Abstract
Mutations of RUNX1 are detected in patients with myelodysplastic syndrome (MDS). In particular, C-terminal truncation mutations lack a transcription regulatory domain and have increased DNA binding through the runt homology domain. The expression of the runt homology domain, RUNX1(41-214), in mouse hematopoietic cells induced progression to MDS and acute myeloid leukemia. Analysis of premyelodysplastic animals found expansion of c-Kit+Sca-1+Lin− cells and skewed differentiation to myeloid at the expense of the lymphoid lineage. These abnormalities correlate with the phenotype of Runx1-deficient animals, as expected given the reported dominant-negative role of C-terminal mutations over the full-length RUNX1. However, MDS is not observed in Runx1-deficient animals. Gene expression profiling found that RUNX1(41-214) c-Kit+Sca-1+Lin− cells have an overlapping yet distinct gene expression profile from Runx1-deficient animals. Moreover, an unexpected parallel was observed between the hematopoietic phenotype of RUNX1(41-214) and aged animals. Genes deregulated in RUNX1(41-214), but not in Runx1-deficient animals, were inversely correlated with the aging gene signature of HSCs, suggesting that disruption of the expression of genes related to normal aging by RUNX1 mutations contributes to development of MDS. The data presented here provide insights into the mechanisms of development of MDS in HSCs by C-terminal mutations of RUNX1.
Introduction
Runx1 is essential for the specification of definitive HSCs.1 Homozygous germline deletion of Runx1 leads to embryonic lethality at E12.5 because of hemorrhaging in the central nervous system and lack of definitive hematopoiesis.2,3 The significance of Runx1 in adult hematopoiesis has been studied in conditional Runx1 knockout mice.4-7 Surprisingly, Runx1 was not essential for hematopoiesis in the adult hematopoietic compartment.6,7 However, further studies reported the importance of Runx1 in the homeostasis of hematopoietic cells. c-Kit+Sca-1+Lin− (KSL) cells accounted for an enlarged share of cells lacking Runx1.4-7 Megakaryocytic differentiation was severely impaired, resulting in decreased platelet numbers in peripheral blood (PB).4,5 Numbers of both B and T lymphocytes in PB decreased, as did those of common lymphoid progenitors (CLP).4 Moreover, in aged Runx1 conditional knockout mice, the expansion of the stem cell compartment is no longer observed, resulting in stem cell exhaustion.8
The dysfunction of RUNX1 is strongly correlated to hematologic disorders. Point mutations of RUNX1 were first described in familial platelet disorder/acute myeloid leukemia (AML)9 and de novo AML,10,11 and later in patients with chronic myelomonocytic leukemia12,13 and myelodysplastic syndrome (MDS).14 The mutations are rarely overlapping and are dispersed throughout RUNX1, but, interestingly, 2 functionally distinct classes can be defined: N-terminal mutations, located within the runt homology domain (RHD), which disrupt DNA binding, and C-terminal mutations, which increase DNA binding but disrupt transcriptional activity.14
Previous studies reported that progressive deletion of regions other than RHD increased the binding of Runx1 in an inversely proportional manner.15 In this report, we selected 2 examples of C-terminal mutations, RUNX1(Ala224fsTer228) and RUNX1(41-214), for further characterization with the use of mouse models. RUNX1(Ala224fsTer228) is the shortest C-terminal mutation reported in patients with MDS14 ; RUNX1(41-214), consisting almost entirely of RHD, has a 69-fold higher affinity than the full-length protein,15 the highest affinity to DNA among reported mutations of RUNX1. The expression of RUNX1(41-214) induced MDS/AML with higher frequency in mouse models of hematopoietic cell transduction/transplantation. To gain insights into the process of development of MDS by RUNX1(41-214), the hematopoietic compartment at the premyelodysplastic phase were analyzed in RUNX1(41-214) animals. RUNX1(41-214) induces a number of alterations in the hematopoietic compartment before the appearance of detectable disease. Because of their combination of higher DNA binding and lack of transcription activation, it has been proposed that C-terminal truncation mutations of RUNX1 have a dominant-negative effect over the transcriptional activity of RUNX1. The phenotype detected in RUNX1(41-214) animals is partly shared with the hematopoietic phenotype of mice deficient for Runx1,4-7 which supports the claim of its dominant-negative effect over the full-length RUNX1. In addition, an unexpected similarity was observed between the hematopoietic phenotype of RUNX1(41-214) animals and aged animals.16-19
To understand the molecular mechanisms of C-terminal mutations of RUNX1 in the development of MDS, gene expression profiling was performed on KSL cells from RUNX1(41-214) animals, Runx1-deficient animals (Runx1floxed/floxed MxCre+/−), and control animals (Runx1floxed/floxed MxCre−/−). The majority of genes (59 of 83) with significant change in expression in Runx1-deficient animals was also represented in RUNX1(41-214) animals compared with controls, showing the same direction of change but higher differential expression. An additional 268 genes were significantly deregulated in RUNX1(41-214) cells but not in Runx1-deficient cells compared with controls. Furthermore, gene set enrichment analysis (GSEA) indicated an inverse correlation between the RUNX1(41-214) gene expression profile and the HSC aging signature.20
Together, these results suggest that C-terminally truncated mutants of RUNX1, because of their increased affinity to DNA, not only induce a Runx1-deficient state in HSCs but also deregulate additional target genes. The GSEA suggests at least partial involvement of deregulation of aging-related genes in this process. The data presented here provide insights into the mechanisms of development of MDS in the HSC by the expression of C-terminal mutations of RUNX1.
Methods
Animals
All experimental protocols were approved by the Institutional Animal Care and Use Committee of the University of California, San Diego (UCSD). Wild-type C57B6/J mice and Runx1 conditional knockout (Runx1floxed/floxed MxCre+/−) mice were housed and bred at the vivarium of the Moores Cancer Center at UCSD. The Runx1 conditional knockout animals were kindly provided by Dr Nancy Speck (University of Pennsylvania).4
Retroviral transduction and primary bone marrow transplantation
For production of retrovirus, 10 μg of retroviral vector plasmid were cotransfected with 10 μg of Ecopac (pIK6.1MCV.ecopac.UTd) in confluent 10-cm plates of HEK293T by Ca3(PO4)2 precipitation. The virus-containing cell culture medium was collected 48 hours after transfection. Hematopoietic cells from liver of E16.5 mice were collected in aseptic conditions and were cultured in IMDM with 15% FBS, 1% penicillin-streptomycin, 1% glutamine, and 4% each of conditioned media from the BHK-MKL cell line (source of SCF) and X63AG-653 (source of IL-3), at the concentration of 2 × 106 cells/mL. The cells were prestimulated overnight, and retroviral infection was performed by adding retroviral supernatant to the media (35% of the total volume), with 8 μg/mL polybrene. Cells were spinoculated at 1200g for 3 hours at 32°C in an Allegrea-12R centrifuge with a SX4750 rotor (Beckman Coulter). The procedure was repeated on the next day. One day after the last retroviral infection, the percentage of enhanced green fluorescent protein-positive (EGFP+) cells in the population was measured by flow cytometry, and the concentration of EGFP+ cells was adjusted to 20% with the use of mock-transduced fetal liver cells. The cells were resuspended in PBS at the concentration of 1 × 107 cells/mL, and 2 × 106 cells per mouse were transplanted by tail vein injection into recipient animals irradiated with 8.5 Gy or 9 Gy total body irradiation. After transplantation, the animals were kept with acidic water (pH 3) for 10 days on regular housing environment. One month after transplantation, PB was collected by retro-orbital bleeding. Hematologic profiles were analyzed with Hemavet HV950FS (Drew Scientific), and PB smears were stained by Wright-Giemsa solution for cytologic analysis. Percentage of EGFP+ white blood cells was measured on blood lysed with ammonium chloride solution (150mM NH4Cl, 0.1mM EDTA, buffered with KHCO3 to pH 7.2-7.6). This procedure was repeated monthly on animals that received a transplant. In addition, the overall health status of the animals was examined daily, and animals showing signs of morbidity (dehydration, decrease in activity, pale membranes, and hunched posture) were humanely killed for analysis. PB, bone marrow (BM), and spleen samples were collected for histopathologic, flow cytometric, and cytologic analyses.
Inverse PCR
For details, see supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Flow cytometry
For details, see supplemental Methods.
Limiting dilution analysis
A previously published protocol was used with minor modifications.6 Limiting numbers of EGFP+ total BM cells from animals transplanted with RUNX1(41-214) or MigR1 were transplanted together with 2 × 105 total BM cells from C57BL/6J mice and injected into lethally irradiated 9.5 Gy recipient mice. Reconstitution was evaluated 4 months after transplantation. Mice were considered positive when the percentage of chimerism (EGFP+ cells) was > 1% with expression of myeloid and lymphoid markers. The frequency of long-term engrafting cells was calculated with the L-Calc Version 1.1.1 software (StemCell Technologies).
Homing assay
Sorted EGFP+ KSL cells (2.5 × 104) from animals transplanted with MigR1 or RUNX1(41-214) were injected into recipients irradiated with a lethal dose of irradiation 9 Gy 3 hours before injection. Animals were killed 16 hours after injection, and the number of EGFP+ cells was analyzed by flow cytometry.
Gene expression profiling and statistical analysis
KSL cells (c-Kit+/Sca-1+/Lin−/IL7Ra−) from the Runx1-deficient (Runx1floxed/floxed MxCre+/−, 3 months after poly IC treatment) group, the control (Runx1floxed/floxed MxCre−/−, 3 months after poly IC treatment) group, and the pre-MDS RUNX1(41-214) group, each one containing 4-20 animals, were sorted and total RNA was collected. Three independent experiments were performed. Transcription and labeling were performed according to the manufacturer's recommendations (Affymetrix) and hybridized on the Affymetrix mouse 430A microarray (Gene Expression Omnibus no. GSE40155). dChip Version 2008 was used for microarray analysis.21 Data were normalized with the invariant set method22 and were filtered with an F-statistic with P < .02, based on an analysis of variance comparison among the 3 groups. Differential expression between groups was evaluated with nominal P < .05, based on a 2-sided t test, a fold change > 2.0, and a present call of ≥ 33%. No adjustment was made for multiple comparisons. Hierarchical clustering was performed with a Pearson correlation distance metric and centroid linkage. GSEA was performed with GenePattern 2.0 software from the Broad Institute.23 The correlation of the Norddhal aging gene set with a phenotype was considered significant,20 based on a nominal P < .01, and the enrichment score (ES) is reported.
For other experiments that compared continuous measures, a 2-sided t test was performed and considered significant at the .05 level. Survival curves were estimated with the method of Kaplan and Meier, calculated from the date of transplantation to the date of death and compared with a log-rank test.
Quantitative RT-PCR and immunoblotting
For details, see supplemental Methods.
Results
MDS and AML are observed in mice transplanted with RUNX1(41-214)–expressing cells
Point mutations in RUNX1 resulting in frameshifts and C-terminally truncated proteins are reported in patients with MDS. Two examples of C-terminal mutations of RUNX1, the RUNX1(Ala224fsTer228) and RUNX1(41-214) (Figure 1A), were selected for retroviral transduction/transplantation in mice models, with MigR1 vector as control (supplemental Figure 1). The full-length RUNX1b would be another desirable control in this experiment. However, as reported previously,24 hematopoietic cells expressing the full-length RUNX1 engrafted poorly in recipients of transplants, for reasons not well understood.
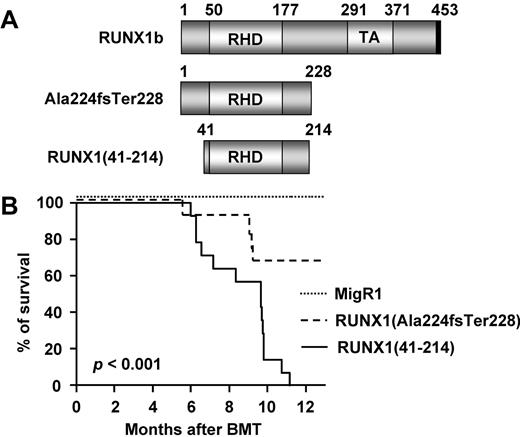
Retroviral transduction/transplantation assays with the use of RUNX1(Ala224fsTer228) or RUNX1(41-214). (A) Schematic representation of full-length RUNX1b, RUNX1(Ala224fsTer228), and RUNX1(41-214). Numbers represent amino acid numbering, based on the human RUNX1 isoform b. (B) Kaplan-Meier survival curve of animals transplanted with control vector (MigR1; n = 17), RUNX1(Ala224fsTer228) (n = 12), and RUNX1(41-214) (n = 15) of 4 independent sets of experiments. TA indicates transcriptional activation domain; and BMT, bone marrow transplantation.
Retroviral transduction/transplantation assays with the use of RUNX1(Ala224fsTer228) or RUNX1(41-214). (A) Schematic representation of full-length RUNX1b, RUNX1(Ala224fsTer228), and RUNX1(41-214). Numbers represent amino acid numbering, based on the human RUNX1 isoform b. (B) Kaplan-Meier survival curve of animals transplanted with control vector (MigR1; n = 17), RUNX1(Ala224fsTer228) (n = 12), and RUNX1(41-214) (n = 15) of 4 independent sets of experiments. TA indicates transcriptional activation domain; and BMT, bone marrow transplantation.
Periodical analysis of PB of animals that received a transplant showed that RUNX1(41-214) and RUNX1(Ala224fsTer228) animals had chronic thrombocytopenia (supplemental Figure 2). White blood cell count and hemoglobin levels did not differ significantly from controls, but mean corpuscular volume increased sharply from 8 months after transplantation.
Signs of morbidity and death were observed from 6 months after transplantation in RUNX1(41-214) animals (Figure 1B). By 11 months, all animals in the RUNX1(41-214) group were dead because of disease (mean survival time, 259 days). In animals transplanted with RUNX1(Ala224fsTer228), the penetrance of disease was lower, and at the termination of the experiment (13 months) 8 of 12 animals were still alive without any obvious signs of disease. Therefore, subsequent studies were focused on the analysis of RUNX1(41-214) animals.
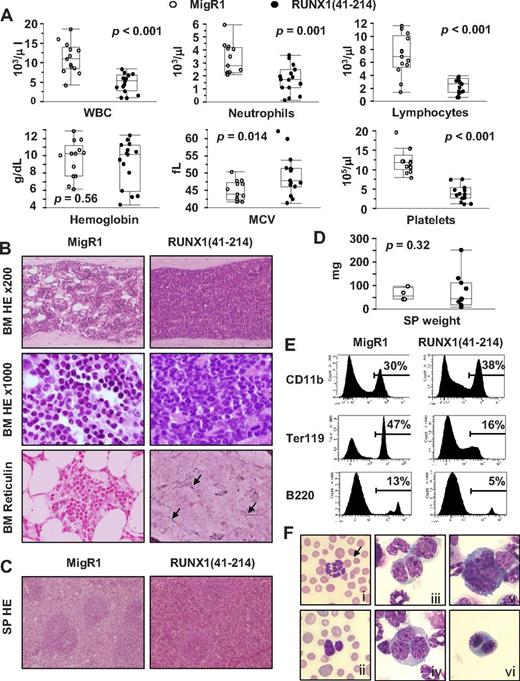
RUNX1(41-214) animals showing lethargy and a hunched posture were killed for analysis of the hematopoietic compartment. PB analysis of moribund animals showed leucopenia with marked lymphopenia and severe thrombocytopenia. Hemoglobin levels were decreased in some animals but were close to normal in others, and this difference was not significant when RUNX1(41-214) animals were analyzed as a group. A significant increase in mean corpuscular volume or macrocytosis was also observed, which was suggestive of dyserythropoiesis25 (Figure 2A; supplemental Table 3). BM of moribund RUNX1(41-214) animals was hypercellular, with dysmegakaryopoiesis and a predominant myeloid population. In addition, in some animals, fibrosis was observed, which was confirmed by reticulin staining (Figure 2B). Histologically, the splenic architecture of moribund RUNX1(41-214) animals was preserved, but the red pulp was expanded, suggesting extramedullary hematopoiesis (Figure 2C). The spleens were slightly enlarged, but the difference was not significant compared with control animals (Figure 2D). Differential count of BM cells showed an increase in blasts in RUNX1(41-214) animals compared with controls (6.4 ± 1.0, n = 4 vs 1.9 ± 0.4, n = 3, respectively). Flow cytometric analysis of BM cells confirmed the predominance of cells of the myeloid lineage by increased expression of CD11b in addition to a pronounced decrease in cells of erythroid lineage (Ter119+ cells). Moreover, cells of the B-cell lineage (B220+ cells) were also decreased in BM of RUNX1(41-214) animals (Figure 2E). Cytologic analysis of BM cytospins showed a number of dysplastic features in RUNX1(41-214) animals. Hypersegmented neutrophils and the Pelger-Huët anomaly were observed in BM and PB (Figure 2Fi-ii). Polychromasia, anysocytosis, and Howell-Jolly bodies were detected on morphologic analysis of red blood cells (see red blood cells in Figure 2Fi-ii). Binucleated and trinucleated promyelocytes (Figure 2Fiii-iv), micromegakaryocytes (Figure 2Fv), and binucleated prorubricytes (Figure 2Fvi) were observed in BM. On the basis of the findings of neutropenia and thrombocytopenia in PB and trilineage dysplasia in BM, the moribund RUNX1(41-214) animals were diagnosed as MDS, according to the Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice.26 In addition, a small number of RUNX1(41-214) animals (3 of 15) presented with AML. Clinical findings for these animals are described in detail in supplemental Tables 4 and 5 and supplemental Figure 3. Splenomegaly and an increase in the percentage of blasts in BM were observed. Surface marker expression of leukemic cells showed an increase in an abnormal population expressing c-Kit in addition to mature lineage markers (CD11b, Gr-1, and B220). The analysis of retroviral insertion sites in MDS samples from RUNX1(41-214) animals did not detect any common integration sites that would justify the frequency of MDS observed in animals that received a transplant (supplemental Table 6). Moreover, the MDS observed in Ala224fsTer228 animals (data not shown) was comparable with that in RUNX1(41-214) animals. The results described in this section indicate that the expression of RUNX1(41-214) in mouse BM induces MDS and AML, in accordance with observation of C-terminal deletion mutations of RUNX1 in patients with MDS.
MDS and AML is observed in animals transplanted with RUNX1(41-214)–expressing cells. (A) Peripheral blood parameters of moribund animals transplanted with RUNX1(41-214)–expressing cells (●) at the time of death. Values of healthy MigR1 animals (○), killed at the same time, are shown as control. (B) Histopathologic analysis of BM of MigR1 and RUNX1(41-214) animals. HE staining of BM is shown at original magnification of ×200 and ×1000. Reticulin staining of BM is shown at original magnification of ×200. Arrows point to reticulin fibers stained in black. (C) Histopathologic analysis of SP of MigR1 and RUNX1(41-214) animals. HE staining of SP is shown at original magnification of ×100. (D) SP weight of MigR1 (○) and RUNX1(41-214) animals (●) at the time of death. (E) Representative flow cytometric analysis of total BM of animals that received a transplant with MigR1 and RUNX1(41-214). (F) Cytologic analysis (Wright-Giemsa staining) of RUNX1(41-214) animals: (Fi) hypersegmented neutrophil in peripheral blood; arrow points to Howell-Jolly body in red blood cell; (Fii) Perger-Huët anomaly in peripheral blood, also note polychromasia and anisocytosis in red blood cells; (Fiii) binucleated neutrophilic metamyelocyte; (Fiv) trinucleated neutrophilic metamyelocyte; (Fv) micromegakaryocyte; (Fvi) binucleated prorubricyte (original magnification, ×1000). WBC indicates white blood cells; MCV, mean corpuscular volume; HE, hematoxylin-eosin; and SP, spleen. All images were acquired with BX51 microscope with objective lenses 10× PlanFLN/0.3, 20× PlanFLN/1.42, 100× PlanFLN/1.3, equipped with DP71 digital camera (Olympus).
MDS and AML is observed in animals transplanted with RUNX1(41-214)–expressing cells. (A) Peripheral blood parameters of moribund animals transplanted with RUNX1(41-214)–expressing cells (●) at the time of death. Values of healthy MigR1 animals (○), killed at the same time, are shown as control. (B) Histopathologic analysis of BM of MigR1 and RUNX1(41-214) animals. HE staining of BM is shown at original magnification of ×200 and ×1000. Reticulin staining of BM is shown at original magnification of ×200. Arrows point to reticulin fibers stained in black. (C) Histopathologic analysis of SP of MigR1 and RUNX1(41-214) animals. HE staining of SP is shown at original magnification of ×100. (D) SP weight of MigR1 (○) and RUNX1(41-214) animals (●) at the time of death. (E) Representative flow cytometric analysis of total BM of animals that received a transplant with MigR1 and RUNX1(41-214). (F) Cytologic analysis (Wright-Giemsa staining) of RUNX1(41-214) animals: (Fi) hypersegmented neutrophil in peripheral blood; arrow points to Howell-Jolly body in red blood cell; (Fii) Perger-Huët anomaly in peripheral blood, also note polychromasia and anisocytosis in red blood cells; (Fiii) binucleated neutrophilic metamyelocyte; (Fiv) trinucleated neutrophilic metamyelocyte; (Fv) micromegakaryocyte; (Fvi) binucleated prorubricyte (original magnification, ×1000). WBC indicates white blood cells; MCV, mean corpuscular volume; HE, hematoxylin-eosin; and SP, spleen. All images were acquired with BX51 microscope with objective lenses 10× PlanFLN/0.3, 20× PlanFLN/1.42, 100× PlanFLN/1.3, equipped with DP71 digital camera (Olympus).
Expansion of the stem/progenitor cell compartment in RUNX1(41-214) animals
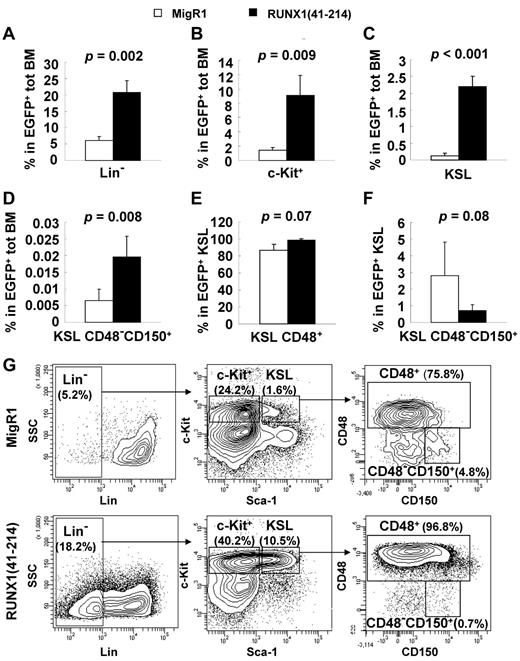
The expression of RUNX1(41-214) in primary BM cells induced MDS in animals that received a transplant after a relatively long latency. To search for possible clues that could explain the progression to MDS by the expression of RUNX1(41-214), premyelodysplastic RUNX1(41-214) animals without any signs of morbidity, at 4-6 months after transplantation, were used for analysis of the hematopoietic compartment. Because the percentage of EGFP-expressing cells can vary across animals in a retroviral transplantation model, the EGFP+ total BM is used for comparison of different hematopoietic compartments between RUNX1(41-214) and control MigR1 animals. RUNX1(41-214) and MigR1 animals did not differ significantly in total number of BM cells (data not shown). In RUNX1(41-214)–expressing BM cells, the lineage-negative compartment was significantly expanded (Figure 3A). The myeloid progenitor compartment (c-Kit+) was expanded 6-fold in RUNX1(41-214) animals compared with controls (Figure 3B), and the KSL compartment was expanded 16-fold compared with controls in RUNX1(41-214) BM (Figure 3C). Although the percentage of KSL CD48−CD150+ cells, or long-term HSCs (LT-HSCs), in total BM was significantly increased in RUNX1(41-214) animals (Figure 3D), careful analysis of the KSL compartment found an imbalance within its subpopulations. In the RUNX1(41-214) KSL compartment, the percentage of CD48+ cells, which are mostly composed of multipotential progenitors, was elevated in the KSL compartment (Figure 3E). Accordingly, the percentage of LT-HSCs was lower, accounting for 0.72% ± 0.35% of cells in the KSL compartment of RUNX1(41-214), in contrast to 2.8% ± 2.0% in control animals (Figure 3F-G).
Analysis of the premyelodysplastic hematopoietic compartment in RUNX1(41-214) animals. (A-F) Percentage of various hematopoietic populations analyzed on flow cytometry. Average and SD values of MigR1 (□; n = 4) and RUNX1(41-214) (■; n = 5) animals are shown. Animals were analyzed 4-6 months after transplantation. Percentage represents values within EGFP+/RUNX1(41-214) cells. (G) Representative flow cytometric analysis of the stem/progenitor cell compartment. Lin− indicates EGFP+/Lin−/IL7Rα−; c-Kit+, EGFP+/Lin−/IL7Rα−/c-Kit+/Sca-1−; and KSL, EGFP+/Lin−/IL7Rα−/c-Kit+/Sca-1+.
Analysis of the premyelodysplastic hematopoietic compartment in RUNX1(41-214) animals. (A-F) Percentage of various hematopoietic populations analyzed on flow cytometry. Average and SD values of MigR1 (□; n = 4) and RUNX1(41-214) (■; n = 5) animals are shown. Animals were analyzed 4-6 months after transplantation. Percentage represents values within EGFP+/RUNX1(41-214) cells. (G) Representative flow cytometric analysis of the stem/progenitor cell compartment. Lin− indicates EGFP+/Lin−/IL7Rα−; c-Kit+, EGFP+/Lin−/IL7Rα−/c-Kit+/Sca-1−; and KSL, EGFP+/Lin−/IL7Rα−/c-Kit+/Sca-1+.
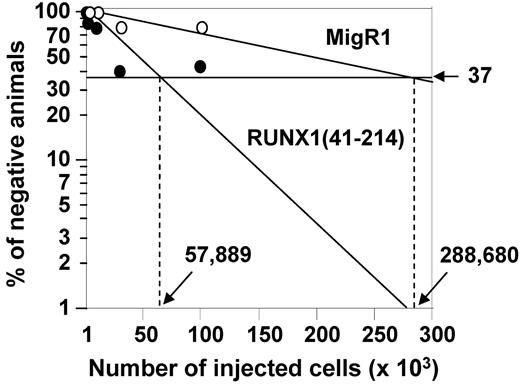
To determine the quantity of functionally defined HSCs in RUNX1(41-214) animals, the engraftment potential of HSCs from RUNX1(41-214) animals was examined with limiting dilution transplantation assays. As shown in Figure 4 and Table 1, the proportion of long-term engrafting cells was 5-fold higher in RUNX1(41-214) BM than in controls. Comparing this result with the values defined by surface marker expression, a 16-fold increase in KSL frequency and approximately a 3-fold increase in the frequency of KSL CD48−CD150+ cells in RUNX1(41-214) cells, we concluded that the KSL CD48−CD150+ marker best defined the population of long-term engrafting cells in RUNX1(41-214) BM. The increase in KSL population, on the other hand, was not proportional to the increase in long-term engrafting cells, possibly because of the expansion of cells with limited long-term engraftment ability, resulting in a dilution of the “true” long-term engrafting cells within the KSL compartment.
Limiting dilution analysis based on competitive repopulation. Graphical representation of results from limiting dilution analysis of EGFP+ total BM cells from animals transplanted with cells expressing MigR1 (○) and RUNX1(41-214) (●).
Limiting dilution analysis based on competitive repopulation. Graphical representation of results from limiting dilution analysis of EGFP+ total BM cells from animals transplanted with cells expressing MigR1 (○) and RUNX1(41-214) (●).
Limiting dilution analysis of RUNX1(41-214) BM
| . | MigR1* . | RUNX1(41-214)† . | ||
|---|---|---|---|---|
| Responses . | Tested . | Responses . | Tested . | |
| Cell dose | ||||
| 1 000 | 0 | 7 | 0 | 6 |
| 3 000 | 0 | 8 | 1 | 7 |
| 10 000 | 0 | 9 | 2 | 9 |
| 30 000 | 2 | 9 | 6 | 10 |
| 100 000 | 2 | 9 | 4 | 7 |
| . | MigR1* . | RUNX1(41-214)† . | ||
|---|---|---|---|---|
| Responses . | Tested . | Responses . | Tested . | |
| Cell dose | ||||
| 1 000 | 0 | 7 | 0 | 6 |
| 3 000 | 0 | 8 | 1 | 7 |
| 10 000 | 0 | 9 | 2 | 9 |
| 30 000 | 2 | 9 | 6 | 10 |
| 100 000 | 2 | 9 | 4 | 7 |
Frequency was 1 in 288 680.
Frequency was 1 in 57 889.
A similar phenomenon was observed in BM of aged animals. It is well-known that HSCs are expanded in aged BM; however, the per-cell efficiency of long-term engraftment of KSLs seemed to be lower than in young animals.16 Detailed analysis of KSLs of aged animals found that they contained an expanded CD48+ population that lacked long-term reconstituting ability,19 similarly to what is observed in RUNX1(41-214) animals.
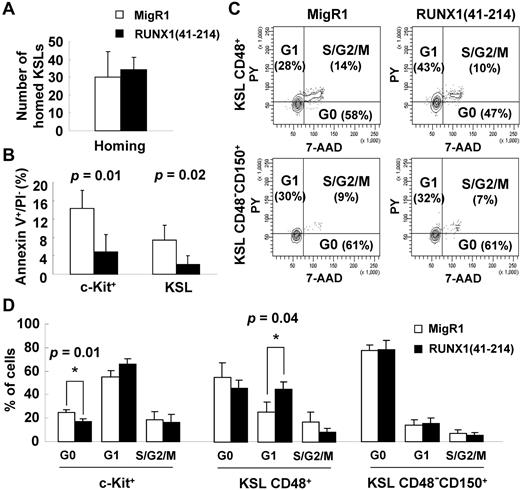
To understand the mechanisms of the abnormal expansion of the stem/progenitor cell compartment, the factors that affect the homeostasis of stem cells were examined. The homing ability of RUNX1(41-214) cells was similar to that of controls (Figure 5A), which excludes the possibility that the increase in engraftment of those cells was because of increased homing to the BM. Apoptosis of RUNX1(41-214) stem/progenitor cells was significantly decreased compared with controls (Figure 5B). With respect to cell cycle distribution, as expected from the previous results, the most immature KSL CD48−CD150+ cells in RUNX1(41-214) were not affected. However, in KSL CD48+ cells, a decrease in cells in the quiescent G0 phase and an increase in cells in the G1 phase of the cell cycle were observed (Figure 5C-D). RUNX1 RHD expression is known to inhibit G1 to S transition.27 This property of RUNX1 may be of particular importance in HSCs, where the G1 phase is a sensitive period in which decisions between self-renewal and differentiation are made.28
Reduced apoptosis and increased cell cycle entry of stem/progenitor cells in RUNX1(41-214) animals. (A) Homing assay of KSL cells from MigR1 (□; n = 3) and RUNX1(41-214) (■; n = 3); numbers represent KSLs homed to bone marrow per 1 million injected cells. (B) Annexin V staining of stem/progenitor cells in MigR1 (□; n = 3) and RUNX1(41-214) (■; n = 3) animals. (C) Representative cell cycle analysis with Pyronin Y/7-AAD in MigR1 and RUNX1(41-214) animals. (D) Cell cycle analysis of stem/progenitor cells of in MigR1 (□) and RUNX1(41-214) (■) animals that received a transplant. Results are average of 3 independent experiments. Three to 6 animals per group were used in each experiment. Only statistically significant P values are shown (*P < .05). All analyses were performed at 4-6 months after transplantation. c-Kit+ indicates EGFP+/Lin−/IL7Rα−/c-Kit+/Sca-1−; KSL, EGFP+/Lin−/IL7Rα−/c-Kit+/Sca-1+; PY, pyronin Y; KSL CD48+, EGFP+/Lin−/IL7Rα−/c-Kit+/Sca-1+/CD48+; and KSL CD48−CD150+, EGFP+/Lin−/IL7Rα−/c-Kit+/Sca-1+/CD48−/CD150+.
Reduced apoptosis and increased cell cycle entry of stem/progenitor cells in RUNX1(41-214) animals. (A) Homing assay of KSL cells from MigR1 (□; n = 3) and RUNX1(41-214) (■; n = 3); numbers represent KSLs homed to bone marrow per 1 million injected cells. (B) Annexin V staining of stem/progenitor cells in MigR1 (□; n = 3) and RUNX1(41-214) (■; n = 3) animals. (C) Representative cell cycle analysis with Pyronin Y/7-AAD in MigR1 and RUNX1(41-214) animals. (D) Cell cycle analysis of stem/progenitor cells of in MigR1 (□) and RUNX1(41-214) (■) animals that received a transplant. Results are average of 3 independent experiments. Three to 6 animals per group were used in each experiment. Only statistically significant P values are shown (*P < .05). All analyses were performed at 4-6 months after transplantation. c-Kit+ indicates EGFP+/Lin−/IL7Rα−/c-Kit+/Sca-1−; KSL, EGFP+/Lin−/IL7Rα−/c-Kit+/Sca-1+; PY, pyronin Y; KSL CD48+, EGFP+/Lin−/IL7Rα−/c-Kit+/Sca-1+/CD48+; and KSL CD48−CD150+, EGFP+/Lin−/IL7Rα−/c-Kit+/Sca-1+/CD48−/CD150+.
Skewing to myelopoiesis at the expense of lymphopoiesis in RUNX1(41-214) animals
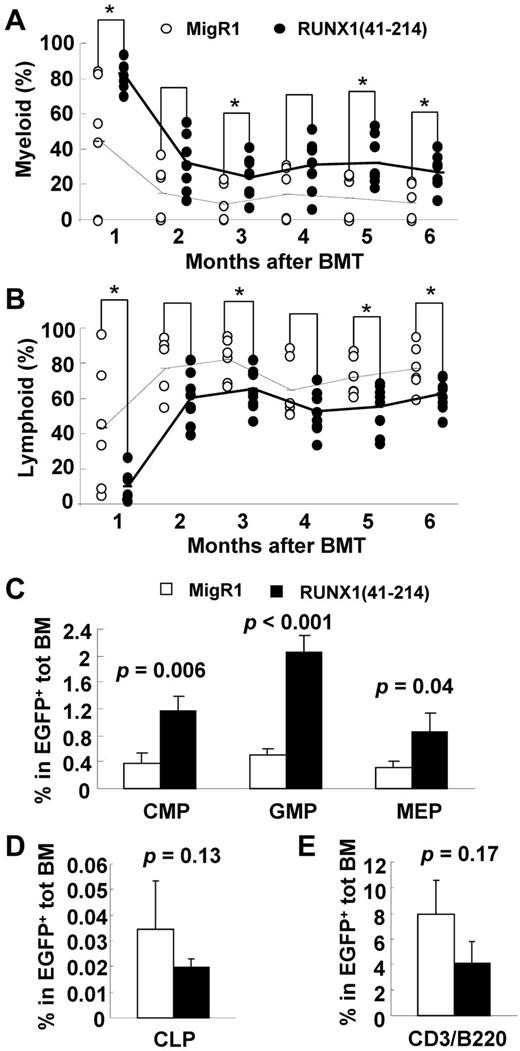
The PB of RUNX1(41-214) animals showed an increase in the percentage of myeloid and a decrease in the percentage of lymphoid cells compared with controls (Figure 6A-B). To determine the origin of this imbalance, the progenitors of those lineages were examined in the BM. As shown in Figure 6C, in accordance with the expansion of the c-Kit+ myeloid progenitor compartment (Figure 3C), all myeloid progenitors, including common myeloid progenitors, granulocyte-macrophage progenitors, and megakaryocyte-erythrocyte progenitors, were expanded in RUNX1(41-214) BM, and among them, the granulocyte-macrophage progenitors showed the most pronounced expansion. The percentage of CLP cells was lower in RUNX1(41-214) animals than in controls, as was the number of more mature lymphocytes in BM. However, in neither population was the difference statistically significant. These results indicated that in RUNX1(41-214)–expressing hematopoietic cells the skewing to the myeloid lineage at the expense of the lymphoid occurs at the early stages of lineage commitment from the HSCs, with pronounced myeloid expansion. Deviation from normal ratios of differentiation into myeloid and lymphoid lineages has also been observed in aged HSCs.18,29
Skewing to myelopoiesis in RUNX1(41-214) animals. Flow cytometric analysis of percentage of (A) myeloid (CD11b or Gr-1) and (B) lymphoid (CD3 or B220) populations in peripheral blood of MigR1 (○) or RUNX1(41-214) (●) animals. Dashed lines connect average values for MigR1 animals, and solid lines connect average values of RUNX1(41-214) animals. *P < .05. BMT indicates bone marrow transplantation. Flow cytometric analysis of (C) myeloid progenitors, (D) common lymphoid progenitors, and (E) CD3- or B220-expressing cells in bone marrow of MigR1 (□) or RUNX1(41-214) (■) animals. Values represent average and SD within EGFP+ population. Experiments were performed 4-6 months after transplantation; n = 3 animals were used for each group. CMP, indicates common myeloid progenitor; GMP, granulocyte-macrophage progenitor; MEP, megakaryocyte-erythrocyte progenitor; and CLP, common lymphoid progenitor.
Skewing to myelopoiesis in RUNX1(41-214) animals. Flow cytometric analysis of percentage of (A) myeloid (CD11b or Gr-1) and (B) lymphoid (CD3 or B220) populations in peripheral blood of MigR1 (○) or RUNX1(41-214) (●) animals. Dashed lines connect average values for MigR1 animals, and solid lines connect average values of RUNX1(41-214) animals. *P < .05. BMT indicates bone marrow transplantation. Flow cytometric analysis of (C) myeloid progenitors, (D) common lymphoid progenitors, and (E) CD3- or B220-expressing cells in bone marrow of MigR1 (□) or RUNX1(41-214) (■) animals. Values represent average and SD within EGFP+ population. Experiments were performed 4-6 months after transplantation; n = 3 animals were used for each group. CMP, indicates common myeloid progenitor; GMP, granulocyte-macrophage progenitor; MEP, megakaryocyte-erythrocyte progenitor; and CLP, common lymphoid progenitor.
Gene expression signature of RUNX1(41-214) overlapped with, yet was distinct from, that of Runx1-deficient KSL cells
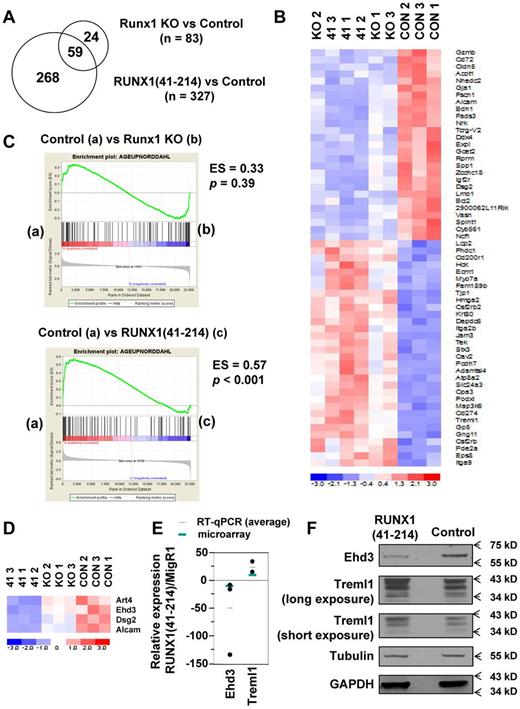
It has been proposed that C-terminally truncated RUNX1 proteins negatively regulate Runx1 function because of their high affinity to DNA and lack of transcriptional activity. We observed expansion of stem/progenitor cells and skewing toward myelopoiesis in RUNX1(41-214)–expressing mice, phenomena that have also been reported in Runx1-deficient mice.4,6,7 However, these abnormal phenotypes are more severe in RUNX1(41-214) mice than that in Runx1-deficient mice. Furthermore, RUNX1(41-214) animals developed MDS/AML, which was not observed in Runx1-deficient mice. These observations suggest that expression of RUNX1(41-214) may affect not only genes normally regulated by Runx1 but also genes that are independent of Runx1. To verify this, gene expression profiling was performed on KSL cells from Runx1-deficient and predisease stage RUNX1(41-214) animals. With a > 2-fold cutoff, the number of differentially expressed genes was higher in RUNX1(41-214) KSL cells versus control (n = 327) than in Runx1-deficient KSL cells versus control (n = 83) (Figure 7A; supplemental Figures 4-5; supplemental Tables 7-8). Fifty-nine differentially expressed genes were found in common between Runx1-deficient versus control and RUNX1(41-214) versus control (Figure 7B; supplemental Table 9). These common genes showed the same direction of change in both groups, with the RUNX1(41-214) group generally showing a larger change. Among these 59 genes are known targets of Runx1 and leukemic translocations that involve Runx1 (Csf2rb,30 Gja1,31 Hmga2,32 Hck,33 Bcl2,34 and Cd200r135 ).
Gene expression profiling and GSEA of KSLs. (A) Venn diagram showing numbers of genes differentially expressed between RUNX1(41-214) versus control and Runx1-deficient versus control and the number of genes in common between the 2 groups. (B) Hierarchical clustering of genes in common between RUNX1(41-214) and Runx1-deficient cells (nominal P < .05; fold change > 2). (C) GSEA of correlation between aging signature genes and genes differentially expressed in RUNX1(41-214) versus Runx1-deficient cells. (D) Genes from the aging signature gene set show differential expression between RUNX1(41-214) versus Runx1-deficient cells. (E) Analysis of relative gene expression by quantitative RT-PCR in RUNX1(41-214) KSL cells compared with control MigR1 KSL cells. (F) Immunoblot analysis of bone marrow lineage-negative cells expressing RUNX1(41-214). 41 indicates RUNX1(41-214) KSL; Runx1 KO, Runx1floxed/floxed MxCre+/− KSL; and CON, Runx1floxed/floxed MxCre−/− KSL.
Gene expression profiling and GSEA of KSLs. (A) Venn diagram showing numbers of genes differentially expressed between RUNX1(41-214) versus control and Runx1-deficient versus control and the number of genes in common between the 2 groups. (B) Hierarchical clustering of genes in common between RUNX1(41-214) and Runx1-deficient cells (nominal P < .05; fold change > 2). (C) GSEA of correlation between aging signature genes and genes differentially expressed in RUNX1(41-214) versus Runx1-deficient cells. (D) Genes from the aging signature gene set show differential expression between RUNX1(41-214) versus Runx1-deficient cells. (E) Analysis of relative gene expression by quantitative RT-PCR in RUNX1(41-214) KSL cells compared with control MigR1 KSL cells. (F) Immunoblot analysis of bone marrow lineage-negative cells expressing RUNX1(41-214). 41 indicates RUNX1(41-214) KSL; Runx1 KO, Runx1floxed/floxed MxCre+/− KSL; and CON, Runx1floxed/floxed MxCre−/− KSL.
To examine if the gene expression profile of RUNX1(41-214) animals correlates with gene alterations observed in CD34+ cells in patients with MDS, the genes found to be most significantly deregulated in microarray studies36-42 were compared with genes significantly affected by RUNX1(41-214). The genes involved in angiogenesis, Vegfc and Tek (Tie2), were among the deregulated genes.43 Two cathepsin family genes, Ctsl and Ctsh, are among the differentially expressed genes in RUNX1(41-214). Proteolytic enzymes are known to be involved in stem cell mobilization and may be involved in the alterations in the stem cell niche in MDS.44 Tjp1, a representative gene of the tight junction pathway, was also differentially expressed in patients with MDS.38 Dntt37,39 was found to be consistently down-regulated in patients and in RUNX1(41-214). Dntt is a DNA polymerase with restricted expression in normal pre-B and pre-T lymphocytes during early differentiation.
Because the expression of RUNX1(41-214) induced a phenotype that is reminiscent of aged hematopoietic cells, it is possible that RUNX1(41-214) is involved in the deregulation of genes related to aging. GSEA was used to verify this hypothesis.23 A gene set based on genes up-regulated with age in the report by Norddahl et al of gene expression profiling of HSCs of aged versus young animals was used in GSEA to examine the correlation with the expression profile of RUNX1(41-214) KSLs.20 As shown in Figure 7C top, the Norddahl aging signature gene set did not show correlation with control versus Runx1-deficient KSL data (ES = 0.33; nominal P = .39; Figure 7C top); however, when the gene expression data from control versus RUNX1(41-214) (ES = 0.57; nominal P < .001) and Runx1-deficient versus RUNX1(41-214) (ES = 0.63; nominal P < .001) were analyzed, a significant inverse correlation was observed (Figure 7C bottom; supplemental Figure 6A). Because the correlation with the aging signature was only observed in RUNX1(41-214) KSLs, genes specifically deregulated by RUNX1(41-214) may be the potential causative genes of the observed phenotypes. Of the 93 probe sets included in the gene set, 4 were differentially expressed in RUNX1(41-214) versus Runx1-deficient KSLs (supplemental Table 10) and were among the top-ranked 10 in the core enrichment set by GSEA: Dsg2, Art4, Alcam, and Ehd3. All 4 of these genes were down-regulated in RUNX1(41-214) (Figure 7D).
To validate our findings on the gene expression profiling of RUNX1(41-214) animals, some genes were chosen on the basis of significance and expression level for further analysis. Quantitative analysis of mRNA expression was performed on KSL cells sorted from RUNX1(41-214) animals (supplemental Figure 6B). Alcam is a cell adhesion molecule that has attracted attention because of its differential expression in several human tumors.45 Alcam was found consistently down-regulated in RUNX1(41-214) animals by analysis of quantitative RT-PCR and flow cytometry (supplemental Figure 6B-C). Finally, 2 genes, Ehd3 and Treml1, attracted our attention because of the magnitude of differential expression in mRNA from RUNX1(41-214) animals (Figure 7E). Ehd3 was one of the top-ranked age-related genes on GSEA. Ehd3 belongs to a family of newly characterized proteins involved in endocytic transport.46 Treml1 is highly expressed in megakaryocytes and platelets and is involved in platelet activation.47 Analysis of protein expression found that both genes showed the same direction of deregulation at both the mRNA and protein levels (Figure 7F).
Discussion
C-terminal deletions of RUNX1 are often reported in patients with MDS. The mutations are known to increase DNA binding through the RHD, possibly acting as a dominant-negative form over the full-length RUNX1. In this work we used RUNX1(41-214), a representative RUNX1 mutation that corresponds to the RHD of RUNX1, to examine the pathologic and molecular effects of C-terminally truncated RUNX1 mutations on hematopoiesis. Animals transplanted with RUNX1(41-214) died of MDS/AML. These results are in agreement with the observations of previous reports that used RUNX1 C-terminal mutations detected in patients with MDS.48 To gain insights into the process of development of MDS, RUNX1(41-214) animals at the premyelodysplastic phase were analyzed. In vivo, an expansion of KSL and myeloid progenitors was observed. However, the expansion of the subpopulations within the KSL compartment was not uniform, with expansion of long-term engrafting HSCs less pronounced than that of KSLs. The differentiation of RUNX1(41-214) cells was skewed to myelopoiesis at the expense of lymphopoiesis, with significant expansion of myeloid progenitors in BM. Chronic thrombocytopenia was also observed in animals that received a transplant. The hematopoietic defects observed in RUNX1(41-214) animals were partially in agreement with the phenotype observed in Runx1-deficient animals, supporting the thesis of a dominant-negative effect of C-terminally truncated Runx1 proteins over the full-length Runx1. However, the long-term consequence of RUNX1(41-214) expression is progression to MDS, which is not observed in Runx1-deficient animals. Moreover, a parallel has been found between the alterations observed in RUNX1(41-214) animals and the phenotypes observed in the hematopoietic system of aged animals.
Gene expression profiling corroborated our findings on phenotypic analysis. Most of the genes deregulated in Runx1-deficient cells were also found even more strongly deregulated in RUNX1(41-214) cells. By contrast, in RUNX1(41-214), most of the deregulated genes were unique to this mutation. The RUNX1(41-214) mutation lacks the generally recognized RUNX1 transactivation domain but has nuclear localization (supplemental Figure 7A). Under special assay conditions, we also detected RUNX1(41-214) as a transcription activator and its synergistic cooperation with C/EBPα (supplemental Figure 7B-C). On the basis of these observations, some hypothetical mechanisms of gene regulation by RUNX1(41-214) can be outlined, apart from the dominant-negative role over the full-length Runx1. First, the higher affinity to DNA may allow binding of RUNX1(41-214) to RUNX1 consensus sequence sites that are not normally targets of RUNX1. Second, the RHD of RUNX1(41-214), through recruitment of cofactors, may be sufficient to drive expression or repression of Runx1-regulated genes. Third, the higher binding to DNA may alter the timing of the activation/repression of the gene. Finally, because RUNX family genes share significant homology at the RHD, and bind the same consensus sequence on the DNA, competition in DNA binding with Runx2 and Runx3 may also contribute to the gene deregulation observed in RUNX1(41-214). The 3 RUNX family proteins (RUNX1, RUNX2, RUNX3) share a highly homologous RHD and recognize the same consensus DNA binding sequence. However, N-terminal and C-terminal regions of these proteins differ considerably, suggesting that the 3 RUNX proteins may have both common and unique target genes via their interaction with other transcription regulators. In adult HSCs, Runx1 is the best studied RUNX family gene, partly because of its requirement for specification of hematopoiesis in the embryo. However, little is known about the role of Runx2 and Runx3 in adult HSCs. Expression of Runx249 and Runx322 have been reported in hematopoietic cells.
Premyelodysplastic RUNX1(41-214) HSCs showed a phenotype similar to aged HSCs. The aging of the hematopoietic compartment has been characterized in detail in mouse models. The lineage potential of the HSC shifts from lymphopoiesis toward myelopoiesis with aging,18,29 as has been observed in RUNX1(41-214) animals. HSCs, as determined by KSL marker, expand considerably in aged animals,16-18 but their long-term engraftment potential is substantially reduced in aged animals (1 in 78 in aged versus 1 in 5 in young animals). Analysis of stem cell populations within the KSL compartment found that, in aged animals, most of the cells expressed CD48, a marker of cells with limited engraftment potential. The number of true long-term engrafting cells, as defined by the expression of CD150, was significantly lower in old than in young animals.19 A similar phenomenon was observed in RUNX1(41-214)–expressing HSCs.
To examine whether this correlation also holds at the gene expression level, GSEA has been used to compare the expression profile of RUNX1(41-214) HSCs with an age-specific gene set. The set of genes up-regulated with aging was found to be inversely correlated with the gene expression profile of RUNX1(41-214) HSCs; in other words, some of genes up-regulated in healthy old animals were down-regulated in RUNX1(41-214) HSCs. However, HSCs from Runx1-deficient cells did not show correlation with the same gene set. A recent report, analyzing HSCs from Runx1-deficient cells, found that essentially all abnormalities observed in RUNX1(41-214) HSCs are also present in Runx-1–deficient HSCs.7 On the basis of this, we speculate that genes correlated with the aged gene set are more probably related to the progression of MDS and not specifically with the HSC phenotype of normal aging, although they may contribute to intensification of the HSC phenotype.7 The observation of an inverse correlation was an unexpected finding, and the reasons for this are not yet clear. Is Runx1 C-terminal mutation-associated MDS a process of “abnormal aging,” which involves deregulation of genes involved in aging, but in an abnormal fashion? What are the critical mechanisms affected by deregulation of those genes? These are interesting questions that need to be addressed in future studies.
Risk of myeloid diseases is known to increase with aging. In MDS in particular, the single greatest risk factor for development of disease is advanced age. Although the incidence of MDS in the United States is estimated at 3.8 per 100 000 people, rates are lowest for people younger than 40 years, at 0.14 per 100 000, and rise with age, to 36 per 100 000 for patients 80 years and older. The median age at diagnosis is 71 years.50 The phenotypic similarity observed between RUNX1(41-214)–expressing cells and aged hematopoietic cells with subsequent induction of MDS in RUNX1(41-214) animals indicates a possible involvement of mechanisms of abnormal aging in the evolution to MDS.
This study showed effects of the disruption of the homeostasis of the hematopoietic compartment by expression of RUNX1(41-214). The expression of RUNX1(41-214) induces MDS/AML. Moreover, a number of alterations in the hematopoietic compartment were detected in premyelodysplastic animals, which phenotypically resemble the hematopoietic compartment of Runx1-deficient and aged animals. Gene expression analysis of KSLs of RUNX1(41-214) animals found a gene expression profile overlapping with yet distinct from that of Runx1-deficient animals, indicating a unique property of these mutations in the deregulation of genes related to the homeostasis of the hematopoietic system. Genes deregulated exclusively in RUNX1(41-214) animals correlated with the gene signature of aged HSCs. The data presented here provide insights into the mechanisms of development of MDS in the HSC by the expression of C-terminal mutations of RUNX1.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Nancy Speck (University of Pennsylvania) for providing conditional Runx1-deficient mice and all members of the Zhang laboratory, especially Luke F. Peterson, for valuable suggestions. The authors are grateful for the generosity of Dr Daniel W. McVicar, National Cancer Institute, who provided the anti-TREML1 antibody and Dr Hamid Band, University of Nebraska Medical Center, who provided the anti-Ehd3 antibody.
This work was supported by funding from the National Institutes of Health (P01DK080665 and R01CA096735). S.M. is a recipient of the Ruth L. Kirschstein National Research Service Award (F32HL091641).
National Institutes of Health
Authorship
Contribution: S.M. planned and performed most experiments and wrote the manuscript; Y.K. performed retrovirus integration site analysis and other experiments; K.E.S. conducted gene expression data analysis; J.R.B. performed gene expression profiling-related studies; K.L. confirmed gene expression data; T.T., M.-C.L., X.C., and M.Y. helped with experiments; D.S.N. supervised microarray data analysis; and D.-E.Z. designed experimental approach, analyzed data, and supervised manuscript preparation.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for S.M. is Boston University School of Medicine, Whitaker Cardiovascular Institute, Boston, MA.
Correspondence: Dong-Er Zhang, Moores UCSD Cancer Center, University of California, San Diego, 3855 Health Sciences Drive, La Jolla, CA 92093; e-mail: d7zhang@ucsd.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal