24 hours), MM1.S cells had begun to enter S phase based on BrdU pulse-labeling (30 minutes) and DNA content per cell, as had CAG and KMS12-PE HMCLs (Figure 1B; supplemental Figure 1A-B). The cell cycle progressed synchronously, leading to a prominent increase in the proportion of S-phase cells, to 67% at 18 hours as they entered G2 (Figure 1B). Induction of pG1 by PD 0332991 for more than 24 hours led to a gradual increase in apoptosis but was reversible even after 5 days (supplemental Figure 1C-D), reinforcing the exceptional precision and reversibility with which PD 0332991 inhibits CDK4/CDK6 and cell-cycle progression.
Inhibition of CDK4/CDK6 induces early G1 arrest and cell-cycle synchronization. (A) Schema for reversible inhibition of CDK4/CDK6 by PD 0332991 (PD). D indicates cyclin D. (B) MM1.S cells were cultured with PD for 24 hours (h) and released into fresh media for the time indicated. *BrdU was added 30 minutes before cell harvest for FACS analysis of BrdU uptake and DNA content per cell. Number in the FACS profile indicates the percentage of BrdU-positive cells. (C) Immunoblotting and (D) quantitative RT-PCR analysis of cells cultured as in panel B. pSRb indicates Rb phosphorylated on serine 807/811. (E) Hierarchical clustering analysis of gene expression in MM1.S cells cultured with PD for 12 hours (12hPD) or 24 hours (24hPD), or released into fresh medium for 4 hours (4hpo24hPD) or 18 hours (18hpo24hPD) after 24 hours of PD treatment. The 2797 genes included in this analysis have a log2(treated/control) value > 1 in at least 1 of 3 independent arrays and a P value < .05. (F) Scatter plot of Log2 (12hPD/Cntl) and Log2(24hPD/Cntl) after normalization to the expression of ACTB (β-actin). The middle line indicates no variation, and the outer 2 lines indicate a 2-fold variation between the log ratio values. (G) Heat maps of genes from panel E. Data are representative of 6 independent experiments.
Inhibition of CDK4/CDK6 induces early G1 arrest and cell-cycle synchronization. (A) Schema for reversible inhibition of CDK4/CDK6 by PD 0332991 (PD). D indicates cyclin D. (B) MM1.S cells were cultured with PD for 24 hours (h) and released into fresh media for the time indicated. *BrdU was added 30 minutes before cell harvest for FACS analysis of BrdU uptake and DNA content per cell. Number in the FACS profile indicates the percentage of BrdU-positive cells. (C) Immunoblotting and (D) quantitative RT-PCR analysis of cells cultured as in panel B. pSRb indicates Rb phosphorylated on serine 807/811. (E) Hierarchical clustering analysis of gene expression in MM1.S cells cultured with PD for 12 hours (12hPD) or 24 hours (24hPD), or released into fresh medium for 4 hours (4hpo24hPD) or 18 hours (18hpo24hPD) after 24 hours of PD treatment. The 2797 genes included in this analysis have a log2(treated/control) value > 1 in at least 1 of 3 independent arrays and a P value < .05. (F) Scatter plot of Log2 (12hPD/Cntl) and Log2(24hPD/Cntl) after normalization to the expression of ACTB (β-actin). The middle line indicates no variation, and the outer 2 lines indicate a 2-fold variation between the log ratio values. (G) Heat maps of genes from panel E. Data are representative of 6 independent experiments.
Inhibition of CDK4/CDK6 halts gene expression in early G1
Further confirming that inhibition of CDK4/CDK6 by PD 0332991 is specific and reversible, removal of PD 0332991 restored the CDK4/CDK6 activity as indicated by the ratios of CDK4/CDK6-specific phosphorylation of Rb (pSRb) to total Rb (Figure 1C). This led to a transient increase in CDK4 mRNA along with CDK6 and CCND2 mRNAs, which were already elevated in pG1 as programmed, and a gradual decline in progress through S phase. It was also accompanied by scheduled expression of CDK2, CCNE1, and CCNA2 mRNAs, which were virtually undetectable in early G1 arrest (Figure 1D). The protein level largely paralleled that of mRNAs, although the cyclin E protein level did not vary appreciably and Rb was modestly reduced in pG1 despite constant mRNA expression (Figure 1C-D). The p27 protein, but not mRNA (CDKN1B), accumulated in pG1 and fell in pG1-S, inversely correlating with the expression of Skp2 mRNA and protein, which is known to mediate ubiquitin-proteasome degradation of p2733 (Figure 1C-D). Inhibition of CDK4/CDK6 thus halts gene expression in early G1 and prevents the expression of Skp2 required for p27 degradation as well as CDK2 and cyclin A necessary for progression to S phase. Removal of CDK4/CDK6 inhibition permits a synchronous progression to S phase by restoring scheduled expression of cell-cycle genes.
Global gene expression profiling further revealed that, for some genes, the up- or down-regulation in early G1 (12-hour PD) relative to asynchronously growing MM cells was maintained in prolonged early G1 arrest (24-hour PD and 36-hour PD) and reversed after the removal of the G1 block (4-, 18-, or 24-hour PD; Figure 1E-F; supplemental Figure 2). For others, the changes were profoundly amplified with the duration of pG1. This included progressive suppression of genes required for DNA synthesis, checkpoint control and mitosis, and genes encoding transcription factors, such as FOXM1,34,35 a target of CDK4/CDK6 activation36 (Figure 1G; supplemental Figure 2). The expression of cell-cycle and checkpoint genes was restored as scheduled in pG1-S (4-, 18-, or 24-hour PD). However, the expression of many others was not. For example, arginine-succinate synthetase (ASS1) and stearoyl-CoA desaturase (SCD) remained suppressed and F2R and P2RY6 were persistently activated in pG1-S (Figure 1G). Prolonged inhibition of CDK4/CDK6 thus halts gene expression in early G1 and prevents the expression of genes scheduled for other cell-cycle phases, with incomplete restoration despite cell-cycle progression to S phase.
Prolonging early G1 arrest sensitizes myeloma cells to cytotoxic killing
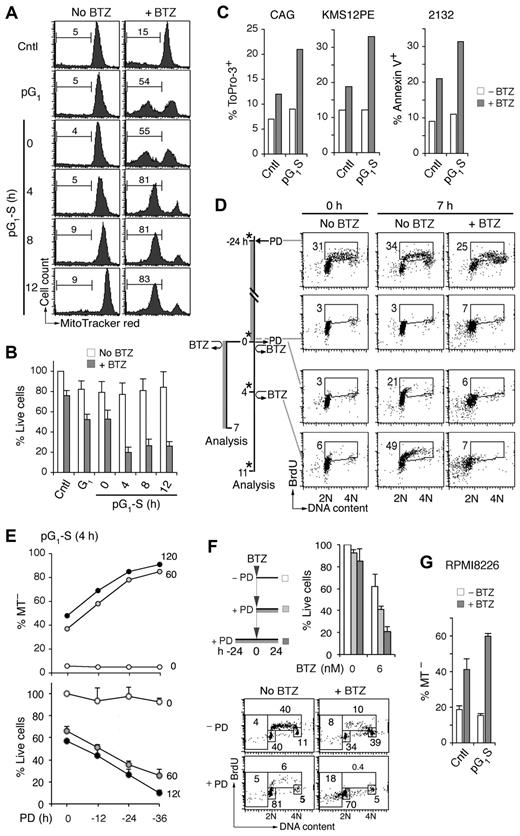
To determine whether prolonged halting of gene expression in early G1 sensitizes myeloma cells to cytotoxic killing, MM1.S cells in pG1 (24-hour PD) or at various times in pG1-S were pulsed with bortezomib (60nM) for 1 hour to mimic its rapid turnover in humans. Cytotoxic killing was profoundly enhanced in pG1 compared with the asynchronous population, as evidenced by the loss of viability and the increase in mitochondrial outer membrane permeabilization by 7 hours of bortezomib pulsing (Figure 2A-B; and data not shown). This enhancement in killing was markedly amplified in synchronous S phase entry and progression in MM1.S and other Rb-positive HMCLs, including CAG, KMS12-PE, and 2132 (Figure 2A-C).
Sensitization of myeloma cells to cytotoxic killing in pG1 and pG1-S. (A) MM1.S cells prolonged arrest in early G1 (pG1) by PD (24 hours), synchronized into S-phase for the time indicated (pG1-S), or without PD treatment (Cntl), were pulsed with bortezomib (BTZ, 60nM) for 1 hour or left untreated (No BTZ). Cell death was determined by the loss of MitoTracker Red (MT−) at 7 hours from BTZ pulsing. (B) “% Live cells” indicates the number of live cells relative to untreated Cntl cells determined after BTZ pulsing. (C) HMCL CAG and KMS12PE cells were pulsed with BTZ (120nM) for 1 hour in pG1-S (4 hours after PD withdrawal), and cell death was determined by ToPro-3 staining at 18 hours after BTZ pulsing. The 2132 cells were cultured similarly except that BTZ was used at 350nM and cell death was determined by annexin V staining. (D) FACS analysis of BrdU uptake and DNA content in pG1 and pG1-S MM1.S cells cultured as indicated in the schema. (E) MM1.S cells were pulsed with BTZ (1 hour, 60 or 120nM) at 4 hours after release from PD pretreatment for the time indicated. The percentages of live and dead cells were determined as in panel B, using cells untreated by PD or BTZ as a reference. (F) MM1.S cells were cultured with low BTZ with or without PD pretreatment as diagrammed, and analyzed for live cells as in panel E and BrdU uptake as in panel D. (G) RPMI8226 cells were cultured with 0.5μM PD for 24 hours and released into fresh media for 12 hours. The cells were then treated with BTZ (20nM) for 8 hours, and the percentage of MT− cells was determined. Data are representative of 5 independent experiments.
Sensitization of myeloma cells to cytotoxic killing in pG1 and pG1-S. (A) MM1.S cells prolonged arrest in early G1 (pG1) by PD (24 hours), synchronized into S-phase for the time indicated (pG1-S), or without PD treatment (Cntl), were pulsed with bortezomib (BTZ, 60nM) for 1 hour or left untreated (No BTZ). Cell death was determined by the loss of MitoTracker Red (MT−) at 7 hours from BTZ pulsing. (B) “% Live cells” indicates the number of live cells relative to untreated Cntl cells determined after BTZ pulsing. (C) HMCL CAG and KMS12PE cells were pulsed with BTZ (120nM) for 1 hour in pG1-S (4 hours after PD withdrawal), and cell death was determined by ToPro-3 staining at 18 hours after BTZ pulsing. The 2132 cells were cultured similarly except that BTZ was used at 350nM and cell death was determined by annexin V staining. (D) FACS analysis of BrdU uptake and DNA content in pG1 and pG1-S MM1.S cells cultured as indicated in the schema. (E) MM1.S cells were pulsed with BTZ (1 hour, 60 or 120nM) at 4 hours after release from PD pretreatment for the time indicated. The percentages of live and dead cells were determined as in panel B, using cells untreated by PD or BTZ as a reference. (F) MM1.S cells were cultured with low BTZ with or without PD pretreatment as diagrammed, and analyzed for live cells as in panel E and BrdU uptake as in panel D. (G) RPMI8226 cells were cultured with 0.5μM PD for 24 hours and released into fresh media for 12 hours. The cells were then treated with BTZ (20nM) for 8 hours, and the percentage of MT− cells was determined. Data are representative of 5 independent experiments.
Simultaneous analysis of BrdU uptake and DNA content further revealed that, in asynchronous cultures, bortezomib preferentially killed cells in early S phase and virtually killed all cells synchronously entering S phase (Figure 2D). The 5-fold increase in bortezomib killing in pG1-S, however, vastly exceeded the approximately 2-fold enrichment in S-phase cells (Figures 1B and 2A-B), and the magnitude of killing increased markedly with time of prior G1 arrest (12-36 hours of PD 0332991 treatment; Figure 2E). Compared with simultaneous addition of PD 0332991, pretreatment with PD 0332991 for 24 hours similarly enhanced the killing of HMCLs by continuous exposure to bortezomib at low dose (6nM; Figure 2F). This led to virtual eradication of HMCL MM1.R cells in pG1 and enhanced killing of RPMI 8226 cells in pG1-S (Figure 2G; supplemental Figure 3). Induction of pG1 therefore sensitizes myeloma cells to cytotoxic killing, which is amplified in pG1-S.
Inhibition of CDK4/CDK6 is the basis for sensitization to bortezomib killing in pG1
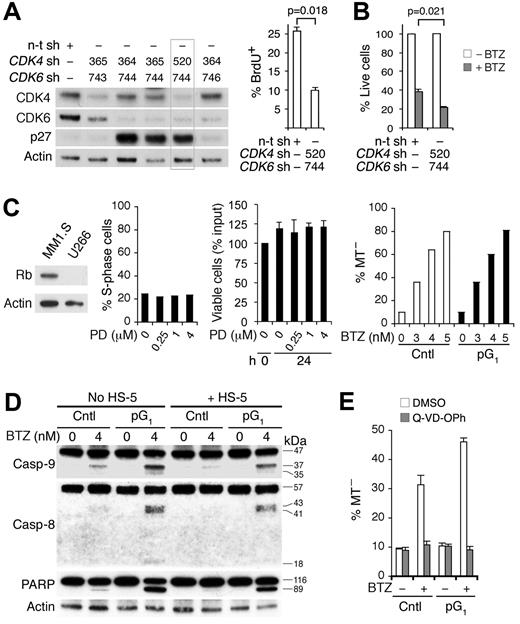
Confirming that sensitization by PD 0332991 is the result of inhibition of CDK4/CDK6, knocking down both CDK4 and CDK6, but not each alone, by shRNA-lentivirus induced G1 arrest as shown by the prominent increase in p27 and the reduction in BrdU uptake, and enhanced the loss of viability in response to bortezomib (Figure 3A-B). Moreover, induction of G1 arrest by PD 0332991 requires Rb, the substrate of CDK4 and CDK6. PD 0332991 failed to induce G1 arrest or enhance bortezomib killing in Rb-deficient U266, even at a 16-fold or higher concentration and REC-1 MCL cells (Figure 3C; supplemental Figure 4) or 293T cells in which Rb is sequestered by adenovirus E1A (data not shown). pG1 apparently enhanced apoptosis induced by bortezomib because it led to cooperative activation of caspase-9 and caspase-8, and poly ADP-ribose polymerase cleavage despite protection by human BMSCs and was abolished by the pan-caspase inhibitor Q-VD-OPh (Figure 3D-E).
Inhibition of CDK4/CDK6 is the basis for sensitization to cytotoxic killing. MM1.S cells were infected with CDK4, CDK6, or a nontargeting (n-t) shRNA (sh) lentivirus. (A) Immunoblotting and BrdU+ uptake at 66 hours after infection. (B) Percentage of live cells was determined at 17 hours of BTZ (4nM) treatment starting at 72 hours after infection, using the n-t shRNA lentivirus-infected cells without treatment as a control. (C) Left: Rb protein expression in MM1.S and U266 cells. Middle: Percentage of S phase and viable U266 cells after culturing with PD for 24 hours relative to input. Right: MT− U266 cells after BTZ treatment (24 hours) in the absence (Cntl) or presence of PD pretreatment (pG1; 0.5μM, 24 hours). (D) Caspase and poly ADP-ribose polymerase cleavage in MM1.S cells after BTZ treatment (12 hours) in pG1 (PD, 24 hours) or left untreated (Cntl), in the presence or absence of HS-5 BMSCs. (E) Q-VD-OPh (20μM) or DMSO was added to MM1.S cells in pG1 or left untreated for 1 hour before BTZ pulsing (120nM). MT− cells were determined at 7 hours from BTZ pulsing. Data are representative of 3 independent experiments.
Inhibition of CDK4/CDK6 is the basis for sensitization to cytotoxic killing. MM1.S cells were infected with CDK4, CDK6, or a nontargeting (n-t) shRNA (sh) lentivirus. (A) Immunoblotting and BrdU+ uptake at 66 hours after infection. (B) Percentage of live cells was determined at 17 hours of BTZ (4nM) treatment starting at 72 hours after infection, using the n-t shRNA lentivirus-infected cells without treatment as a control. (C) Left: Rb protein expression in MM1.S and U266 cells. Middle: Percentage of S phase and viable U266 cells after culturing with PD for 24 hours relative to input. Right: MT− U266 cells after BTZ treatment (24 hours) in the absence (Cntl) or presence of PD pretreatment (pG1; 0.5μM, 24 hours). (D) Caspase and poly ADP-ribose polymerase cleavage in MM1.S cells after BTZ treatment (12 hours) in pG1 (PD, 24 hours) or left untreated (Cntl), in the presence or absence of HS-5 BMSCs. (E) Q-VD-OPh (20μM) or DMSO was added to MM1.S cells in pG1 or left untreated for 1 hour before BTZ pulsing (120nM). MT− cells were determined at 7 hours from BTZ pulsing. Data are representative of 3 independent experiments.
Bim enhances bortezomib-induced apoptosis in the absence of Noxa in pG1 and in cooperation with Noxa in pG1-S
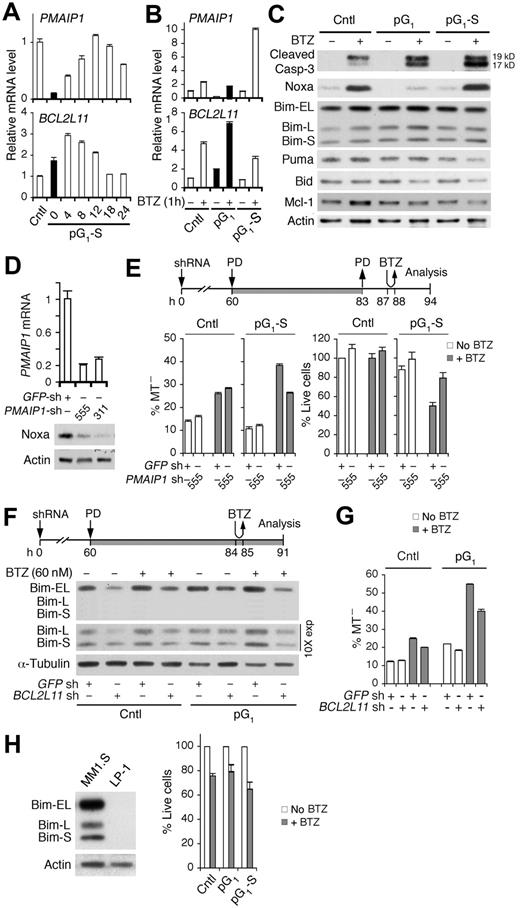
This led us to investigate the role of proapoptotic PMAIP1 (Noxa) and BCL2L11 (Bim), previously known to mediate bortezomib-induced apoptosis in myeloma cells,31,37 in cell-cycle sensitization to bortezomib killing. PMAIP1 mRNA expression was silenced in pG1 but was restored with time after release of the G1 block, inversely correlating with BCL2L11 mRNA expression (Figure 4A). Bortezomib up-regulated PMAIP1 and BCL2L11 mRNAs as expected. However, the up-regulation of PMAIP1 mRNA by bortezomib was severely attenuated in pG1 and profoundly enhanced (10-fold) in pG1-S, whereas the up-regulation of BCL2L11 mRNA was most prominent in pG1 (Figure 4B). Thus, PMAIP1 and BCL2L11 are reciprocally regulated by the cell cycle, and pG1 represses PMAIP1 expression and antagonizes induction of PMAIP1 by bortezomib.
Bim enhances bortezomib-induced apoptosis in the absence of Noxa in pG1 and in cooperation with Noxa in pG1-S. (A) Quantitative RT-PCR analysis of PMAIP1 and BCL2L11 mRNAs in MM1.S cells cultured as in Figure 1B. (B-C) Quantitative RT-PCR analysis of Bcl-2 family genes and immunoblotting of Bcl-2 family proteins and cleaved caspase-3 at 7 hours from BTZ pulsing (60nM) in control, pG1 or pG1-S (12 hours after PD withdrawal) cells. (D) Noxa mRNA and protein expression in MM1.S cells at 72 hours after infection with PMAIP1 or GFP shRNA lentivirus. (E) PMAIP1 or GFP shRNA (sh) lentivirus-infected MM1.S cells were treated with PD and BTZ (60nM) as indicated. MT− and live cells were determined at 7 hours from BTZ pulsing. (F-G) BCL2L11 or GFP shRNA (sh) lentivirus-infected MM1.S cells were treated with PD and BTZ (60nM) as indicated. Bim protein levels and the percentages of MT− cells were determined as indicated. (H) Immunoblotting of Bim in MM1.S and LP-1 myeloma cells (left). LP-1 cells were pulsed with BTZ (120nM) in pG1 (0.5μM PD, 24 hours) or pG1-S (4 hours after PD withdrawal). The percentage of live cells relative to non-BTZ–treated cells was determined 18 hours from BTZ pulsing (right). Data are representative of 3 independent experiments.
Bim enhances bortezomib-induced apoptosis in the absence of Noxa in pG1 and in cooperation with Noxa in pG1-S. (A) Quantitative RT-PCR analysis of PMAIP1 and BCL2L11 mRNAs in MM1.S cells cultured as in Figure 1B. (B-C) Quantitative RT-PCR analysis of Bcl-2 family genes and immunoblotting of Bcl-2 family proteins and cleaved caspase-3 at 7 hours from BTZ pulsing (60nM) in control, pG1 or pG1-S (12 hours after PD withdrawal) cells. (D) Noxa mRNA and protein expression in MM1.S cells at 72 hours after infection with PMAIP1 or GFP shRNA lentivirus. (E) PMAIP1 or GFP shRNA (sh) lentivirus-infected MM1.S cells were treated with PD and BTZ (60nM) as indicated. MT− and live cells were determined at 7 hours from BTZ pulsing. (F-G) BCL2L11 or GFP shRNA (sh) lentivirus-infected MM1.S cells were treated with PD and BTZ (60nM) as indicated. Bim protein levels and the percentages of MT− cells were determined as indicated. (H) Immunoblotting of Bim in MM1.S and LP-1 myeloma cells (left). LP-1 cells were pulsed with BTZ (120nM) in pG1 (0.5μM PD, 24 hours) or pG1-S (4 hours after PD withdrawal). The percentage of live cells relative to non-BTZ–treated cells was determined 18 hours from BTZ pulsing (right). Data are representative of 3 independent experiments.
Sequential pG1 and pG1-S appear to lower the threshold for caspase-activation because activation of caspase-3 by bortezomib was enhanced in pG1 and further in pG1-S. (Figure 4C). Concomitantly, the Noxa protein was elevated by bortezomib in pG1-S, which paralleled the increase in PMAIP1 mRNA and was specific given the decrease in Puma, Bid, and Mcl-1 proteins and the marginal variation in the Bim protein levels (Figure 4C). These data suggest that Noxa mediates bortezomib-induced apoptosis in pG1-S. Indeed, shRNA knockdown of Noxa attenuated bortezomib killing and increased the recovery of live cells in pG1-S (Figure 4D-E). Noxa, therefore, is repressed in early G1 and selectively up-regulated by bortezomib in pG1-S to induce apoptosis.
Bim is thought to mediate bortezomib killing by neutralizing Mcl-1 and Bcl-2.38 Although the Bim protein levels in pG1 did not match the increase in BCL2L11 mRNA by bortezomib (Figure 4B-C), the association of Bim with Mcl-1 or Bcl-2, but not Bcl-xL, was augmented by bortezomib (supplemental Figure 4). Partial knockdown of Bim attenuated bortezomib-induced apoptosis as well as the enhancement in pG1 (Figure 4F-G), and the absence of Bim in HMCL LP-1 cells abolished the enhancement of bortezomib killing in both pG1 and pG1-S (Figure 4H). Collectively, these data strongly suggest that Bim mediates the enhancement of bortezomib killing in pG1 in the absence of Noxa, and in pG1-S in cooperation with Noxa.
Cell cycle–coupled loss of IRF4 mediates bortezomib killing
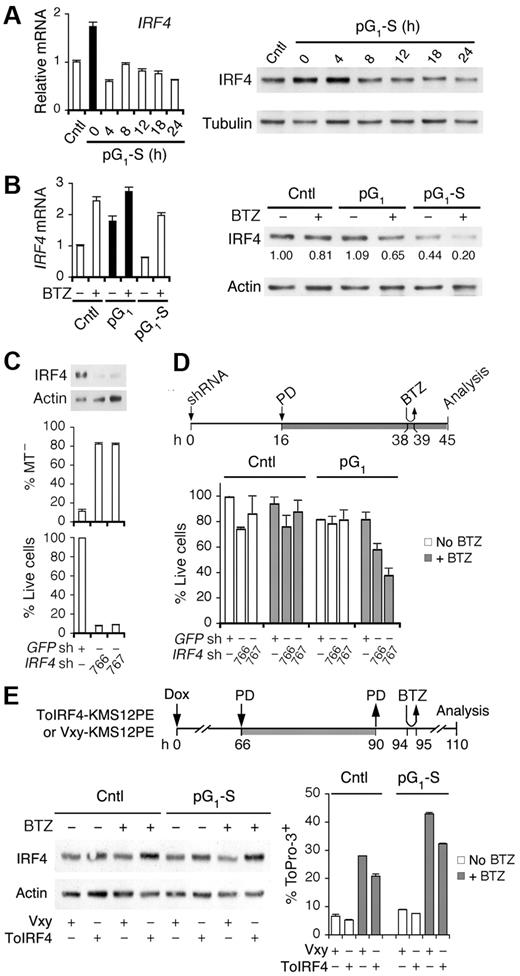
Cell-cycle enhancement of bortezomib killing may also be the result of loss of survival functions, in particular IRF4, an essential survival factor of myeloma cells.29 The IRF4 mRNA and IRF4 protein levels were coordinately elevated in pG1 by PD 0332991 treatment for 24 hours and reduced in synchronous progression through S phase (Figure 5A). Bortezomib up-regulated IRF4 mRNA independent of the cell cycle but reduced the IRF4 protein, more markedly in pG1 (to 65%) and drastically in pG1-S (to 20%; Figure 5B). These data provide the first direct evidence that IRF4 expression is coupled to the cell cycle and that bortezomib up-regulates IRF4 mRNA but reduces the IRF4 protein in cooperation with the cell cycle, greater in pG1 and further in pG1-S.
Cooperative regulation of IRF4 by the cell cycle and bortezomib. Quantitative RT-PCR analysis and immunoblotting of IRF4 expression in MM1.S cells (A) cultured as in Figure 1B, and (B) at 7 hours from pulsing with BTZ (1 hour, 120nM) in pG1 (PD, 24 hours) or in pG1-S (4 hours after PD withdrawal). (C) IRF4 protein level, MT− and live cells were determined in MM1.S cells at 72 hours after infection with IRF4 or the control GFP shRNA lentivirus. (D) MM1.S cells were treated with PD and pulsed with BTZ (60nM) after lentivirus infection as indicated. Percentage of live cells was determined using cells infected with the GFP shRNA lentivirus as a control. (E) KMS12PE cells infected with a retrovirus expressing human IRF4 (ToIRF4) or the control virus (Vxy) were treated with doxycycline (Dox, 20 ng/mL), PD (0.3μM), or BTZ (250nM) as indicated. The IRF4 protein level was determined by immunoblotting and cell death by ToPro-3 staining. Data are representative of 3 independent experiments.
Cooperative regulation of IRF4 by the cell cycle and bortezomib. Quantitative RT-PCR analysis and immunoblotting of IRF4 expression in MM1.S cells (A) cultured as in Figure 1B, and (B) at 7 hours from pulsing with BTZ (1 hour, 120nM) in pG1 (PD, 24 hours) or in pG1-S (4 hours after PD withdrawal). (C) IRF4 protein level, MT− and live cells were determined in MM1.S cells at 72 hours after infection with IRF4 or the control GFP shRNA lentivirus. (D) MM1.S cells were treated with PD and pulsed with BTZ (60nM) after lentivirus infection as indicated. Percentage of live cells was determined using cells infected with the GFP shRNA lentivirus as a control. (E) KMS12PE cells infected with a retrovirus expressing human IRF4 (ToIRF4) or the control virus (Vxy) were treated with doxycycline (Dox, 20 ng/mL), PD (0.3μM), or BTZ (250nM) as indicated. The IRF4 protein level was determined by immunoblotting and cell death by ToPro-3 staining. Data are representative of 3 independent experiments.
The reduction in IRF4 protein by bortezomib correlated with the marked increase in caspase-3 activation in pG1 and pG1-S (Figure 4C), suggesting that IRF4 protects MM cells from apoptosis. Indeed, knocking down IRF4 expression by shRNA drastically reduced viability through mitochondrial outer membrane permeabilization in 72 hours (Figure 5C). Preceding this, partial reduction of IRF4 at 45 hours enhanced bortezomib killing within 6 hours, but only after pG1 sensitization (Figure 5D). Furthermore, ectopic expression of human IRF4 in a recombinant retrovirus (ToIRF4), but not of the control virus (Vxy), by doxycycline induction in KMS12PE cells compromised bortezomib killing, more markedly in pG1-S (Figure 5E). On this basis, we conclude that IRF4 protects myeloma cells from apoptosis and that cooperative cell cycle–coupled reduction of IRF4 sensitizes myeloma cells to bortezomib-induced apoptosis in pG1 in coordination with Bim, in the absence of Noxa in pG1 and together with Noxa in pG1-S.
Inhibition of CDK4/CDK6 sensitizes primary myeloma cells to bortezomib killing
Although long-term proliferation of primary human myeloma cells in vitro is not yet attainable, proliferation of freshly isolated CD138+ BM myeloma cells can be extended transiently in coculture with HS-5 BMSCs.23 Using this system, we have demonstrated that PD 0332991 inhibited CDK4/CDK6 and accelerated early G1 arrest in primary CD138+ human myeloma cells.23
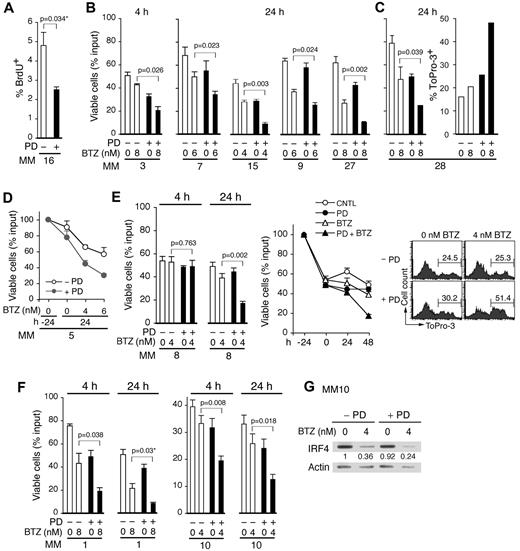
Acceleration of early G1 arrest by PD 0332991 treatment for 4 hours or more (Figure 6A) exacerbated the loss of viability in response to bortezomib in a dose-dependent manner in 24 hours in the majority of MM cases (17 of 27; Figure 6B-D; supplemental Figure 6; Table 1). In some cases (MM 8), primary MM cells remained refractory to bortezomib ex vivo but became increasingly sensitive to bortezomib with time of PD 0332991 pretreatment, as indicated by DNA fragmentation (Figure 6E). Further investigation confirmed that sensitization by PD 0332991 was time-dependent (MM1 and MM10) and associated with a greater loss of IRF4 before DNA fragmentation in bortezomib killing (Figure 6E-F). Of the primary BM myeloma cells that failed to be sensitized by PD 0332991 (10 of 27), some were completely refractory to bortezomib (MM18). Others were already highly responsive to bortezomib (MM13; supplemental Figure 6B). Moreover, although preliminary, it appears that sensitization by pG1 in vitro preferentially correlates with proliferation (Ki67-positive) of MM cells in vivo in these late-stage diseases (Table 1). Collectively, these data suggest that acceleration of early G1 arrest by CDK4/CDK6 inhibition sensitizes the majority of primary BM myeloma cells to bortezomib-induced apoptosis in the presence of BMSCs in part through loss of IRF4.
CDK4/CDK6 inhibition accelerates G1 arrest and enhances bortezomib killing of primary myeloma cells. Primary CD138+ BM myeloma cells isolated from individual patients (MM, referred to by the number at the bottom of each graph) were pretreated with 0.5μM PD for 4 or 24 hours in the HS-5 BMSC coculture before addition of BTZ and cultured for an additional 24 hours unless otherwise indicated. (A) BrdU uptake in MM cells treated with PD for 16 hours, with BrdU presented in the last 13 hours. (B-D) Viability of MM cells after BTZ addition. (E) Viability of MM cells after culturing as indicated (left); immediately after isolation (−24), at 24 hours after incubation with PD (0), and at 24 and 48 hours after further incubation with BTZ (4nM) and PD (middle), and ToPro-3 analysis at 48 hours of BTZ treatment (right). (F) Viability after BTZ addition in MM cells pretreated with PD for time indicated. (G) Immunoblotting of IRF4 at 12 hours after BTZ addition in MM10 (as shown in panel F) with or without 24-hour PD pretreatment. Numbers indicate the relative level of IRF4 compared with cells left untreated by PD or BTZ and corrected for loading by the actin signal. Data represent mean ± SD in triplicate. P value was determined by 2-tailed or 1-tailed (*) t test.
CDK4/CDK6 inhibition accelerates G1 arrest and enhances bortezomib killing of primary myeloma cells. Primary CD138+ BM myeloma cells isolated from individual patients (MM, referred to by the number at the bottom of each graph) were pretreated with 0.5μM PD for 4 or 24 hours in the HS-5 BMSC coculture before addition of BTZ and cultured for an additional 24 hours unless otherwise indicated. (A) BrdU uptake in MM cells treated with PD for 16 hours, with BrdU presented in the last 13 hours. (B-D) Viability of MM cells after BTZ addition. (E) Viability of MM cells after culturing as indicated (left); immediately after isolation (−24), at 24 hours after incubation with PD (0), and at 24 and 48 hours after further incubation with BTZ (4nM) and PD (middle), and ToPro-3 analysis at 48 hours of BTZ treatment (right). (F) Viability after BTZ addition in MM cells pretreated with PD for time indicated. (G) Immunoblotting of IRF4 at 12 hours after BTZ addition in MM10 (as shown in panel F) with or without 24-hour PD pretreatment. Numbers indicate the relative level of IRF4 compared with cells left untreated by PD or BTZ and corrected for loading by the actin signal. Data represent mean ± SD in triplicate. P value was determined by 2-tailed or 1-tailed (*) t test.
Multiple myeloma cases in this study
| MM no. . | Figure no. . | Age, y/sex . | Ig isotype . | Stage* . | No. of prior treatments . | CD138+, % . | Ki67+/CD138+, % . |
|---|---|---|---|---|---|---|---|
| 1 | 6F | 59/F | Gκ | III | 1 | < 5 | NA |
| 2 | NS | 43/F | κ† | II | 1 | 60 | < 1 |
| 3 | 6B | 46/F | Gκ | II | 0 | 30 | 2 |
| 4 | S6A | 36/F | Gκ | III | 2 | 60 | 12 |
| 5 | 6D | 57/F | κ† | III | 7 | 90 | 49 |
| 6 | S6A | 44/F | Gκ | III | 1 | < 5 | < 1 |
| 7 | 6B | 55/F | Gκ | III | 0 | 80 | 42 |
| 8 | 6E | 63/M | κ† | III | 3 | 30 | 16 |
| 9 | 6B | 60/M | Gκ | III | 2 | 30 | 8 |
| 10 | 6F-G | 55/F | Gκ | III | 2 | 90 | 71 |
| 13 | S6B | 72/F | Aκ | III | 1 | 80 | 22 |
| 14 | NS | 82/F | Gλ | III | 0 | 50 | 4 |
| 15 | 6B | 44/M | λ† | II | 0 | 80 | 8 |
| 16 | 6A | 68/M | λ† | II | 1 | 10 | 3 |
| 17 | NS | 58/M | Gκ | II | 0 | 30 | 2 |
| 18 | S6B | 74/M | Gκ | II | 0 | 25 | 2 |
| 20 | S6A | 71/F | Gκ | III | 7 | 90 | 2 |
| 21 | NS | 58/M | Gκ | III | 6 | 90 | 14 |
| 22 | NS | 76/F | Gλ | III | 10 | 95 | 25 |
| 23 | NS | 85/M | κ† | II | 4 | 80 | 6 |
| 24 | NS | 60/M | Gκ | III | 7 | 5 | NA |
| 25 | S6A | 76/F | Gκ | III | 0 | 40 | 12 |
| 26 | NS | 61/F | Gκ | III | 8 | 90 | 25 |
| 27 | 6B | 46/F | λ† | II | 8 | 100 | 35 |
| 28 | 6C | 71/F | Gλ | III | 1 | NA | NA |
| MM no. . | Figure no. . | Age, y/sex . | Ig isotype . | Stage* . | No. of prior treatments . | CD138+, % . | Ki67+/CD138+, % . |
|---|---|---|---|---|---|---|---|
| 1 | 6F | 59/F | Gκ | III | 1 | < 5 | NA |
| 2 | NS | 43/F | κ† | II | 1 | 60 | < 1 |
| 3 | 6B | 46/F | Gκ | II | 0 | 30 | 2 |
| 4 | S6A | 36/F | Gκ | III | 2 | 60 | 12 |
| 5 | 6D | 57/F | κ† | III | 7 | 90 | 49 |
| 6 | S6A | 44/F | Gκ | III | 1 | < 5 | < 1 |
| 7 | 6B | 55/F | Gκ | III | 0 | 80 | 42 |
| 8 | 6E | 63/M | κ† | III | 3 | 30 | 16 |
| 9 | 6B | 60/M | Gκ | III | 2 | 30 | 8 |
| 10 | 6F-G | 55/F | Gκ | III | 2 | 90 | 71 |
| 13 | S6B | 72/F | Aκ | III | 1 | 80 | 22 |
| 14 | NS | 82/F | Gλ | III | 0 | 50 | 4 |
| 15 | 6B | 44/M | λ† | II | 0 | 80 | 8 |
| 16 | 6A | 68/M | λ† | II | 1 | 10 | 3 |
| 17 | NS | 58/M | Gκ | II | 0 | 30 | 2 |
| 18 | S6B | 74/M | Gκ | II | 0 | 25 | 2 |
| 20 | S6A | 71/F | Gκ | III | 7 | 90 | 2 |
| 21 | NS | 58/M | Gκ | III | 6 | 90 | 14 |
| 22 | NS | 76/F | Gλ | III | 10 | 95 | 25 |
| 23 | NS | 85/M | κ† | II | 4 | 80 | 6 |
| 24 | NS | 60/M | Gκ | III | 7 | 5 | NA |
| 25 | S6A | 76/F | Gκ | III | 0 | 40 | 12 |
| 26 | NS | 61/F | Gκ | III | 8 | 90 | 25 |
| 27 | 6B | 46/F | λ† | II | 8 | 100 | 35 |
| 28 | 6C | 71/F | Gλ | III | 1 | NA | NA |
Ig indicates immunoglobulin; NS, data not shown in those MM cases in which PD 0332991 does not significantly enhance bortezomib killing of MM cells in vitro; NA, data not available; S6A, supplemental Figure 6A; and S6B, supplemental Figure 6B.
Durie-Salmon staging.39 Double immunohistochemical staining of CD138 and Ki67 was performed on core bone marrow biopsies as previously described. % CD138+ means the percentage of BM cells that are CD138 positive and % Ki67/CD138+ means the percentage of Ki67 positive cells among the CD138 positive cells.19
Absence of Ig heavy chains.
Reversible inhibition of CDK4/CDK6 enhances tumor suppression by bortezomib
The cell cycle–dependence of antitumor activity was then investigated by serial noninvasive bioluminescence imaging (BLI) in a NOD/SCID xenograft human myeloma model, in which disseminated tumors develop after injection of Luc+GFP+MM1.S cells stably expressing the HSV-TK-eGFP-luciferase fusion protein.32 Previously, we have established, in a related Luc+GFP+CAG model, that tumor development was markedly suppressed by day 14 of PD 0332991 treatment.23
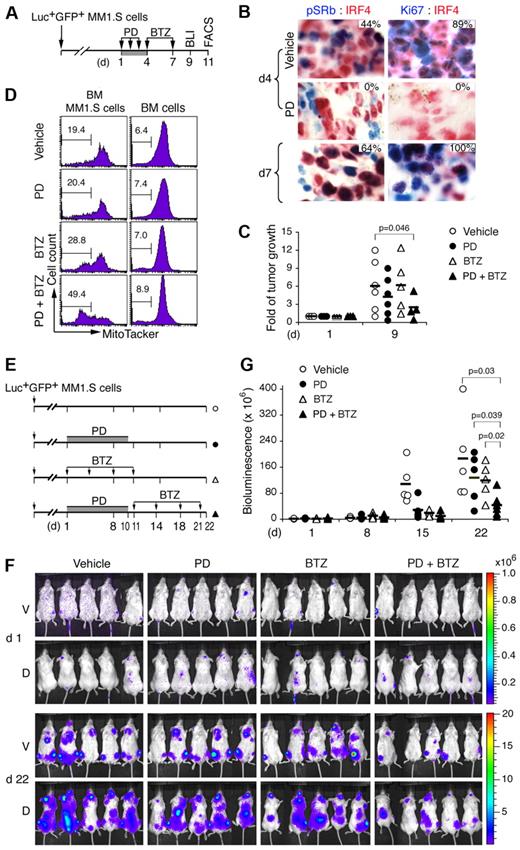
We now show, by immunohistochemistry in mice developing aggressive tumor, that the same daily PD 0332991 treatment (150 mg/kg) for 3 days completely inhibited CDK4/CDK6 (pSRb, from 44% to 0%) and proliferation (Ki67, from 89% to 0%) in BM myeloma cells (IRF4+; Figure 7A-B). The cell cycle resumed synchronously after discontinuation of PD 0332991 on day 7, as evidenced by the marked increase in the proportion of myeloma cells expressing pSRb (66%) and Ki67 (100%; Figure 7B). To determine whether induction of sequential pG1 and pG1-S augments tumor suppression by bortezomib, mice were given a subtherapeutic dose of bortezomib (0.25 mg/kg) in pG1 on day 4, and again on day 7 when the cell cycle had resumed. BLI analysis on day 9 demonstrated cooperative tumor suppression by the combination therapy compared with PD 0332991 or bortezomib treatment alone (Figure 7C; supplemental Figure 5). As a corollary, prolonged treatment with PD 0332991 at half the dose (80 mg/kg, daily) for 10 days followed by bortezomib at the same low dose (0.25 mg/kg, on days 11, 14, 18, and 21) reduced tumor growth to 20% of the vehicle-treated mice by day 22, compared with 50% by PD 0332991 or bortezomib alone (Figure 7E-G).
Induction of pG1 and pG1-S enhances tumor suppression by bortezomib. (A) A schema for treatment of NOD/SCID mice developing aggressive tumor after injection with Luc+GFP+MM1.S cells with PD (150 mg/kg) and BTZ (0.25 mg/kg), and the time (day) of BLI and MT− (FACS) analyses. (B) IHC of IRF4 (red) and phospho-Rb (Ser807/811; pSRb, blue) or Ki67 (blue) in MM1.S cells from BM of mice treated as indicated. The percentages of pSRb+ or Ki67+ cells in IRF4+ myeloma cells are indicated. (C) Fold of tumor growth represents BLI on day 9 relative to that on day 1 of the same mice. (D) BM GFP+ MM1.S cells and BM cells were flushed from femurs on day 11, stained with MitoTracker Red, and analyzed by FACS. Number indicates the percentage of MT− cells (mean ± SD). (E) Schema for treatment of myeloma-developing NOD/SCID mice with PD (80 mg/kg) and BTZ (0.25 mg/kg). (F) BLI of tumors in mice on days 1 and 22. (G) Bioluminescence representing tumor mass (photons/s/cm2/steradian) on days indicated. V indicates ventral; and D, dorsal. P value was determined by 2-tailed or 1-tailed (*) t test. Data are representative of 3 independent experiments.
Induction of pG1 and pG1-S enhances tumor suppression by bortezomib. (A) A schema for treatment of NOD/SCID mice developing aggressive tumor after injection with Luc+GFP+MM1.S cells with PD (150 mg/kg) and BTZ (0.25 mg/kg), and the time (day) of BLI and MT− (FACS) analyses. (B) IHC of IRF4 (red) and phospho-Rb (Ser807/811; pSRb, blue) or Ki67 (blue) in MM1.S cells from BM of mice treated as indicated. The percentages of pSRb+ or Ki67+ cells in IRF4+ myeloma cells are indicated. (C) Fold of tumor growth represents BLI on day 9 relative to that on day 1 of the same mice. (D) BM GFP+ MM1.S cells and BM cells were flushed from femurs on day 11, stained with MitoTracker Red, and analyzed by FACS. Number indicates the percentage of MT− cells (mean ± SD). (E) Schema for treatment of myeloma-developing NOD/SCID mice with PD (80 mg/kg) and BTZ (0.25 mg/kg). (F) BLI of tumors in mice on days 1 and 22. (G) Bioluminescence representing tumor mass (photons/s/cm2/steradian) on days indicated. V indicates ventral; and D, dorsal. P value was determined by 2-tailed or 1-tailed (*) t test. Data are representative of 3 independent experiments.
Of note, myeloma cells were more apoptotic than the surrounding nontransformed BM cells and were selectively killed by bortezomib, more prominently after cell-cycle synchronization with PD 0332991 (Figure 7D). Thus, reversible inhibition of CDK4/CDK6 induces early G1 arrest followed by synchronous cell-cycle progression in vivo. This leads to cooperative tumor suppression in response to cytotoxic agents, such as bortezomib, at low doses by amplifying tumor-specific killing in the native BM niches.
Discussion
Through specific and reversible inhibition of CDK4/CDK6 using the small molecule PD 0332991, we have developed a novel strategy to both inhibit the cell cycle in tumor cells and sensitize them to cytotoxic killing. We demonstrate that selective inhibition of the catalytic activities of CDK4 and CDK6 leads to cell-cycle arrest in early G1 with precision. This permits continuous expression of genes programmed for early G1 but prevents the expression of those scheduled for other phases of the cell cycle when early G1 arrest is prolonged. The cell cycle progresses synchronously to S phase and mitosis after removal of the CDK4/CDK6 inhibitor because of timely restoration of gene expression necessary for DNA synthesis, cell-cycle checkpoint control, and mitosis.
The expression of many other genes, however, was not restored on schedule. This was exemplified by sustained suppression of genes, such as ASS1 required for arginine biosynthesis, and SCD, which catalyzes the rate-limiting step in the biosynthesis of monounsaturated fatty acid from saturated fatty acid. By contrast, the expression of selective G-protein coupled receptor genes, such as F2R and P2RY6, the latter specifically responds to extracellular UDP for chemokine induction and monocyte recruitment, remained elevated in the early G1 state despite cell-cycle progression.
Tumor cells differ from untransformed cells in macromolecule synthesis, metabolism, and bioenergetics.40-42 On this basis, it is tempting to postulate that simultaneous suppression of amino acid and fatty acid synthesis may lead to a metabolic imbalance. Alone, it may not cause significant death in pG1 but could sensitize tumor cells to cytotoxic killing, more prominently in pG1-S when DNA synthesis and mitosis impose a greater demand on macromolecule synthesis and metabolism. This hypothesis may help to explain why tumor cells are more apoptotic and sensitive to bortezomib killing than the nontransformed BM cells in the human myeloma xenografts and why PD 0332991 is well tolerated in MCL28 and myeloma patients while effectively inhibiting CDK4/CDK6 and sensitizing tumor cells to cytotoxic killing in clinical studies (R.N. and S.C.-K., unpublished data).
We have demonstrated that inhibition of CDK4/CDK6 is the basis for induction of pG1 and sensitization to bortezomib killing by PD 0332991. CDK4 and CDK6 are essential for expansion of hematopoietic progenitors but dispensable for development or for cell-cycle activation by serum stimulation in mouse embryonic fibroblasts in vitro because of functional compensation by CDK2.43 Thus, it seems paradoxical that inhibition of CDK4 and CDK6 is sufficient to induce G1 arrest. However, unlike genetic ablation of CDK4 and CDK6 or pharmacologic intervention of CDK4 and CDK6 synthesis, inhibition of the CDK4/CDK6 activities prevents the synthesis of CDK2, cyclin E, and cyclin A by halting gene expression in early G1 and thus cannot be compensated for by CDK2.
Our data further suggest that pG1 and pG1-S lower the threshold for caspase activation and that loss of IRF4 plays a key role in this process. IRF4 was found to be essential for myeloma cell survival in an siRNA screen.29 We demonstrated that IRF4 protects myeloma cells from apoptosis and that the IRF4 function is determined by cooperative transcriptional and posttranscriptional regulation of IRF4 synthesis by the cell cycle and bortezomib. To summarize our findings: (1) IRF4 synthesis is higher in pG1 and lower during progression through S phase; (2) bortezomib up-regulates IRF4 mRNA independent of the cell cycle but reduces the IRF4 protein, greater in pG1 and further in pG1-S; (3) loss of IRF4 protein is associated with enhanced caspase-3 activation by bortezomib in both pG1 and pG1-S; and (4) ectopic IRF4 expression attenuates bortezomib killing in pG1-S. Little is known about posttranscriptional regulation of IRF4. In response to lenalidomide, IRF4 appears to be downstream of cereblon,44 an E3 ligase that mediates thalidomide action,45 and is also subject to translational control in myeloma cells.46 Our findings highlight the complexity of IRF4 regulation and suggest a central role for IRF4 in linking the cell cycle to apoptosis in myeloma cells.
Loss of IRF4 apparently sensitizes myeloma cells to bortezomib killing in cooperation with cell cycle–dependent gain of Noxa and Bim. Noxa is selectively required to enhance bortezomib killing in pG1-S but not in pG1, which dominantly suppresses the synthesis and up-regulation of Noxa by bortezomib. These results complement the finding that Noxa expression is induced by cell-cycle activation in primary B cells and required for apoptotic control of cycling B cells and iPCs, the rapidly cycling and apoptotic plasma cell precursors.47 By contrast, Bim mRNA expression and up-regulation by bortezomib are most prominent in pG1, and Bim functions to enhance bortezomib killing in both pG1 in the absence of Noxa and in pG1-S in cooperation with Noxa. These data reveal a previously unappreciated specificity with which the cell-cycle controls the expression and function of BH3-only proapoptotic genes.
The exceptional selectivity and reversibility make PD 0332991 an ideal reagent for cell-cycle synchronization and for determining the cell-cycle targeting specificity of cytotoxic agents. We have demonstrated that bortezomib preferentially targets myeloma cells in early S phase over in pG1, which provides a framework for optimizing bortezomib killing by sequential combination with PD 0332991. We further show that it is feasible to induce sequential pG1 and pG1-S in vivo with PD 0332991, which markedly enhanced tumor-specific killing by a subtherapeutic dose of bortezomib. This result corroborated our earlier observation that PD 0332991 prolonged the survival of mice treated with bortezomib in the 5T mouse myeloma model,27 and revealed that cell-cycle sensitization to bortezomib killing is tumor-specific. Most importantly, our data provide the first evidence that acceleration of early G1 arrest by inhibition of CDK4/CDK6 sensitizes the majority of primary human BM myeloma cells isolated from late stages of the disease to bortezomib killing in vitro despite BMSC protection, and this is associated with loss of IRF4.
As a therapeutic agent, PD 0332991 is orally bioavailable, highly specific, and low in toxicity.21,23 Based on this study and the promising clinical response in a single-agent study in MCL,28 we have implemented our strategy to target CDK4/CDK6 with PD 0332991 in combination with bortezomib in myeloma and MCL. Preliminary data have confirmed that PD 0332991 preferentially and completely inhibits CDK4 and CDK6 and halts the cell cycle in early G1 in tumor cells, within tolerable doses and with encouraging clinical response (R.N. and S.C.-K., unpublished data). Mechanism-based targeting of CDK4/CDK6 in sequential combination therapy may therefore have a significant therapeutic benefit for myeloma and MCL, and potentially other cancers. Elucidating how inhibition of CDK4/CDK6 controls apoptosis in the context of clinical responses should provide unique and invaluable insights into targeting the cell cycle in human cancer.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Lee M. Kiang, Peter Toogood, Beatrice Knudsen, and Catherine Pellat-Deceunynck for helpful discussions and David Chiron, Adriana Rossi, and Jackson Harvey for a critical reading of the manuscript.
This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research (A.L.S. and L.M.S.; NCI R01 120531, S.C.-K.), a Leukemia & Lymphoma Society Translational Research Program grant (S.C.-K.), and a Starr Cancer Consortium grant (W.C.H. and S.C.-K.).
National Institutes of Health
Authorship
Contribution: X.H., M.D., J.B., M.A.S.M., and S.C.-K. designed the experiments; X.H., M.D., D.J., J.L., S.E., and T.L. performed the experiments; A.L.S., I.C., S.R., W.C.H., L.M.S., and R.N. provided essential reagents and important input to the experiments performed; X.H., M.D., J.B., M.A.S.M., and S.C.-K. analyzed the data; and X.H., M.D., and S.C.-K. prepared the manuscript.
Conflict-of-interest disclosure: S.R. is employed by Pfizer. I.C. was employed by Pfizer. The remaining authors declare no competing financial interests.
The current affiliation for J.L. is Department of Science, Borough of Manhattan Community College, City University of New York, New York, NY.
Correspondence: Selina Chen-Kiang, Department of Pathology and Laboratory Medicine, Weill Cornell Medical College, 1300 York Ave, C-338, New York, NY 10065; e-mail: sckiang@med.cornell.edu.
References
Author notes
X.H. and M.D. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal