Abstract
miRs play a critical role in tumor pathogenesis as either oncogenes or tumor-suppressor genes. However, the role of miRs and their regulation in response to proteasome inhibitors in multiple myeloma (MM) is unclear. In the current study, miR profiling in proteasome inhibitor MLN2238-treated MM.1S MM cells shows up-regulation of miR33b. Mechanistic studies indicate that the induction of miR33b is predominantly via transcriptional regulation. Examination of miR33b in patient MM cells showed a constitutively low expression. Overexpression of miR33b decreased MM cell viability, migration, colony formation, and increased apoptosis and sensitivity of MM cells to MLN2238 treatment. In addition, overexpression of miR33b or MLN2238 exposure negatively regulated oncogene PIM-1 and blocked PIM-1 wild-type, but not PIM-1 mutant, luciferase activity. Moreover, PIM-1 overexpression led to significant abrogation of miR33b- or MLN2238-induced cell death. SGI-1776, a biochemical inhibitor of PIM-1, triggered apoptosis in MM. Finally, overexpression of miR33b inhibited tumor growth and prolonged survival in both subcutaneous and disseminated human MM xenograft models. Our results show that miR33b is a tumor suppressor that plays a role during MLN2238-induced apoptotic signaling in MM cells, and these data provide the basis for novel therapeutic strategies targeting miR33b in MM.
Introduction
Multiple myeloma (MM), a fetal cancer of the plasma cells in the BM, remains the leading cause of death among patients with hematologic malignancy in the United States.1 The development of novel therapeutics, in particular rational combinations of therapeutics, have considerably improved patient outcome,2 but a cure is still elusive.
miRs are 19- to 25-nucleotide-long noncoding RNA molecules. RNA polymerase II transcribes miR genes to a long primary transcripts (pri-miRs) in the nucleus. Drosha processes the pri-miR to yield hairpin precursors (pre-miRs) consisting of approximately 70 nt. Sequentially, the pre-miR hairpins are exported to the cytoplasm by Exportin-5 and are processed into approximately 22-nt mature miRs by Dicer. miRs regulate gene expression at the level of both mRNA degradation and translation. They are able to silence gene expression posttranscriptionally by binding to partially complementary target sites in the 3′untranslated region (UTR) of targeting mRNAs, leading to repression of translation or reduction of mRNA.3-5 To date, approximately 700 miRs have been discovered in humans. Although studies about the identification of druggable targets and biomarkers have thus far mainly focused on protein-coding genes, increasing data indicate that miRs regulate major biologic process such as development, apoptosis, cell proliferation, and cell differentiation.6 More importantly, emerging evidence shows that miRs play a critical role in tumor pathogenesis by functioning either as oncogenes or tumor-suppressor genes.7,8 Nevertheless, little is known about miR regulation in MM. Several recent studies in MM have shown that genome-wide miR expression patterns are correlated with distinct genetic subgroups, drug resistance, and prognosis.9 For example, the transcription of miR21 is regulated by IL-6 through a STAT-3 mechanism in the IL-6–dependent INA-6 and XG-1 MM cell lines.10 Furthermore, miR15a and miR16 regulate proliferation, migration, angiogenesis, and growth of MM cells in vitro and in vivo by inhibiting the AKT/ribosomal-protein-6 and MAPK pathways.1 Therefore, the identification of miRs and delineation of their function in MM may provide novel therapeutic targets.
MLN2238, the hydrolyzed, biologically active form of MLN9708, is a selective, orally bioavailable proteasome inhibitor. It is currently being tested in clinical studies and has demonstrated preclinical antitumor activity in both solid-tumor and hematological xenograft models. MLN2238 has improved pharmacokinetics, pharmacodynamics, and antitumor activity compared with bortezomib.11 Our previous study showed that MLN2238 inhibits growth and triggers apoptosis in MM cells resistant to conventional and bortezomib therapies without affecting the viability of normal cells. In a human plasmacytoma xenograft model, MLN2238 was well tolerated, repressed tumor growth, and prolonged survival and was associated with significantly reduced tumor recurrence. Mechanistic studies have indicated that activation of caspases, the p53 pathway, and endoplasmic reticulum stress and inhibition of NF-κB are associated with MLN2238-induced MM cell death.12 Nonetheless, the role of miRs and their regulation in response to MLN2238 treatment in MM is undefined.
In the present study, we performed miR profiling in MM.1S MM cells after MLN2238 treatment and identified miR33b as one the target of MLN2238. We further delineated the role of miR33b in MM-cell pathogenesis and during MLN2238-induced cell death. Our findings provide the rationale for the development of a novel therapeutic strategy of targeting miR33b to improve patient outcome in MM.
Methods
Cell culture and drug treatment
The MM.1S, H929, ANBL-6, INA-6 (IL-6–dependent), RPMI-8226, and ARP-1 human MM cell lines; the human myeloid leukemia cell line K562; the human acute lymphoblastic leukemia cell line CCRF-CEM; the human mantle cell lymphoma cell lines Mino and Jerko-1; and the human diffuse large B-cell lymphoma cell line Toledo were cultured in complete RPMI 1640 medium supplemented with 10% FBS, 100 units/mL of penicillin, 100 μg/mL of streptomycin, and 2mM l-glutamine. CD138+ cells were freshly isolated and purified from MM patients or healthy donor BM by CD138+ selection using the AutoMACS magnetic cell sorter (Miltenyi Biotec). PBMCs from healthy donors were also maintained in the culture medium. BM stroma cells (BMSCs) were derived from CD138− cells from MM patients and cultured in DMEM containing 20% FBS. Informed consent was obtained from all patients in accordance with the Declaration of Helsinki. MLN2238 (MLN9708 rapidly hydrolyzes to its biologically active form, MLN2238) was from Millennium Pharmaceuticals; suberoyl anilide hydroxamic acid (SAHA) and SGI-1776 were from Selleck Chemicals; and dexamethasone and actinomycin D (Act-D) were from Sigma-Aldrich.
RNA extraction, miR array, and real-time PCR
Total RNA was extracted using TRIzol reagent (Invitrogen). RNA quantity and quality were determined with a spectrophotometer (ND-1000; NanoDrop Technologies). miR profiling was performed with TaqMan Array Human MiRNA A-Card Set Version 2.0. First, a reverse transcription was done to convert total RNA to cDNA with an miR-specific primer, and then miR was quantified by real-time PCR with TaqMan probes. The data were analyzed using dCHIP.13 Quantitative RT-PCR (qRT-PCR) was used to determine miR and gene expression according to the manufacturer's instructions (Applied Biosystems). Briefly, expression of primary miR miR33b and the mature miRs miR33b, miR15a, miR200a, and miR200 were measured with a TaqMan probe. Precursor miR33b, PIM-1, and HDM2 mRNA expression were quantified with the SYBR Green assay. The expression of mature miRs was calculated relative to RNU48 and the expression of pri-miR and pre-miR and mRNA was calculated relative to GAPDH (mRNA) using the 2−ΔΔCt method.14,15
Transient transfections
MM.1S cells were transiently transfected with either pre-miR33b or a control probe (Ambion) using the Cell Line Nucleofector Kit V according to the manufacturer 's instructions (Amaxa Biosystems). After the indicated times of transfection, various functional studies were performed, as described previously.12
Stable cell-line generation
miR33b-overexpressing MM.1S cell line was generated by a lentivector-based miR overexpression system (LV500A-1; SBI).16 Briefly, 293T cells were transfected with the miR33b precursor expression construct (PMIRH-33b-PA-1, SBI)/empty construct pMIRNA1 (CDS11B-1, SBI) using pPACKH1 Lentivector Packing Kit (LV100A-1, SBI) and PureFection Transfection Reagent (LV750A-1,SBI). The pseudoviral particles were harvested at 48 and 72 hours after transfection. Then miR33b-overexpressing stable cell line was established with virus infection using TransDux (LV850A-1; SBI) followed by green fluorescent protein– positive (GFP+) sorting with flow cytometry. The MM.1S cell line stably overexpressing PIM-1 (pWZL Neo Myr Flag PIM1; Addgene) without the 3′-UTR region (MM.1S-PIM1) or vector were established using retroviral infection followed by G418 selection.
Cell viability, apoptosis, colony formation, and migration assays
Cell viability in cell lines and primary patient samples were assessed by 3-(4,5-dimethylthiozol-2-yl)-2,5- diphenyltetrazolium bromide (MTT; Chemicon International), CellTiter-Glo (Promega) and thymidine incorporation assays, according to the manufacturers' instructions. An annexin V–FITC/propidium iodide apoptosis detection kit (BD Biosciences) was used to quantify apoptosis and data were analyzed with a FACSCalibur flow cytometer (BD Biosciences).17 Colony formation was measured by the soft agar method as described previously. Briefly, 2 × 104/mL of GFP-miR33b–transfected MM.1S cells or vector-transfected MM.1S cells (5 mL) were used to form a cell layer and then colonies were stained with MTT and counted.18 Migration was performed using 24-well Transwell plate (Millipore) in the presence of 10% FBS and the migration of cells was quantified by measuring the intensity of fluorescence.19
Western blotting
Cells were lysed in cold RIPA buffer (50mM Tris-HCl, pH 7.4, 150mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS) supplemented with protease inhibitor cocktail tablets (Roche). Equal amounts of proteins were resolved by 4%-12% SDS-PAGE and transferred onto nitrocellulose membranes. Membranes were blocked by incubation in 5% nonfat dry milk in PBST (0.05% Tween-20 in PBS) and probed with anti-PARP (BD Biosciences/BD Pharmingen), anti–PIM-1, and anti–phospho-Bad Abs (Cell Signaling Technology). Blots were then developed by enhanced chemiluminescence (Amersham).
Reporter assay
pmiR-Report plasmids for the miR33b putative target PIM-1 were constructed. The sequences for creating pmiR-pim1 (pmiR-pim1-WT) and pmiR-pim1-mutant (pmiR-pim1-MT) were as follows: pmiR-pim1-WT, 5′CGCGTAAGCCAAGACCTCACACACACAAAAAATGCACAAACAATGCAATCAACAGAAAAGCTGTAAAGGATCCA-3′ (forward) and 5′ AGCTTGGATCCTTTACAGCTTTTCTGTTGATTGCATTGTTTGTGCATTTTTTGTGTGTGTGAGGTCTTGGCTTA-3′ (reverse); and pmiR-pim1-MT, 5′ CGCGTAAGCCAAGACCTCACACACACAAAAAATGCACAAATCAACAGAAAAGCTGTAAAGGATCCA-3′ (forward) and 5′- AGCTTGGATCCTTTACAGCTTTTCTGTTGATTTGTGCATTTTTTGTGTGTGTGAGGTCTTGGCTTA-3′ (reverse). The oligonucleotides were annealed and inserted into the pmiR-Report vector (Ambion). The vector (pmiR0) alone and pmiR-pim1-MT were used as a blank and negative control, respectively.16,20,21 MM.1S and 293T cells were cotransfected with reporter plasmids, miR (miR33b or control miR), and Renilla luciferase with the Cell Line Nucleofector Kit V or Lipofectamine 2000. Ten hours after transfection, cells were subjected to the luciferase reporter assay using the Dual-Luciferase Reporter Assay System (Promega). Luciferase activities were analyzed as relative activity of firefly to Renilla.
Human xenograft model
To establish subcutaneous or disseminated human MM xenograft models, 5 × 106 or 2 × 106/100μl of GFP-miR33b–transfected MM.1S cells or vector-transfected MM.1S cells were injected subcutaneously or intravenously into NOD.CB17-Prkdcscid/J mice (The Jackson Laboratory; 5 mice/group).22,23 All experiments involving animals were approved by an institutional animal care and use committee. The mice started to develop subcutaneous tumors after approximately day 25 after injection. Tumor size was monitored and measured every 3 days in 2 dimensions using calipers, and tumor volume was calculated using the following formula: V = 0.5a Xb2, where “a” and “b” are the long and short diameter of the tumor, respectively. Animals were euthanized when tumors reached 2 cm3. Survival was evaluated from the first day of tumor injection until death. Hind limb paralysis was used as an end point in the disseminated disease model. Tumor progression was monitored by imaging (LAS-4000 Luminescent Imager Analyzer; Fujifilm).
Statistical analysis
The Student t test (2 tailed) was used to determine significance in all in vitro experiments and in the in vivo tumor burden study. The log-rank test was used to evaluate the significance of survival of mice, and the survival curve was derived using the Prism Version 5 analysis software (GraphPad). P < .05 was considered statistically significant for all experiments.
Results
MLN2238 induces miR33b and MM patient tumor cells express low constitutive levels of miR33b
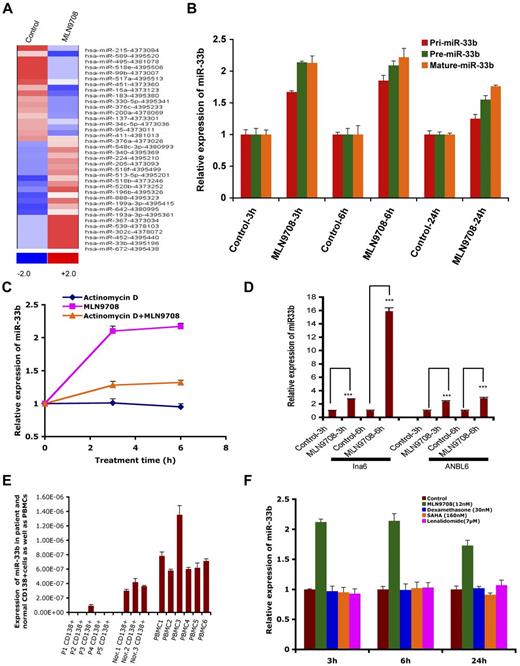
miRs have emerged as important posttranscriptional regulators in various types of cancer, and are therefore now considered a new class of targets for therapeutic intervention. The role of miRs and their regulation in response to proteasome inhibitor treatment in MM is unclear. Therefore, to gain a comprehensive understanding of the effect of MLN2238 on miR profile, the expression of 381 miRs was assessed in the MM.1S MM cell line, and data were analyzed using dCHIP.13 Treatment of MM.1S cells with MLN2238 (12nM) for 3 hours changed 36 miRs; 19 were up-regulated and 17 were down-regulated. miR33b was one of the most up-regulated miRs in response to MLN2238 treatment (Figure 1A). To further validate the miR array data, we examined the expression of miR33b in various MM cell lines after exposure to MLN2238 using qRT-PCR. qRT-PCR confirmed that MLN2238 induces miR33b expression in MM cells (Figure 1B and D).
MLN2238-inducedmiRprofiling in MM cells and expression of miR33b in MM and normal cells. (A) MM.1S MM cells was treated with vehicle or MLN2238 (12nM) for 3 hours and cells were harvested. RNAs were isolated for miR profiling using ABI Gene Card A Version 2. The data were analyzed by dCHIP and a minimum difference of ≥ 1.5 ΔΔCt value of a miR between control and test samples is shown. Blue represents down-regulated and red represents up-regulated miRs. (B) MM.1S cells were treated with vehicle or MLN 2238 (12nM) for indicated times and then cells were harvested. RNAs were isolated and subjected to qRT-PCR to examine the expression of pri-miR33b, pre-miR33b, and mature miR33b. (C) MM.1S cells were treated with Act-D (5 μg/mL), MLN2238, or Act-D + MLN 2238 (12nM) for the indicated times. The cells were harvested and RNAs were prepared to examine the expression of miR33b. (D) INA6 and ANBL6 MM cells were treated with vehicle or MLN 2238 (12nM) for the indicated times and then harvested. RNAs were isolated and subjected to qRT-PCR to examine the expression of miR33b. (E) RNAs were isolated from purified patient MM cells, normal CD138+ cells derived from BM, and PBMCs from healthy donors, followed by analysis of basal expression level of miR33b using qRT-PCR. (F) MM.1S cells were treated with vehicle, dexamethasone (30nM), lenalidomide (7μM), or SAHA (160nM) for the indicated times and then cells were harvested. RNAs were isolated and subjected to qRT-PCR to examine the expression of miR33b. Results shown are means ± SD (n = 3). ***P ≤ .001.
MLN2238-inducedmiRprofiling in MM cells and expression of miR33b in MM and normal cells. (A) MM.1S MM cells was treated with vehicle or MLN2238 (12nM) for 3 hours and cells were harvested. RNAs were isolated for miR profiling using ABI Gene Card A Version 2. The data were analyzed by dCHIP and a minimum difference of ≥ 1.5 ΔΔCt value of a miR between control and test samples is shown. Blue represents down-regulated and red represents up-regulated miRs. (B) MM.1S cells were treated with vehicle or MLN 2238 (12nM) for indicated times and then cells were harvested. RNAs were isolated and subjected to qRT-PCR to examine the expression of pri-miR33b, pre-miR33b, and mature miR33b. (C) MM.1S cells were treated with Act-D (5 μg/mL), MLN2238, or Act-D + MLN 2238 (12nM) for the indicated times. The cells were harvested and RNAs were prepared to examine the expression of miR33b. (D) INA6 and ANBL6 MM cells were treated with vehicle or MLN 2238 (12nM) for the indicated times and then harvested. RNAs were isolated and subjected to qRT-PCR to examine the expression of miR33b. (E) RNAs were isolated from purified patient MM cells, normal CD138+ cells derived from BM, and PBMCs from healthy donors, followed by analysis of basal expression level of miR33b using qRT-PCR. (F) MM.1S cells were treated with vehicle, dexamethasone (30nM), lenalidomide (7μM), or SAHA (160nM) for the indicated times and then cells were harvested. RNAs were isolated and subjected to qRT-PCR to examine the expression of miR33b. Results shown are means ± SD (n = 3). ***P ≤ .001.
miR15a, miR215, and miR200a are linked to inhibition of MM cell proliferation, whereas our miR array data show that MLN2238 treatment down-regulates these miRs after a short time (3 hours) treatment of MM cells. Therefore, we further evaluated MLN2238 effects on miR15a, miR215, and miR200a by measuring the time-dependent alterations in these miRs in MLN2238-treated cells using qRT-PCR. Similar to our miR array data, qRT-PCR analysis showed that 3-hour treatment of cells with MLN2238 down-regulated miR15a and miR215; however, their expression was significantly up-regulated at 6 hours and maintained a steady-state level even after 24 hours of treatment. Furthermore, miR200a was only slightly down-regulated after 3 hours of treatment with ML2238, but was significantly up-regulated after 24 hours of treatment (supplemental Figure 1B). These data suggest that the initial down-regulation of miR15a and miR215 (at 3 hours) in response to MLN2238 treatment is a transient phenomenon that may be a component of MLN2238-induced stress response signaling in MM cells. Nonetheless, MLN2238-induced up-regulation of miR15a, miR215, and miR200a at later time points (6 and 24 hours) is consistent with previous reports showing MM cell growth–inhibitory activity of these miRs and their potential modulation during MLN2238-induced apoptosis. Furthermore, similar dynamic alterations in miRs have been reported in other cell systems on treatment with various stress inducers.24 MLN2238 triggers an early and more sustained induction of miR33b compared with miR15a, miR215, or miR200a. It is also likely that different miRs are activated in response to distinct apoptotic stimuli and may also depend on cancer cell type. Our ongoing studies are further evaluating these possibilities.
miR expression is controlled at the level of transcription, processing, subcellular localization, and stability. To evaluate the possible mechanisms of up-regulation of miR33b after short-time exposure to MLN2238, we investigated whether the MLN2238-induced up-regulation of miR33b is due to the induction of transcriptional and maturation changes in miR33b. Treatment of MM.1S cells with MLN2238 (12nM) for the indicated times induced both pri-miR33b and pre-miR33b (Figure 1B). To examine the stability of miR33b, cells were treated with MLN2238 in the presence or absence of transcriptional inhibitor Act-D for 3 and 6 hours, followed by analysis of mature miR33b expression using qRT-PCR. Although cells treated with only MLN2238 showed a significant increase in miR33b, the addition of Act-D significantly blocked MLN2238-induced miR33b (Figure 1C). A residual increase in mature miR33b in MLN2238 + Act-D–treated versus control (DMSO) cells may have been due to regulation of miR33b at the miR-processing level. As a control, Act-D treatment significantly blocked transcription of an irrelevant gene, HDM2 (supplemental Figure 1A).
Predicted targets of miR33b include genes involved in ubiquitination, cell-cycle regulation, and tumorigenesis; therefore, we hypothesized that miR33b may play a role in MM pathogenesis. Examination of the basal expression level of miR33b in MM patient tumor cells showed that miR33b expression was significantly lower in normal plasma cells and PBMCs from healthy donors (Figure 1E). These data provide the rationale for further investigation of the function of miR33b in MM.
To determine whether up-regulation of miR33b is specific to MLN2238, we examined the effect of other anti-MM agents (dexamethasone, lenalidomide, and the HDAC inhibitor SAHA) compared with the proteasome inhibitor MLN2238 on miR33b expression. In contrast to MLN2238, no other drug triggered induction of miR33b (Figure 1F).
miR33b regulates cell viability, colony formation, and migration of MM cells and sensitizes MM cells to MLN2238 treatment
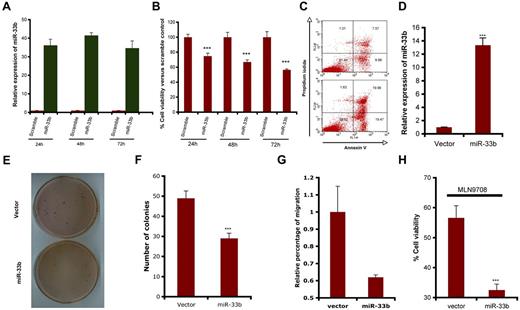
To determine the functional role of miR33b in MM cells and during MLN2238-induced MM cell death, pre-miR33b or control probes were transiently transfected into MM.1S cells and cell viability and apoptosis were measured. miR33b expression was markedly increased after transfection (Figure 2A), and overexpression of miR33b triggered significant cell death and apoptosis in MM.1S cells (Figure 2B-C). Stable overexpression of miR33b in MM.1S cells (Figure 2D) significantly decreased the number of colonies and the number of migrating cells induced by serum (Figure 2E-G). To identify the role of miR33b during MLN2238 treatment, we examined the response to MLN2238 in miR33b-overexpressing MM.1S cells. Our data show that MLN2238 triggered more cell death in the miR33b overexpressing MM.1S stable cell line compared with MM.1S cells transfected with empty vector (Figure 2H). This indicates that miR33b sensitizes MM cells to MLN2238 treatment.
Role ofmiR33b asatumor suppressor in MM.1S MM cells. (A-C) MM.1S cells were transiently transfected with either pre-miR33b or scrambled probe using the Cell Line Nucleofector Kit V. The transfected cells were examined for miR33b expression by qRT-PCR (A), for cell viability using the CellTiter-Glo assay (B); and for apoptosis by annexin V–FITC/propidium iodide double staining (C). (D) RNA was isolated from GFP vector controls or miR33b-overexpressing MM.1S cells, and miR33b expression was analyzed by qRT-PCR. (E-F) 2 × 104/mL of MM.1S cells stably expressing either vector or miR33b (5mL) were used in forming the cell layer; colonies were stained with MTT (E), and counted 4 weeks later (F). (G) MM.1S cells stably expressing either vector or miR33b were analyzed for their effect on serum-induced migration ability. The migration assay was performed using a 24-well Transwell plate. The migration cells were quantified by measuring the intensity of fluorescence. (H) MM.1S cells stably expressing either vector or miR33b were treated with MLN2238 (12nM) for 24 hours, and cell viability was examined with the CellTiter-Glo assay. Results shown are representative or means ± SD (n = 3). The experiments were repeated at least 3 times. ***P ≤ .001.
Role ofmiR33b asatumor suppressor in MM.1S MM cells. (A-C) MM.1S cells were transiently transfected with either pre-miR33b or scrambled probe using the Cell Line Nucleofector Kit V. The transfected cells were examined for miR33b expression by qRT-PCR (A), for cell viability using the CellTiter-Glo assay (B); and for apoptosis by annexin V–FITC/propidium iodide double staining (C). (D) RNA was isolated from GFP vector controls or miR33b-overexpressing MM.1S cells, and miR33b expression was analyzed by qRT-PCR. (E-F) 2 × 104/mL of MM.1S cells stably expressing either vector or miR33b (5mL) were used in forming the cell layer; colonies were stained with MTT (E), and counted 4 weeks later (F). (G) MM.1S cells stably expressing either vector or miR33b were analyzed for their effect on serum-induced migration ability. The migration assay was performed using a 24-well Transwell plate. The migration cells were quantified by measuring the intensity of fluorescence. (H) MM.1S cells stably expressing either vector or miR33b were treated with MLN2238 (12nM) for 24 hours, and cell viability was examined with the CellTiter-Glo assay. Results shown are representative or means ± SD (n = 3). The experiments were repeated at least 3 times. ***P ≤ .001.
The BM microenvironment confers resistance to many anti-MM agents. To determine whether there is an interaction between miR33b and the BM microenvironment, we first examined the effect of BMSCs on MLN2238-induced miR33b expression. GFP+ MM.1S cells were cocultured with BMSCs for 21 hours and then treated with either vehicle control or MLN2238 for an additional 3 hours. MM.1S cells were harvested by GFP-selective sorting, total RNA was isolated, and miR33b expression was analyzed. The results show that MLN2238 triggered up-regulation of miR33b even in the presence of BMSCs (supplemental Figure 2A). In addition, we examined whether miR33b introduction diminishes protection from BMSCs. MM.1S cells stably expressing either vector or miR33b were cultured alone or with BMSCs. Forty-eight hours later, cell proliferation was measured with the thymidine incorporation assay. We found that proliferation of miR33b-overexpressing MM.1S cocultured with BMSCs was slightly lower than the corresponding MM.1S control vector–transfected cells (supplemental Figure 2B), suggesting that introduction of miR33b modestly diminishes protection of stroma cells.
Our prior study showed that ML2238-induced apoptosis in MM cells is associated with caspase activation and that biochemical inhibition of caspases can block MLN2238-induced cell death. To examine whether a caspase inhibitor could affect MLN2238-induced miR33b expression, MM.1S cells were pretreated with pan caspase inhibitor, followed by MLN2238 treatment. Our results show that the pan-caspase inhibitor failed to block MLN2238-induced induction of miR33b (supplemental Figure 2C). The possible reason for this might be that caspase activation is a late and effector event for MLN2238-induced apoptosis, and miR33b induction occurs very early (3 hours).
Overexpression of miR33b and MLN2238 treatment negatively regulates PIM1 expression
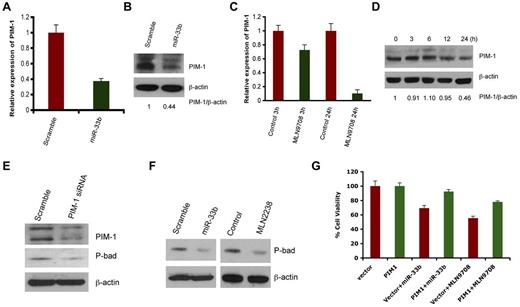
Predicted target of miR33b includes ubiquitin-, cell cycle-, apoptosis-, and tumorigenesis-related genes such as USP6, USP32, ccnd1, cdk6, cdk8, and PIM-1. However, PIM1 appeared as a putative target predicted by 3 different algorithms: Targetscan, miRDB, and DIANA-microT Version 3.0. Nonetheless, we delineated the correlation of the expression of all predicted genes with miR33b overexpression or MLN2238 treatment. No changes in mRNA or protein expression was observed on USP6, USP32, ccnd1, cdk6, or cdk8 in response to overexpression of miR33b (data not shown), whereas transient overexpression of miR33b significantly inhibited PIM-1 at both the gene and protein level (Figure 3A-B). Because MLN2238 up-regulates miR33b, we next examined the effect of MLN2238 on PIM-1 expression by qRT-PCR and Western blotting. Similar to miR33b overexpression, MLN2238 treatment also decreased PIM-1 expression in MM.1S cells (Figure 3C-D).
MLN2238 oroverexpression ofmiR33b negatively regulates PIM-1 signaling. (A-B) MM.1S cells were transiently transfected with either pre-miR33b or scrambled probe using the Cell Line Nucleofector Kit V, and cells were harvested 24h after transfection, followed by analysis of PIM-1 transcripts (A) and PIM-1 protein levels (B) by qRT-PCR and Western blot analysis, respectively. (C-D) MM.1S cells were treated with MLN2238 (12nM) for the indicated times, and cells were harvested. Total RNA and protein extracts were subjected to analysis of PIM-1 transcripts (C) and PIM-1 protein levels (D) using qRT-PCR and Western blotting, respectively. The relative expression of PIM1 to β-actin was quantified using ImageJ Version 1.38x software. (E) MM.1S cells were transfected with either scramble siRNA or PIM-1 siRNA. Total protein lysates were subjected to immunoblot analysis using Abs specific against PIM-1, phospho-Bad, and actin. (F) MM.1S cells overexpressing miR33b or MLN2238-treated MM.1S cells were harvested and protein lysates were subjected to immunoblot analysis using Abs specific against phospho-Bad and actin. (G) MM.1S cells stably expressing pWzl-neo retroviral vector or PIM1 (Addgene) were established by retrovirus infection. The stable cell lines were then transfected with miR33b or treatment with MLN9708 (12nM) for 24 hours. The cell viability was measured with the MTT assay. Results are shown as means ± SD (n = 3).
MLN2238 oroverexpression ofmiR33b negatively regulates PIM-1 signaling. (A-B) MM.1S cells were transiently transfected with either pre-miR33b or scrambled probe using the Cell Line Nucleofector Kit V, and cells were harvested 24h after transfection, followed by analysis of PIM-1 transcripts (A) and PIM-1 protein levels (B) by qRT-PCR and Western blot analysis, respectively. (C-D) MM.1S cells were treated with MLN2238 (12nM) for the indicated times, and cells were harvested. Total RNA and protein extracts were subjected to analysis of PIM-1 transcripts (C) and PIM-1 protein levels (D) using qRT-PCR and Western blotting, respectively. The relative expression of PIM1 to β-actin was quantified using ImageJ Version 1.38x software. (E) MM.1S cells were transfected with either scramble siRNA or PIM-1 siRNA. Total protein lysates were subjected to immunoblot analysis using Abs specific against PIM-1, phospho-Bad, and actin. (F) MM.1S cells overexpressing miR33b or MLN2238-treated MM.1S cells were harvested and protein lysates were subjected to immunoblot analysis using Abs specific against phospho-Bad and actin. (G) MM.1S cells stably expressing pWzl-neo retroviral vector or PIM1 (Addgene) were established by retrovirus infection. The stable cell lines were then transfected with miR33b or treatment with MLN9708 (12nM) for 24 hours. The cell viability was measured with the MTT assay. Results are shown as means ± SD (n = 3).
PIM-1 is a target for proviral activation in murine leukemia virus–induced T-cell lymphomas,25 and is also implicated in solid-tumor and hematologic malignancies.26 PIM-1 phosphorylates its downstream target, Bad, which prevents the association of Bad with Bcl2 and Bcl-xl, thereby reversing Bad-induced cell apoptosis.27-29 Knockdown of PIM-1 by siRNA down-regulated phospho-Bad (Figure 3E), confirming the PIM-1/Bad signaling axis in MM.1S MM cell line. Furthermore, both miR33b overexpression and MLN2238 treatment led to decreases in phospho-Bad in the MM.1S cell line (Figure 3F).
To establish a more definitive functional link between PIM-1 kinase and miR33b/MLN2238, we established a stable MM.1S cell line overexpressing PIM-1 without the 3′-UTR region (MM.1S-PIM1) using retroviral infection. MM.1S-PIM1 or MM.1S cells expressing vector alone were transfected with miR33b or treated with MLN2238 for 24 hours and then analyzed for cell viability using the MTT assay. The results showed that overexpression of miR33b induced approximately 30% cell death in the vector alone–transfected MM.1S cell line, but failed to induce significant cell death in PIM-1–overexpressing MM.1S-PIM1 cells. In parallel to overexpression of miR33b, treatment of MM.1S-PIM1 with MLN2238 triggers less cell death compared with vector control cells (Figure 3G). These data suggest that PIM-1, at least in part, mediates miR33b- or MLN2238-induced cell death.
PIM-1 is a direct target of miR33b
To confirm that miR33b targets PIM-1 directly, pmiR-PIM1-WT and pmiR-PIM1-MT constructs were generated using pmiR-Report vector, and the mutation was created by deletion of 7 nucleotides matched with miR33b seed sequence at the PIM1 3′-UTR region (Figure 4A). We were then able to examine whether miR33b can inhibit PIM-1 in 293T cells and MM.1S cells. The results showed that miR33b overexpression in both cell lines decreased pmiR-PIM1-WT, but not pmiR-PIM1-MT reporter activity (Figure 4B-C). In addition, reflecting the overexpression study results, MLN2238 treatment also decreased pimR-PIM1-WT, but not pmiR-PIM1-MT reporter activity (Figure 4D). These data demonstrate that miR33b directly targets PIM-1 kinase.
miR33b directly targets PIM-1. (A) Sequence alignment of the miR33b seed sequence with pim1 3′-UTR. Matched nuclear acid base pairs were labeled in red and deletions are labeled as “−.” (B). 293T cells were transiently cotransfected with reporter plasmids (pmiR-vector, pmiR-PIM1-WT, and pmiR-PIM1-MT), miR (miR33b or control miR), and Renilla luciferin with Lipofectamine 2000, and cells were harvested to measure the fluorescence of firefly and Renilla luciferin accordingly. (C) MM.1S cells were transiently cotransfected with reporter plasmids (pmiR-vector, pmiR-PIM1-WT, and pmiR-PIM1-MT), miR (miR33b or control miR), and Renilla luciferin using the Cell Line Nucleofector Kit V. Cells were harvested and lysed 10 hours after transfection, followed by measurement of relative fluorescence intensity of firefly and Renilla luciferin. (D) MM.1S cells were cotransfected with reporter plasmids (pmiR-vector, pmiR-PIM1-WT, and pmiR-PIM1-MT) and Renilla luciferin and then treated with MLN2238 (12nM). Cells were lysed and subjected to luciferase reporter assay using the Dual-Luciferase Reporter Assay System. Luciferase activities were analyzed as the relative activity of firefly to Renilla. Results shown are means ± SD (n = 3). ***P ≤ .001.
miR33b directly targets PIM-1. (A) Sequence alignment of the miR33b seed sequence with pim1 3′-UTR. Matched nuclear acid base pairs were labeled in red and deletions are labeled as “−.” (B). 293T cells were transiently cotransfected with reporter plasmids (pmiR-vector, pmiR-PIM1-WT, and pmiR-PIM1-MT), miR (miR33b or control miR), and Renilla luciferin with Lipofectamine 2000, and cells were harvested to measure the fluorescence of firefly and Renilla luciferin accordingly. (C) MM.1S cells were transiently cotransfected with reporter plasmids (pmiR-vector, pmiR-PIM1-WT, and pmiR-PIM1-MT), miR (miR33b or control miR), and Renilla luciferin using the Cell Line Nucleofector Kit V. Cells were harvested and lysed 10 hours after transfection, followed by measurement of relative fluorescence intensity of firefly and Renilla luciferin. (D) MM.1S cells were cotransfected with reporter plasmids (pmiR-vector, pmiR-PIM1-WT, and pmiR-PIM1-MT) and Renilla luciferin and then treated with MLN2238 (12nM). Cells were lysed and subjected to luciferase reporter assay using the Dual-Luciferase Reporter Assay System. Luciferase activities were analyzed as the relative activity of firefly to Renilla. Results shown are means ± SD (n = 3). ***P ≤ .001.
PIM-1 biologic inhibitor SGI-1776 triggers cell death in MM
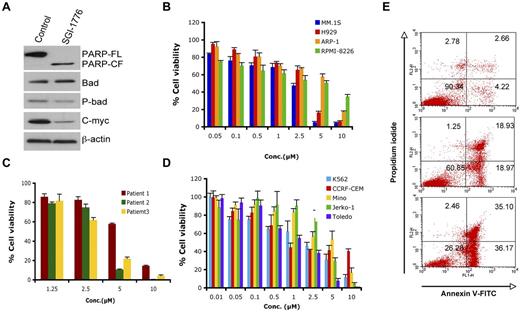
We next used SGI-1776,26 a biologic PIM-1 inhibitor, to elucidate the significance of down-regulation of PIM-1 signaling in MM cells. SGI-1776 markedly blocked PIM-1 signaling, as evidenced by a decreased phospho-Bad and expression of PIM-1–cooperating protein c-Myc30 (Figure 5A). SGI-1776 significantly decreased cell viability in various MM cell lines and primary MM tumor cells, including newly diagnosed and relapsed samples (Figure 5B-C). Recent preclinical studies showed PIM-1 inhibitor (SGI-1776) as a potential therapeutic agent in chronic lymphocytic leukemia (CLL), acute myeloid leukemia (AML) and precursor T-cell lymphoblastic leukemia/lymphoma (pre-T-LBL).31-33 We also found that SGI-1776 triggered cell death in leukemia (k562 and CCRF-CEM) and lymphoma (Mino, Jerko-1, and Toledo) cell lines (Figure 5D). Comparative cytotoxicity analysis showed that the PIM-1 inhibitor SGI-1776 was an equally potent cytotoxic agent in MM cells (5B and 5C), suggesting its potential clinical utility in MM. Finally, treatment of MM.1S cells with SGI-1776 for 24 hours triggered a dose-dependent apoptosis, as determined using annexin V/propidium iodide double staining (Figure 5E).
SGI-1776 triggers MM cell death. (A) MM.1S cells were treated with SGI-1776 (3μM) for 24 hours. Total protein lysates were subjected to immunoblotting with the indicated Abs. (B) MM cell lines were treated with SGI-1776 at indicated the concentrations for 48 hours, followed by assessment of cell viability using the MTT assay. Data presented are means ± SD of 3 independent experiments. (C) Purified CD138+ cells were treated with SGI-1776 at the indicated concentrations for 48 hours, and cell viability was measured using the CellTiter-Glo assay. Data are presented as means ± SD of triplicates. (D) Leukemia and lymphoma cell lines were treated with SGI-1776 at the indicated concentrations for 48 hours and cell viability was measured with the MTT assay. Data are presented as means ± SD of 3 independent experiments. (E) MM.1s cells were treated with vehicle or SGI-1776 at 3 and 5μM for 24 hours. Cells were harvested and analyzed for apoptosis by annexin V-FITC/propidium iodide double staining. Data shown are representative of 3 experiments.
SGI-1776 triggers MM cell death. (A) MM.1S cells were treated with SGI-1776 (3μM) for 24 hours. Total protein lysates were subjected to immunoblotting with the indicated Abs. (B) MM cell lines were treated with SGI-1776 at indicated the concentrations for 48 hours, followed by assessment of cell viability using the MTT assay. Data presented are means ± SD of 3 independent experiments. (C) Purified CD138+ cells were treated with SGI-1776 at the indicated concentrations for 48 hours, and cell viability was measured using the CellTiter-Glo assay. Data are presented as means ± SD of triplicates. (D) Leukemia and lymphoma cell lines were treated with SGI-1776 at the indicated concentrations for 48 hours and cell viability was measured with the MTT assay. Data are presented as means ± SD of 3 independent experiments. (E) MM.1s cells were treated with vehicle or SGI-1776 at 3 and 5μM for 24 hours. Cells were harvested and analyzed for apoptosis by annexin V-FITC/propidium iodide double staining. Data shown are representative of 3 experiments.
miR33b inhibits tumor growth and prolongs survival in human plasmacytoma xenograft models
We next evaluated the effect of miR33b on tumorigenesis and survival using a gain-of-function strategy in an MM xenograft model. Five × 106 or 2 × 106/100 μL of GFP-miR33b–transfected MM.1S cells or vector-transfected MM.1S cells were subcutaneously or IV injected into NOD.CB17-Prkdcscid/J mice to establish a subcutaneous or disseminated model, respectively.23 Tumors were measurable in the vector control group by day 23 after injection and later; 30 days after injection, in the miR33b-overexpressing group. Tumor growth analysis in both groups showed a retarded development of tumors in mice receiving miR33b-overexpressing MM.1S cells (Figure 6A-B). Mice tumors were resected and analyzed for PIM-1 protein expression. Consistent with our in vitro data, PIM-1 was decreased in miR33b-overexpressing tumors compared with vector controls (Figure 6D), confirming that miR-33b targets PIM-1 in vivo. Mice survival was evaluated from the first day of tumor injection until death or paralysis in the subcutaneous and disseminated MM xenograft models, respectively. Paralysis was observed after 50 days of injection of GFP-vector–transfected MM.1S cells, and after 65 days of injection of GFP-miR33b–transfected MM.1S cells. Paralyzed mice developed typical visualized tumors in the skull and backbone (Figure 6E). Mice receiving miR33b-overexpressing MM.1S cells as either subcutaneous or intravenous injections had significantly longer lifespans than mice receiving vector-MM.1S cells (Figure 6C and F).
Overexpression ofmiR33b inhibits tumor growth and prolongs survival in human plasmacytoma xenograft models. 5 × 106 or 2 × 106/100 μL of GFP-miR33b–transfected MM.1S cells or vector-transfected MM.1S cells were subcutaneously or intravenously injected into NOD.CB17-Prkdcscid/J mice (5 mice/group). Tumor volume was calculated using the following formula: V = 0.5a Xb2, where “a” and “b” are the long and short diameter of the tumor, respectively. Survival was evaluated from the first day of tumor injection until death. Representative photos of mice with tumor burden were taken with Imager Analyzer. (A) Representative image of mice with subcutaneous tumors. Left panel shows a mouse injected with vector-transfected MM.1S cells; the right panel shows a mouse injected with miR33b-transfected MM.1S cells. (B) Overexpression of miR33b inhibits tumor growth in a subcutaneous model. (C) Overexpression of miR33b prolongs survival in a subcutaneous model. (D) Mice tumors from the experiment described in panel A were analyzed for PIM-1 protein expression. (E) Representative image from mice receiving MM.1S miR33b-overexpressing cells in a disseminated MM model. Photo shows tumor homing in the backbone and skull region. (F) Overexpression of miR33b prolongs survival of mice in disseminated model. The log-rank test was used to evaluate the significance of survival of mice.
Overexpression ofmiR33b inhibits tumor growth and prolongs survival in human plasmacytoma xenograft models. 5 × 106 or 2 × 106/100 μL of GFP-miR33b–transfected MM.1S cells or vector-transfected MM.1S cells were subcutaneously or intravenously injected into NOD.CB17-Prkdcscid/J mice (5 mice/group). Tumor volume was calculated using the following formula: V = 0.5a Xb2, where “a” and “b” are the long and short diameter of the tumor, respectively. Survival was evaluated from the first day of tumor injection until death. Representative photos of mice with tumor burden were taken with Imager Analyzer. (A) Representative image of mice with subcutaneous tumors. Left panel shows a mouse injected with vector-transfected MM.1S cells; the right panel shows a mouse injected with miR33b-transfected MM.1S cells. (B) Overexpression of miR33b inhibits tumor growth in a subcutaneous model. (C) Overexpression of miR33b prolongs survival in a subcutaneous model. (D) Mice tumors from the experiment described in panel A were analyzed for PIM-1 protein expression. (E) Representative image from mice receiving MM.1S miR33b-overexpressing cells in a disseminated MM model. Photo shows tumor homing in the backbone and skull region. (F) Overexpression of miR33b prolongs survival of mice in disseminated model. The log-rank test was used to evaluate the significance of survival of mice.
Discussion
Previous studies showed a role of miRs in MM pathogenesis, supporting miRs as a potential pharmacologic intervention strategy.24,34-36 MLN2238, a second-generation, orally bioavailable proteasome inhibitor, is currently in phase 1/2 clinical trials and has demonstrated improved pharmacokinetics, pharmacodynamics, and antitumor activity with significantly reduced tumor recurrence compared with bortezomib.11,12 However, the role of miRs and their regulation in response to MLN2238 treatment in MM is unclear.
In the present study, we examined MLN2238-induced miR changes in MM cells. We have shown that MLN2238 modulates miR expression of 36 miRs and that miR672 is the most highly up-regulated miR. However, based on miRBase, hsa-miR672 (MI0005522) is absent in the NCBI36 genome assembly, suggesting that this miR sequence is not currently clearly identified in humans. For these reasons, we did not choose miR672 for further validation studies. miR33b was the second highly expressed miR in our array data and its up-regulation was confirmed in various MM cell lines by qRT-PCR.
Alteration of either biogenesis or stability may regulate miR expression. To explore the potential mechanisms of induction of miR33b, we evaluated the biogenesis and stability of miR33b. Our results show the increase of pri-miR33b and pre-miR33b by MLN2238 treatment, whereas mature miR33b was stable within the tested time (6 hours) after blocking transcription with Act-D. These results indicate that MLN2238-induced up-regulation of miR33b is mainly because of transcriptional regulation.
It has been reported that miR15a, miR215, and miR200a play a role in cell growth and proliferation.21-23 Because our miR data showed down-regulation of these miRs after short-time exposure to MLN2238, we measured the dynamic change of these miRs by qRT-PCR. Our results show that 3-hour treatment of cells with MLN2238 down-regulated these miRs; however, their expression was significantly up-regulated after 6 or 24 hours of treatment, which is consistent with previous reports showing the MM cell growth–inhibitory activity of these miRs. These data suggest the kinetic alteration of miR profiling and their potential modulation during MLN2238-induced apoptosis.
Bioinformatics algorithms analysis of the predicted target of miR33b includes ubiquitin-, cell cycle-, apoptosis-, and tumorigenesis-related genes such as USP6, USP32, ccnd1, cdk6, cdk8, and PIM-1. Therefore, we hypothesized that miR33b may play a role in MM pathogenesis and during MLN2238-induced MM cell death. miR33b is embedded in intron 17 of the SREBP-1 gene on chromosome 17 and coexpresses with the host gene to control cholesterol and fatty acid homeostasis. Concurrently, miR33b targets and represses ABCA1, an important regulator of HDL biogenesis. Antisense miR33b up-regulates ABCA1 and elevates plasma HDL in mice and nonhuman primates.37-39 Nevertheless, little is known about miR33b in cancer.
To define the biologic significance of miR33b in MM, we investigated whether miR33b is deregulated in MM patient tumor cells. Our results showed that MM cells had low constitutive miR33b expression compared with normal CD138+ cells and PBMCs from healthy donors. These results provide the rationale to further characterize the function of miR33b in MM. Overexpression studies with miR33b showed significant cell death and apoptosis in MM.1S cells. In addition, miR33b also blocked colony formation and migration of MM cells. These data confirmed the tumor-suppressor function of miR33b in MM cells. We have also shown that miR33b sensitizes MM cells to MLN2238 treatment.
We next explored the molecular mechanisms whereby miR33b or MLN2238 trigger MM cell death. A protooncogene, PIM-1, emerged as a putative target of miR33b by 3 different algorithms, so we examined its expression in response to miR33b overexpression and MLN2238 treatment. As predicted, PIM-1 was down-regulated at both the gene and protein levels either by miR33b overexpression or by MLN2238 treatment. Multiple lines of evidence show that PIM-1 has a role in the pathogenesis of hematologic malignancies and solid cancers. PIM-1 promotes cell proliferation and survival, as well as homing and migration, through modification of cell cycle regulator, apoptosis mediator, and chemokine receptors.26,40 A recent study showed that miR328 impairs colony formation through canonical targeting PIM-1.41 Therefore, down-regulation of PIM-1 contributes, at least in part, to miR33b-induced decreases in cell viability, colony formation, and migration. In addition, PIM-1 downstream signaling is also affected by miR33b and MLN2238 treatment. In the present study, we have demonstrated the existence of PIM-1/Bad signaling in MM. PIM-1 siRNA down-regulated its substrate protein, Bad. Similar to PIM-1 knockdown, overexpression of miR33b and MLN2238 treatment blocked PIM-1 kinase activity, which resulted in the down-regulation of phospho-Bad. Luciferase assays using plasmids harboring the PIM1-WT or PIM1-MT 3′UTR sequence confirmed that PIM-1 is a direct target of miR33b. Cotransfected PIM1-WT plasmid with miR33b precursor or treatment with MLN2238 inhibited luciferase activity, which indicates a direct negative regulation of PIM-1 by miR33b. In addition, the significant differences of the response of PIM1-overexpressing MM.1S-PIM1 and vector controls to miR33b overexpression and MLN2238 treatment suggest that PIM-1 mediates, at least in part, miR33b- or MLN2238-induced cell death.
Recently, PIM-1 inhibitors have been developed, offering great promise as therapeutic agents in CLL, AML, and pre-T-LBL.31-33 Given that PIM-1 is a potential target for chemotherapy, we assessed the significance of PIM-1 inhibition in MM using the PIM biologic inhibitor SGI-1776. SGI-1776 was equally effective in MM, CLL, and AML cells, killing MM cells and leukemia and lymphoma cells at a similar IC50 range. These results provide the basis for targeting PIM-1 inhibition as a potential therapeutic strategy in MM.
Most importantly, the results of the present study demonstrate that overexpression of miR33b inhibits tumor growth and prolongs survival in both subcutaneous and disseminated human MM xenograft models. PIM-1 plays a critical role in tumorigenesis. A PIM1-specific mAb suppresses human and mouse tumor growth by decreasing PIM-1 levels, reducing Akt phosphorylation, and activating apoptosis.42,43 In agreement with these previous findings, we have shown herein that overexpression of miR33b blocks PIM-1 in vivo, indicating that blockade of PIM-1 may partially contribute to the overall antitumor activity of miR33b in MM cells. Our results suggest that miR33b is a tumor suppressor that plays a role during MLN2238-induced apoptotic signaling in MM cells, and these data provide the basis for novel therapeutic strategies targeting miR33b in MM.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This investigation was supported by National Institutes of Health Specialized Programs of Research Excellence (SPORE) grants P50100707, PO1-CA078378, and RO1 CA050947. K.C.A. is an American Cancer Society Clinical Research Professor. J. H. Lin at Dana-Farber Cancer Institute provided normal plasma cells.
National Institutes of Health
Authorship
Contribution: Z.T. designed the research, performed the experiments, and wrote the manuscript; J.Z. provided reagents; Y.-T.T. and P.R. provided patient samples; S.B.A. analyzed the array data;. Y.H. helped in establishing the PIM1-overexpressing cell lines; A.J.B. provided the drug MLN9708 and critically reviewed the manuscript; D.C. designed the research and wrote the manuscript; and K.C.A. wrote the manuscript.
Conflict-of-interest disclosure: P.R. and K.C.A. are scientific board members with Millennium Pharmaceuticals. A.J.B. is an employee of Millenium, The Takeda Oncology Company. The remaining authors declare no competing financial interests.
Correspondence: Dharminder Chauhan or Kenneth C. Anderson, LeBow Institute for Myeloma Therapeutics and Jerome Lipper Myeloma Center, Department of Medical Oncology, Dana-Farber Cancer Institute, Harvard Medical School, Mayer 561, 450 Brookline Ave, Boston, MA 02115; e-mail: Dharminder_Chauhan@dfci.harvard.edu or Kenneth_Anderson@dfci.harvard.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal