Abstract
To assess the frequency of TP53 alterations and their correlation with other genetic changes and outcome in acute myeloid leukemia with complex karyotype (CK-AML), we performed integrative analysis using TP53 mutational screening and array-based genomic profiling in 234 CK-AMLs. TP53 mutations were found in 141 of 234 (60%) and TP53 losses were identified in 94 of 234 (40%) CK-AMLs; in total, 164 of 234 (70%) cases had TP53 alterations. TP53-altered CK-AML were characterized by a higher degree of genomic complexity (aberrations per case, 14.30 vs 6.16; P < .0001) and by a higher frequency of specific copy number alterations, such as −5/5q−, −7/7q−, −16/16q−, −18/18q−, +1/+1p, and +11/+11q/amp11q13∼25; among CK-AMLs, TP53-altered more frequently exhibited a monosomal karyotype (MK). Patients with TP53 alterations were older and had significantly lower complete remission rates, inferior event-free, relapse-free, and overall survival. In multivariable analysis for overall survival, TP53 alterations, white blood cell counts, and age were the only significant factors. In conclusion, TP53 is the most frequently known altered gene in CK-AML. TP53 alterations are associated with older age, genomic complexity, specific DNA copy number alterations, MK, and dismal outcome. In multivariable analysis, TP53 alteration is the most important prognostic factor in CK-AML, outweighing all other variables, including the MK category.
Introduction
Chromosomal abnormalities are found in approximately 55% of adult patients with acute myeloid leukemia (AML) and are among the most important independent prognostic factors.1–3 AMLs exhibiting 3 or more acquired chromosome aberrations in the absence of chromosomal rearrangements listed in the World Health Organization (WHO) 2008 category “AML with recurrent genetic abnormalities” are now defined as AML with complex karyotype (CK-AML).1,2 CK-AMLs account for 10% to 15% of adult AMLs, and the frequency increases with age. CK-AMLs belong to the cytogenetic adverse-risk group because they are associated with very poor outcome when treated with intensive or nonintensive conventional chemotherapy.1,3,4 Recently, a new cytogenetic category was introduced, that is, the monosomal karyotype (MK) defined by the presence of one single autosomal monosomy in association with at least one additional autosomal monosomy or one structural chromosomal abnormality (in the absence of core binding factor AML and acute promyelocytic leukemia).5 This MK category was reported to be associated with a dismal prognosis and to add prognostic information, even in CK-AML.
Complex karyotypes often contain numerous chromosome aberrations that can only be partially or not at all interpreted using standard cytogenetic techniques. Such aberrations include unbalanced translocations with chromosomal material of unknown origin, marker or ring chromosomes, homogeneously staining regions, or double minutes, the latter representing cytogenetic equivalents of high-level DNA amplifications. In general, CK-AMLs are characterized by chromosomal gains and losses, rather than balanced translocations, suggesting distinct mechanisms in leukemogenesis.6
In recent years, molecular cytogenetic and array-based techniques have enabled a more precise characterization of these complex genetic changes. The imbalances most frequently found are losses affecting chromosome 5 or 5q (−5/5q−), −17/17p−, −7/7q−, −18/18q−, −16/16q−, −12/12p−, and gains affecting chromosome 8 or 8q (+8/+8q), +11/+11q, +21/+21q, +22/+22q, and +1/+1p.7 Furthermore, novel potential target genes have been delineated based on the observation that they are located in critical regions of deletions8 or contained in amplicons, such as MYC in 8q24, ETS1 and FLI1 in 11q24, CDX2 in 13q12, and ETS2 and ERG in 21q22.7,9–12
One target located in the commonly deleted region of 17p13 is the tumor suppressor gene TP53. In AML, TP53 alterations (mutations and/or losses, TP53altered) are rare and have been closely associated with CK-AML.13–16 Clinically, TP53 alterations appear to be associated with inferior outcome.15–17 However, these data are based on a small number of studies, and only one of these addressed both TP53 losses and mutations but not the prognostic significance.15
The objectives of our study were: (1) to study a large cohort of CK-AMLs (n = 234) for TP53 mutations; (2) to analyze these cases for DNA copy number alterations (CNAs) using array-based techniques; and (3) to correlate TP53 alterations with specific chromosome abnormalities, CNAs, MK, and clinical outcome.
Methods
Patients
Peripheral blood (PB) and/or bone marrow (BM) samples from 234 adult patients with CK-AML were analyzed. The definition of CK-AML followed recommended criteria.1,2 The diagnosis of AML was based on French-American-British Cooperative Group criteria,18 and after 2004 on WHO criteria.19 A total of 133 patients had de novo AML, 31 secondary AML (s-AML) after myelodysplastic syndrome or myeloproliferative neoplasms, 30 therapy-related (t-AML), and in 40 patients it was unknown. Of these 234 patients, 155 (66%) were treated on consecutive multicenter treatment trials of the German-Austrian AML Study Group (AMLSG) applying age-adjusted intensive chemotherapy: AML HD93 (n = 1),20 AML HD98A (n = 30),21 and AMLSG 07-04 (n = 54; NCT00151242) for younger patients (16-60 years); and AML HD98B (n = 27)22 and AMLSG 06-04 (n = 43; NCT00151255) for elderly patients (> 60 years). All trials were approved by the local ethics committees of all participating institutions; all patients gave informed consent for treatment, cryopreservation of samples, and molecular analyses according to the Declaration of Helsinki. Samples were primarily selected based on availability of sufficient material for genomic profiling and mutational analysis.
Cytogenetic and molecular genetic analysis
For cytogenetic classification, metaphases of sufficient quality could be studied by chromosome banding analysis in 219 patients; karyotypes were described according to the International System for Human Cytogenetic Nomenclature.23
Because of evolving technology that occurred in the course of the study, we switched from array comparative genomic hybridization (CGH) to single nucleotide polymorphism (SNP) array-based genomic profiling. Array CGH (n = 131) using the 2.8k platform and/or the 8.0k platform and unpaired SNP analyses using Affymetrix GeneChip Human Mapping 250K Array (n = 61) were performed as previously described7,24 ; Genome-Wide Human SNP Array 6.0 profiling (n = 42) was performed according to the manufacturer's protocols (Affymetrix). Genotyping Version 2.0 console (Affymetrix) was used for analysis of 6.0 arrays. Microarray data will be available at gene expression omnibus at http://www.ncbi.nlm.nih.gov/geo/ (GEO accession number GSE34542).
TP53 sequence analysis
To identify mutations in exons 4 to 10 of TP53, denaturating high-performance liquid chromatography was performed as previously described.25 Aberrant profiles were verified by bidirectional sequencing and compared with wild-type sequence (GenBank; X54156). Mutations were described using 2 different databases (IARC TP53 Database; www.p53.iarc.fr and The TP53 Web site; www.p53.free.fr).26,27 Subcloning analyses using the TOPO TA Cloning Kit and resequencing were performed according to manufacturer's protocols (Invitrogen).
Statistical analyses
The section on statistical analyses is provided in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Results
TP53 mutation analysis
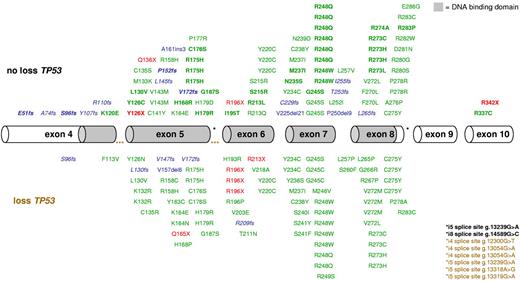
In 141 of the 234 (60%) patients with CK-AML, a total of 168 mutations were identified; 161 (96%) were located in the sequence-specific DNA-binding domain of p53 (residues 102-292). The majority were missense mutations (n = 130), followed by deletions/insertions (n = 21); 17 resulted in premature stop; 4 preserved the open reading frame (g.13149del6, g.13149del9, g.14001del21, and g.13162ins3), nonsense mutations (n = 9), and splice site mutations (n = 8; Figure 1). Seventeen of 25 patients harboring 2 or more TP53 mutations exhibited no TP53 loss; 5 of these showed a homozygous TP53 mutation; and in all remaining patients with available DNA for subcloning (n = 8), compound heterozygous mutations were confirmed. Hemizygous mutations (loss of 1 allele and at least 1 mutation in the remaining allele; 47% [79 of 168]) were more frequent than homozygous mutations (26% [43 of 168]), heterozygous (18% [30 of 168]), including possible compound heterozygous mutations among the 4 patients with more than 1 heterozygous mutation), and compound heterozygous mutations (10% [16 of 168]); 65 (39%) mutations affected common hot spots, such as codons 175, 245, 248, 273, and 275. Mutations affecting codons 175, 248, and 273 were associated with biallelic TP53 alteration compared with all other TP53 mutations (100% [27 of 27] vs 79% [90 of 114], P = .008).
Mapping of 168 TP53 mutations in 141 CK-AMLs. Hemizygous mutations are indicated in the bottom panel, heterozygous, and/or homozygous mutations are marked in the top panel. Exons 4 to 10 are drawn to relative scale; missense mutations (green), nonsense mutations (red), and insertion/deletion mutations (blue) are shown at their approximate location along the exons. Bold represents homozygous mutations, and blue italics, frameshift mutations leading to a premature stop codon.
Mapping of 168 TP53 mutations in 141 CK-AMLs. Hemizygous mutations are indicated in the bottom panel, heterozygous, and/or homozygous mutations are marked in the top panel. Exons 4 to 10 are drawn to relative scale; missense mutations (green), nonsense mutations (red), and insertion/deletion mutations (blue) are shown at their approximate location along the exons. Bold represents homozygous mutations, and blue italics, frameshift mutations leading to a premature stop codon.
CNAs and copy number neutral loss of heterozygosity UPD
In the entire cohort of 234 CK-AML, genomic losses (n = 1.845) were more frequent than gains (n = 778) or high-level DNA amplifications (n = 153). The median number of aberrations per case was 10 (range, 0-51); median numbers of losses, gains, and amplifications per case were 6 (range, 0-43), 2 (range, 0-28), and 0 (range, 0-7), respectively.
Recurrent losses were identified for the following chromosomes: monosomy 5 or losses of 5q (−5/5q−) (n = 147; 63%), −7/7q− (n = 123; 53%), −17/17p− (n = 106; 45%), −16/16q− (n = 66; 28%), −18/18q−, −12/12p− (n = 65 each; 28%), −20/20q− (n = 55; 24%), −3/3p− (n = 54; 23%), and −11/11q− (n = 35; 15%). Most frequent gains were +8/+8q (n = 67; 29%), +11/+11q (n = 61; 26%), +21/+21q (n = 39; 17%), +1/+1p (n = 37; 16%), +22/+22q (n = 33; 14%), +13/+13q (n = 29; 12%), +9/+9p (n = 28; 12%), and +19/+19p (n = 25; 11%). Most frequent high-level DNA amplifications mapped to 21q22, 11q13∼25 (n = 22 each; 9%), and 8q24 (n = 8; 4%).
Usually large, but also submicroscopic, losses (down to 800 kb in size) affecting the TP53 locus on 17p13 were identified in 94 of 234 (40%) cases. Uniparental disomy of 17p [UPD(17p); 8.14-22.50 Mb in size] encompassing the TP53 locus was detected in 15 of 103 (15%) cases analyzed by SNP arrays.
Biallelic TP53 alteration
Combining mutational and microarray findings, 164 of 234 (70%) CK-AMLs exhibited TP53 alteration (mutation and/or loss of TP53); 71 of 164 (43%) cases had biallelic TP53 alteration by hemizygous mutation pattern, and 38 of 70 (54%) cases without TP53 loss exhibited homozygous TP53 mutation caused by UPD(17p) in 15 of 19 SNP-analyzed cases. In the 4 cases lacking evidence for UPD(17p) in SNP profiling, homozygous TP53 mutations possibly resulted from intragenic loss of heterozygosity. Furthermore, 19 of 131 (15%) cases analyzed by array CGH on DNA sequence analysis exhibited homozygous TP53 mutation that are probably caused by UPD(17p) considering the frequency of UPD(17p) found by SNP array analysis. In addition, subcloning analysis confirmed biallelic TP53 alteration by compound heterozygous mutations in 8 patients. Together, at least 117 of 164 (71%) TP53altered CK-AMLs had biallelic TP53 inactivation (not taken into account the 4 patients with potentially compound heterozygous TP53 mutations). Patient 96 exhibited a homozygous missense mutation in exon 6 (p.R213L) and an additional heterozygous frameshift mutation in exon 4 (p.A74fs), suggesting that these had occurred sequentially, with p.R213L being the primary event followed by UPD(17p) resulting in the homozygous mutation pattern, whereas the p.A74fs mutation followed the recombination event (supplemental Figure 1).
Correlation of TP53 alteration with pattern of chromosome abnormalities and CNAs
We correlated TP53 alterations, as assessed by DNA sequence analysis and array profiling, with the pattern of chromosome abnormalities identified by conventional cytogenetics and with the pattern of CNAs detected by array-based analyses (Table 1).
Genetic and clinical characteristics according to TP53 alteration
| . | TP53unaltered . | TP53altered . | P . |
|---|---|---|---|
| Cytogenetics | n = 62 | n = 157 | |
| ≥ 5 aberrations | 38 (61%) | 139 (89%) | < .0001 |
| Marker chromosomes | 29 (47%) | 114 (73%) | .0005 |
| −5/5q− | 20 (32%) | 124 (79%) | < .0001 |
| −7/7q− | 25 (40%) | 81 (52%) | .14 |
| −5/5q− and −7/7q− | 12 (19%) | 70 (45%) | .0006 |
| −20/20q− | 8 (13%) | 44 (28%) | .02 |
| MK | 34 (55%) | 137 (87%) | < .0001 |
| Array-based genomics | n = 70 | n = 164 | |
| Total no. of losses (mean ± SD) | 4.00 ± 4.88 | 9.54 ± 7.49 | < .0001 |
| Total no. of gains (mean ± SD) | 1.94 ± 1.92 | 3.91 ± 3.80 | < .0001 |
| Total no. of amplifications (mean ± SD) | 0.21 ± 0.83 | 0.84 ± 1.31 | .0002 |
| Total no. of genomic aberrations (mean ± SD) | 6.16 ± 5.53 | 14.30 ± 9.41 | < .0001 |
| −3/3p− | 7 (10%) | 47 (29%) | .002 |
| −5/5q− | 20 (29%) | 127 (77%) | < .0001 |
| −7/7q− | 26 (37%) | 97 (59%) | .003 |
| −5/5q− and −7/7q− | 13 (19%) | 87 (53%) | < .0001 |
| −11/11q− | 13 (19%) | 22 (13%) | .32 |
| −12/12p− | 13 (19%) | 52 (32%) | .06 |
| −16/16q− | 5 (7%) | 61 (37%) | < .0001 |
| −18/18q− | 9 (13%) | 56 (34%) | .0008 |
| −20/20q− | 8 (11%) | 47 (29%) | .004 |
| +1/+1p | 3 (4%) | 34 (21%) | .001 |
| +8/+8q | 21 (30%) | 46 (28%) | .75 |
| +9/+9p | 11 (16%) | 17 (10%) | .27 |
| +11/+11q | 7 (10%) | 54 (33%) | .0002 |
| +13/+13q | 3 (4%) | 26 (16%) | .02 |
| +19/+19p | 3 (4%) | 22 (13%) | .04 |
| +21/+21q | 7 (10%) | 32 (20%) | .09 |
| +22/+22q | 6 (9%) | 27 (16%) | .15 |
| amp(8)(q24) | 4 (6%) | 4 (2%) | .24 |
| amp(11)(q13∼25) | 0 (0%) | 22 (13%) | .0004 |
| amp(21)(q22) | 3 (4%) | 19 (12%) | .09 |
| Molecular MK | 16 (23%) | 59 (36%) | .07 |
| Molecular genetics | n = 50 | n = 99 | |
| FLT3-ITD positive | 3 (6%) | 1 (1%) | .11 |
| FLT3-TKD mutation | 3 (6%) | 1 (1%) | .11 |
| NPM1 mutation | 3 (6%) | 0 (0%) | .04 |
| Clinical data | n = 52 | n = 103 | |
| Sex (male/female) | 26 (50%)/26 (50%) | 54 (52%)/49 (48%) | .87 |
| Median age, y | 54 | 61 | .002 |
| AML history | |||
| De novo | 36 (69%) | 76 (74%) | .57 |
| Secondary | 6 (12%) | 9 (9%) | .58 |
| Therapy-related | 9 (17%) | 16 (16%) | .82 |
| Median WBC, × 106/L | 12.9 | 6.5 | .18 |
| Median platelet count, × 106/L | 48 | 41 | .46 |
| Median hemoglobin, g/dL | 9.1 | 8.9 | .38 |
| Median BM blast count, % | 78 | 65 | .04 |
| Median PB blast count, % | 45 | 30 | .18 |
| Median LDH serum level, U/L | 391 | 438 | .25 |
| Response | n = 52 | n = 103 | |
| CR after induction therapy | 26 (50%) | 29 (28%) | .01 |
| RD after induction therapy | 18 (35%) | 53 (51%) | .06 |
| Outcome | n = 52 | n = 103 | |
| OS | |||
| Median, mo | 10.97 | 4.14 | < .0001 |
| 3-y survival rate, % | 28 | 3 | |
| EFS | |||
| Median, mo | 1.94 | 1.12 | .0007 |
| 3-y survival rate, % | 13 | 1 | |
| RFS | |||
| Median, mo | 12.16 | 6.51 | .01 |
| 3-y survival rate, % | 30 | 7 |
| . | TP53unaltered . | TP53altered . | P . |
|---|---|---|---|
| Cytogenetics | n = 62 | n = 157 | |
| ≥ 5 aberrations | 38 (61%) | 139 (89%) | < .0001 |
| Marker chromosomes | 29 (47%) | 114 (73%) | .0005 |
| −5/5q− | 20 (32%) | 124 (79%) | < .0001 |
| −7/7q− | 25 (40%) | 81 (52%) | .14 |
| −5/5q− and −7/7q− | 12 (19%) | 70 (45%) | .0006 |
| −20/20q− | 8 (13%) | 44 (28%) | .02 |
| MK | 34 (55%) | 137 (87%) | < .0001 |
| Array-based genomics | n = 70 | n = 164 | |
| Total no. of losses (mean ± SD) | 4.00 ± 4.88 | 9.54 ± 7.49 | < .0001 |
| Total no. of gains (mean ± SD) | 1.94 ± 1.92 | 3.91 ± 3.80 | < .0001 |
| Total no. of amplifications (mean ± SD) | 0.21 ± 0.83 | 0.84 ± 1.31 | .0002 |
| Total no. of genomic aberrations (mean ± SD) | 6.16 ± 5.53 | 14.30 ± 9.41 | < .0001 |
| −3/3p− | 7 (10%) | 47 (29%) | .002 |
| −5/5q− | 20 (29%) | 127 (77%) | < .0001 |
| −7/7q− | 26 (37%) | 97 (59%) | .003 |
| −5/5q− and −7/7q− | 13 (19%) | 87 (53%) | < .0001 |
| −11/11q− | 13 (19%) | 22 (13%) | .32 |
| −12/12p− | 13 (19%) | 52 (32%) | .06 |
| −16/16q− | 5 (7%) | 61 (37%) | < .0001 |
| −18/18q− | 9 (13%) | 56 (34%) | .0008 |
| −20/20q− | 8 (11%) | 47 (29%) | .004 |
| +1/+1p | 3 (4%) | 34 (21%) | .001 |
| +8/+8q | 21 (30%) | 46 (28%) | .75 |
| +9/+9p | 11 (16%) | 17 (10%) | .27 |
| +11/+11q | 7 (10%) | 54 (33%) | .0002 |
| +13/+13q | 3 (4%) | 26 (16%) | .02 |
| +19/+19p | 3 (4%) | 22 (13%) | .04 |
| +21/+21q | 7 (10%) | 32 (20%) | .09 |
| +22/+22q | 6 (9%) | 27 (16%) | .15 |
| amp(8)(q24) | 4 (6%) | 4 (2%) | .24 |
| amp(11)(q13∼25) | 0 (0%) | 22 (13%) | .0004 |
| amp(21)(q22) | 3 (4%) | 19 (12%) | .09 |
| Molecular MK | 16 (23%) | 59 (36%) | .07 |
| Molecular genetics | n = 50 | n = 99 | |
| FLT3-ITD positive | 3 (6%) | 1 (1%) | .11 |
| FLT3-TKD mutation | 3 (6%) | 1 (1%) | .11 |
| NPM1 mutation | 3 (6%) | 0 (0%) | .04 |
| Clinical data | n = 52 | n = 103 | |
| Sex (male/female) | 26 (50%)/26 (50%) | 54 (52%)/49 (48%) | .87 |
| Median age, y | 54 | 61 | .002 |
| AML history | |||
| De novo | 36 (69%) | 76 (74%) | .57 |
| Secondary | 6 (12%) | 9 (9%) | .58 |
| Therapy-related | 9 (17%) | 16 (16%) | .82 |
| Median WBC, × 106/L | 12.9 | 6.5 | .18 |
| Median platelet count, × 106/L | 48 | 41 | .46 |
| Median hemoglobin, g/dL | 9.1 | 8.9 | .38 |
| Median BM blast count, % | 78 | 65 | .04 |
| Median PB blast count, % | 45 | 30 | .18 |
| Median LDH serum level, U/L | 391 | 438 | .25 |
| Response | n = 52 | n = 103 | |
| CR after induction therapy | 26 (50%) | 29 (28%) | .01 |
| RD after induction therapy | 18 (35%) | 53 (51%) | .06 |
| Outcome | n = 52 | n = 103 | |
| OS | |||
| Median, mo | 10.97 | 4.14 | < .0001 |
| 3-y survival rate, % | 28 | 3 | |
| EFS | |||
| Median, mo | 1.94 | 1.12 | .0007 |
| 3-y survival rate, % | 13 | 1 | |
| RFS | |||
| Median, mo | 12.16 | 6.51 | .01 |
| 3-y survival rate, % | 30 | 7 |
ITD indicates internal tandem duplication; and TKD, tyrosine kinase domain.
Correlation with chromosome abnormalities.
TP53 alterations were identified in 157 of 219 (72%) CK-AMLs that could be analyzed by conventional cytogenetics. TP53altered CK-AMLs had a higher degree of genomic complexity as measured by total number of aberrations (≥ 5 aberrations, P < .0001) and the presence of marker chromosomes (P = .0005). TP53 alterations were correlated with the presence of specific cytogenetic abnormalities, such as −5/5q− (P < .0001), concomitant −5/5q− and −7/7q− (P = .0006), and 20q− (P = .02); we found no correlation with −7/7q− (P = .14; Table 1).
Correlation with CNAs.
TP53 alterations were correlated with the total number of losses (mean ± SD; 9.54 ± 7.49 vs 4.00 ± 4.88, P < .0001), gains (3.91 ± 3.80 vs 1.94 ± 1.92, P < .0001), high-level DNA amplifications (0.84 ± 1.31 vs 0.21 ± 0.83, P = .0002), and genomic complexity as measured by total number of aberrations per case (14.30 ± 9.41 vs 6.16 ± 5.53, P < .0001). Moreover, TP53 alterations were positively correlated with specific genomic aberrations, such as −5/5q− (P < .0001), −7/7q− (P = .003), concomitant −5/5q− and −7/7q− (P < .0001), and also −3/3p− (P = .002), −16/16q− (P < .0001), −18/18q− (P = .0008), and −20/20q− (P = .004); further correlations were identified for +1/+1p (P = .001), +11/+11q (P = .0002), +13/+13q (P = .02), +19/+19p (P = .04), and amplifications in 11q13∼25 [amp(11)(q13∼25)] (P = .0004; Table 1; Figure 2).
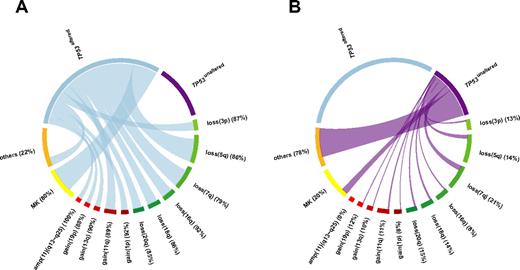
Relative frequencies and pairwise co-occurrences of TP53 alteration-associated genomic aberrations illustrated using Circos Table Viewer Version 0.52.28 The percentages indicate the proportion of each aberration associated with (A) TP53altered CK-AMLs and (B) TP53unaltered CK-AMLs. Unaltered TP53 and amp(11)(q13∼q25) were mutually exclusive. MK is based on cytogenetics analysis.
Relative frequencies and pairwise co-occurrences of TP53 alteration-associated genomic aberrations illustrated using Circos Table Viewer Version 0.52.28 The percentages indicate the proportion of each aberration associated with (A) TP53altered CK-AMLs and (B) TP53unaltered CK-AMLs. Unaltered TP53 and amp(11)(q13∼q25) were mutually exclusive. MK is based on cytogenetics analysis.
Correlation of TP53 alterations with MK
By conventional cytogenetics, 171 of 219 (78%) CK-AMLs fulfilled the MK criteria (CK+/MK+ AML) as previously defined. TP53 alterations were found in 137 of 171 (80%) CK+/MK+ AMLs and in only 20 of 48 (42%) CK+/MK− AMLs (P < .0001; Table 1). Compared with CK+/MK− AMLs, CK+/MK+ AMLs were characterized by a higher degree of genomic complexity determined by cytogenetics: more than or equal to 5 aberrations, 88% (151 of 171) versus 54% (26 of 48), P < .0001; and by genomic profiling as measured by total number of losses (mean ± SD; 9.29 ± 7.40 vs 3.67 ± 5.72; P < .0001) and aberrations per case (13.59 ± 9.61 vs 6.81 ± 6.11, P < .0001).
We subsequently determined MK+ AML based on array data (molMK). The frequency of CK+/molMK+ AML was much lower (75 of 234; 32%) because many monosomies described in chromosome banding analysis were not real monosomies but part of chromosomal material hidden in unbalanced translocation or marker chromosomes. TP53 alterations were found in 59 of 75 (79%) CK+/molMK+ AMLs and in 105 of 159 (66%) CK+/molMK− AMLs (P = .07; Table 1).
Correlation of TP53 alterations with clinical characteristics, response to therapy, and survival
Analyses were restricted to patients enrolled into AMLSG multicenter treatment trials applying age-adjusted intensive chemotherapy (n = 155, median age, 59 years; range, 18-81 years). Because there were no significant differences regarding clinical characteristics, response to therapy, and survival for TP53monoallelic altered and TP53biallelic altered CK-AMLs (supplemental Table 1; supplemental Figure 2), these genotypes were grouped as TP53altered CK-AML for further analyses.
Clinical characteristics.
TP53altered CK-AML patients were older (median 61 vs 54 years, P = .002) and had lower BM blast counts (median 65% vs 78%, P = .04; Table 1).
Response to therapy.
TP53 alterations were associated with resistance to chemotherapy. Response to induction therapy was as follows: complete remission (CR) 28% and 50% (P = .01), refractory disease (RD) 51% and 35% (P = .06), and early/hypoplastic death 21% and 15% (P = .52) for CK+/TP53altered and CK+/TP53unaltered AML, respectively (Table 1). Other variables predicting for poor response to induction therapy were age (P < .0001) and genomic losses affecting 5q (P = .02), 7q (P = .03), and 16q (P = .04). Lactate dehydrogenase (LDH) serum levels, white blood cell count (WBC), s/t-AML, and cytogenetic MK did not impact CR achievement.
For multivariable analysis, a conditional model was used with an age cut-point at 60 years to address the different treatment intensities applied in the different age cohorts. This model revealed as significant factors TP53altered (odds ratio [OR] = 0.55; 95% confidence interval [CI], 0.30-1.00; P = .05) and age (OR for a 10-year difference, 0.67; 95% CI, 0.52-0.87; P = .003). No significant impact on CR achievement was found for the variables WBC, platelet counts, cytogenetic MK, and s/t-AML (Table 2).
Multivariate analyses of outcome
| CK-AML . | Response . | OS . | ||
|---|---|---|---|---|
| OR . | P . | HR . | P . | |
| TP53 alteration | 0.55 | .05 | 2.43 | .0001 |
| Age (difference of 10 y) | 0.67 | .003 | 1.26 | .04 |
| s/t-AML | 0.67 | .24 | 1.05 | .81 |
| Logarithm of WBC | 0.74 | .19 | 1.62 | .004 |
| Logarithm of platelets | 0.76 | .39 | 1.13 | .62 |
| MK* | 0.75 | .43 | 0.87 | .57 |
| CK-AML . | Response . | OS . | ||
|---|---|---|---|---|
| OR . | P . | HR . | P . | |
| TP53 alteration | 0.55 | .05 | 2.43 | .0001 |
| Age (difference of 10 y) | 0.67 | .003 | 1.26 | .04 |
| s/t-AML | 0.67 | .24 | 1.05 | .81 |
| Logarithm of WBC | 0.74 | .19 | 1.62 | .004 |
| Logarithm of platelets | 0.76 | .39 | 1.13 | .62 |
| MK* | 0.75 | .43 | 0.87 | .57 |
Determined by chromosome banding analysis.
Survival analysis.
The median follow-up time for survival in the 155 CK-AML was 36.6 months (95% CI, 29.9-51.4 months); the estimated 3-year event-free survival (EFS), relapse-free survival (RFS), and overall survival (OS) of the entire cohort were 5% (95% CI, 2%-10%), 17% (95% CI, 9%-31%), and 12% (95% CI, 7%-19%), respectively.
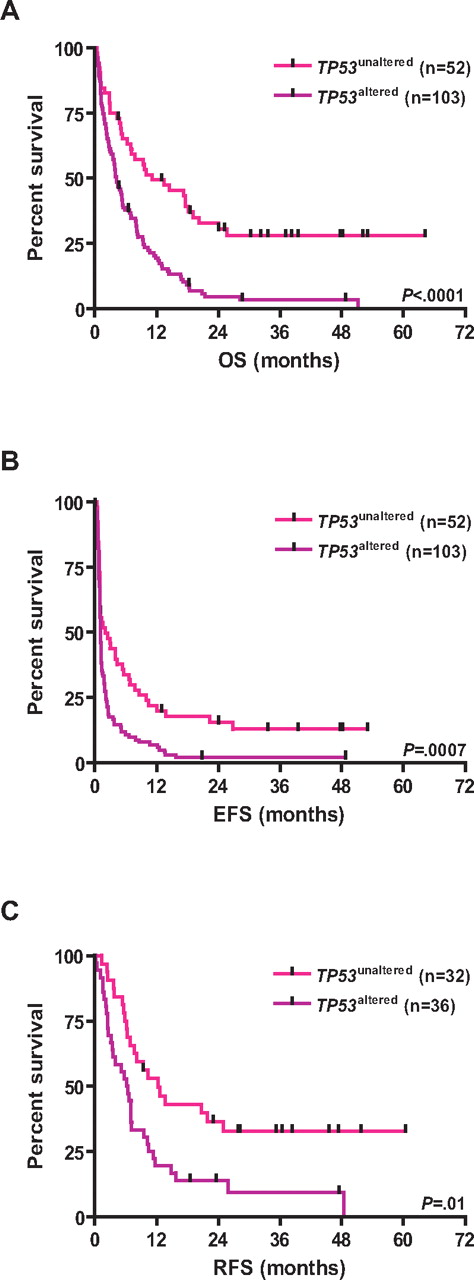
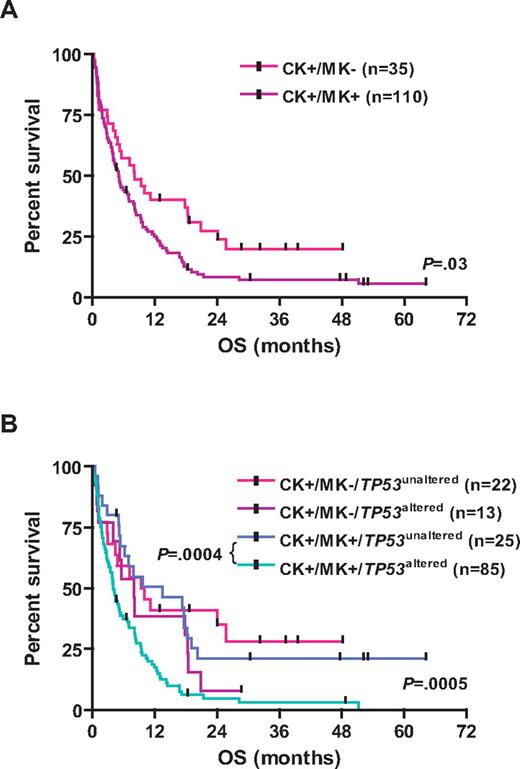
TP53 alterations were associated with inferior survival; the 3-year estimated survival rates for CK+/TP53altered and CK+/TP53unaltered patients were as follows: EFS, 1% versus 13% (log-rank, P = .0007); RFS, 7% versus 30% (P = .01); and OS, 3% versus 28% (P < .0001), respectively (Table 1; Figure 3). Other variables predicting for inferior OS in univariable analysis were age (P < .0001), cytogenetic MK (P = .03), and genomic losses of 5q (P = .03), 7q (P = .003), 16q (P = .0004), and gains of 1p (P = .04), and amp(11)(q13∼25; P = .05). LDH and WBC did not impact OS. Among CK+/MK+ AMLs, those with TP53 alterations had significantly worse OS (P = .0004; Figure 4).
Kaplan-Meier survival estimates according to the TP53 status. Data are shown for TP53unaltered CK-AMLs and TP53altered CK-AMLs. (A) OS. (B) EFS. (C) RFS.
Kaplan-Meier survival estimates according to the TP53 status. Data are shown for TP53unaltered CK-AMLs and TP53altered CK-AMLs. (A) OS. (B) EFS. (C) RFS.
Kaplan-Meier survival estimates according to the cytogenetic status. (A) Data are shown for OS for CK+/MK− AML and CK+/MK+ AML. (B) Data are shown for OS for the subgroups CK+/MK−/TP53unaltered, CK+/MK−/TP53altered, CK+/MK+/TP53unaltered, and CK+/MK+/TP53altered.
Kaplan-Meier survival estimates according to the cytogenetic status. (A) Data are shown for OS for CK+/MK− AML and CK+/MK+ AML. (B) Data are shown for OS for the subgroups CK+/MK−/TP53unaltered, CK+/MK−/TP53altered, CK+/MK+/TP53unaltered, and CK+/MK+/TP53altered.
Multivariable analysis stratified again for age at a cut-point of 60 years revealed TP53altered (hazard ratio [HR] = 2.43; 95% CI, 1.56-3.77; P = .0001), logarithm of WBC (HR = 1.62; 95% CI, 1.17-2.26; P = .004), and age (HR for 10-year difference, 1.26; 95% CI, 1.01-1.56, P = .04) as significant variables; not significant for OS were platelet counts, cytogenetic MK, and s/t-AML (Table 2).
Allogeneic hematopoietic stem cell transplantation in first CR was performed in 30 CK-AML patients. Of those, 14 of 15 TP53altered CK-AML relapsed and died, whereas in TP53unaltered CK-AML 9 of 15 relapsed and died (P = .04). This translated into significantly worse OS for TP53altered CK-AML (P = .04; supplemental Figure 3).
Discussion
In our series of 234 CK-AMLs, TP53 was deleted and/or mutated in 70% of cases, thus representing the most frequently known altered gene in this AML subgroup. TP53 alterations were associated with older age, genomic complexity, specific chromosome abnormalities, monosomal karyotype, specific CNAs, and predicted for dismal outcome.
Loss of TP53 was found in approximately 40% of CK-AMLs by array-based techniques, a value that corresponded well with that of 17p abnormalities found on chromosome banding analysis. By DNA sequence analysis, 60% of cases exhibited TP53 mutations, consistent with previous reports.6,15,16 Of note, at least two-thirds of mutated cases had biallelic TP53 alteration resulting from hemizygous, compound heterozygous, and homozygous mutations commonly as a result of homologous recombination leading to UPD. Thus, when assessing for TP53 mutational status in CK-AML, it will be necessary to include DNA sequence analysis.
TP53altered CK-AMLs were characterized by a significantly higher degree of genomic complexity, as assessed by total number of genomic losses and gains, as well as the frequency of high-level DNA amplifications. This observation fits well into the p53 pathomechanism of genomic instability.29–35 TP53altered CK-AMLs were also associated with specific abnormalities. As previously reported,15,16,36,37 −5/5q− and/or −7/7q− were significantly more frequent among TP53altered CK-AMLs. Because we also applied array-based techniques, we identified additional CNAs associated with TP53altered CK-AML, that is, −3/3p−, −16/16q−, −18/18q−, −20/20q−, and gains or amplifications of 1p, 11q, 13q, and 19p. Such genomic pattern associated with TP53 alterations may pinpoint to candidates cooperating in p53-dependent leukemogenesis.
Recently, the cytogenetic category of “monosomal karyotype” was described, allowing further risk stratification of CK-AML patients.5 Of note, in our study, CK+/MK+ AMLs were significantly associated with TP53 alterations, found in 80% of CK+/MK+ AML compared with only 42% in CK+/MK− AML. Thus, TP53 alterations appear to be one molecular basis for this purely descriptive cytogenetic subset. The association of TP53 alterations with CK+/MK+ AML was lost when TP53 alterations were correlated with CNAs identified by array-based assays. Not unexpectedly, many monosomies described in chromosome banding analysis were not real monosomies but were part of chromosomal material hidden in unbalanced translocations or marker chromosomes.
Little is known about the pathogenesis of CK-AML, but the high frequency of TP53 alteration, and in particular biallelic alteration, suggests an important role of p53 in leukemogenesis. Evidence for this hypothesis comes from several observations in mice and human disease: (1) mouse studies requiring biallelic TP53 inactivation and a concomitant “second hit” for myeloid leukemogenesis38,39 demonstrated that p53lost myeloid progenitors exhibit aberrant self-renewal, thereby promoting AML40 ; (2) in high-risk MDS and/or AML evolving from a 5q− syndrome, the expansion of preexisting TP53 mutated subclones was observed41,42 ; and (3) recently, next-generation sequencing of a therapy-related CK-AML genome identified several acquired genetic lesions and a heterozygous intragenic germline TP53 deletion, becoming homozygous in AML as a result of acquired UPD(17p),43 a mechanism possibly underlying the sequential TP53 inactivation in patient 96 (supplemental Figure 1).
Besides being older and having lower BM blasts, TP53altered CK-AML had no distinct clinical phenotype, possibly because of the complexity of concurrent genetic events and different consequences of TP53 alterations. TP53 losses or mutations entail various tumor phenotypes,44 and mouse models investigating TP53 inactivation identified gain of function for hot spot mutations, such as R175H, R248W, and R273H, as well as increased proliferation related to accelerated tumorigenesis and leukemogenesis, resulting in a more aggressive AML.35,44–46
p53 loss of function has been shown to be related to resistance to chemotherapy, also to cytarabine.46,47 Consistent with this finding, TP53 alterations in our study were associated with resistance to “3 + 7”-based induction chemotherapy (Tables 1 and 2). Refractory disease was observed in 51% of CK+/TP53altered compared with 35% of CK+/TP53unaltered AMLs. In univariable analysis, TP53 alteration also predicted for inferior OS; median survival times for CK+/TP53altered and CK+/TP53unaltered patients were 4.14 and 10.97 months, respectively. In multivariable analysis, TP53 alteration was by far the strongest prognostic factor for OS, followed by logarithm of WBC and age; of note, the cytogenetic category MK completely lost its prognostic impact. Explorative subset analysis suggested that allogeneic hematopoietic stem cell transplantation had no favorable impact on outcome in TP53altered CK-AML.
TP53 alterations are the most common molecular lesions in CK-AML and predict for resistance to conventional chemotherapy and dismal outcome. TP53 alterations correlate with specific CNAs and with the MK category. In CK-AML, TP53 alteration represents the most important prognostic marker, even outweighing the MK category in multivariable analysis. Therefore, TP53 mutational status should be assessed in clinical trials investigating novel agents to identify compounds that may be effective in this subset of patients.
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank all members of the AMLSG for their participation in this study and for providing patient samples. The following AML Study Group institutions and investigators participated in this study: Daniel Oruzio, MD, Klinikum Augsburg, Augsburg, Germany; Dietmar Reichert, MD, Ubbo-Emmius-Klinik Aurich, Aurich, Germany; Jörg Westermann, MD, Charité Berlin, Berlin, Germany; Christian Teschendorf, MD, Klinikum Bochum, Bochum, Germany; Ingo Schmidt-Wolf, MD, Universitätsklinikum Bonn, Bonn, Germany; Florian Lordick, MD, Städtisches Klinikum Braunschweig, Braunschweig, Germany; Bernd Hertenstein, MD, Klinikum Bremen-Mitte, Bremen, Germany; Helga Bernhard, MD, Klinikum Darmstadt, Darmstadt, Germany; Ulrich Germing, MD, Universitätsklinikum Düsseldorf, Düsseldorf, Germany; Carsten Schwänen, MD, Klinikum Esslingen, Esslingen, Germany; Mohammed Wattad, MD, Kliniken Essen Süd, Ev. Krankenhaus Essen-Werden gGmbH, Essen, Germany; Elke Jäger, MD, Krankenhaus Nordwest GmbH, Frankfurt, Germany; Michael Lübbert, MD, Universitätsklinikum Freiburg, Freiburg, Germany; Andrea Distelrath, MD, Klinikum Fulda, Fulda, Germany; Volker Runde, MD, Wilhelm-Anton-Hospital, Goch, Germany; Detlef Haase, MD, Universitätsklinikum Göttingen, Göttingen, Germany; Walter Fiedler, Universität Hamburg, Hamburg, Germany; Hans Salwender, MD, Allgemeines Krankenhaus Altona, Hamburg, Germany; Elisabeth Lange, MD, Klinikum Hamm, Hamm, Germany; Andrea Sendler, MD, Klinikum Hanau, Hanau, Germany; Hartmut Kirchner, MD, Krankenhaus Siloah, Hannover, Germany; Uwe Martens, SLK-Kliniken GmbH Heilbronn, Heilbronn, Germany; Ulrich Kaiser, MD, Bernward-Krankenhaus Hildesheim, Hildesheim, Germany; Michael Pfreundschuh, MD, Universitätsklinikum Homburg/Saar, Homburg, Germany; David Nachbaur, Universitätsklinikum Innsbruck, Austria; Martin Bentz, MD, Städtisches Klinikum Karlsruhe, Karlsruhe, Germany; Heinz A. Horst, MD, Universitätsklinikum Kiel, Kiel, Germany; Stefan Kremers, MD, Caritas-Krankenhaus Lebach, Lebach, Germany; Frank Hartmann, MD, Klinikum Lemgo, Lemgo, Germany; Andreas Petzer, MD, Krankenhaus der Barmherzigen Schwestern Linz, Linz, Austria; Gerhard Heil, MD, Klinikum Lüdenscheid, Lüdenscheid, Germany; Thomas Fischer, MD, Klinikum Magdeburg, Magdeburg, Germany; Thomas Kindler, MD, Universitätsklinikum Mainz, Mainz, Germany; Martin Grießhammer, MD, Klinikum Minden, Minden, Germany; Katharina Götze, MD, Technische Universität München, München, Germany; Ali-Nuri Hünerlitürkoglu, Lukaskrankenhaus Neuss, Neuss, Germany; Claus-Henning Köhne, MD, Klinikum Oldenburg, Oldenburg, Germany; Thomas Südhoff, MD, Klinikum Passau, Passau, Germany; Karin Corduan, MD, Elisabeth Krankenhaus Recklinghausen, Recklinghausen, Germany; Michael Schenk, MD, Krankenhaus der Barmherzigen Brüder, Regensburg, Germany; Artur Wehmeier, MD, Klinikum Remscheid, Remscheid, Germany; Axel Matzdorff, MD, Caritas-Klinik St Theresia, Saarbrücken, Germany; Richard Greil, MD, Salzburger Landeskliniken, Salzburg, Austria; Wolfgang Grimminger, MD, Klinikum Schwäbisch-Gmünd, Mutlangen, Germany; Hans-Günther Mergenthaler, MD, Klinikum Stuttgart, Stuttgart, Germany; Else Heidemann, MD, Diakonie-Klinikum Stuttgart. Stuttgart, Germany; Heinz Kirchen, MD, Krankenhaus der Barmherzigen Brüder, Trier, Germany; Helmut Salih, MD, Universitätsklinikum Tübingen, Tübingen, Germany; Wolfgang Brugger, MD, Klinikum Villingen-Schwenningen, Villingen-Schwenningen, Germany; Elisabeth Koller, MD, Hanuschkrankenhaus, Vienna, Austria; and Aruna Raghavachar, MD, Helios Klinikum Wuppertal, Wuppertal, Germany.
This study was supported in part by the Bundesministerium für Bildung und Forschung (grants 01GS0439 NGFN2 and 01GS0871 NGFNplus).
Authorship
Contribution: F.G.R. designed and performed research, analyzed and interpreted data, performed statistical analysis, and wrote the manuscript; R.F.S. provided study materials or patients, collected data, and analyzed and interpreted data; L.B. collected, analyzed, and interpreted data; S.K., V.T., V.I.G., P.P., G.H., M.v.L.-T., M.L., J.K., B.S., and A.G. provided study materials or patients and collected data; H.K., M.H., and C.-M.K. performed research and collected data; K.H. designed research and collected data; S.F., T.Z., and P.L. analyzed and interpreted data; and K.D. and H.D. designed research, provided study materials or patients, collected data, analyzed and interpreted data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hartmut Döhner, Department of Internal Medicine III, University Hospital of Ulm, Albert-Einstein-Allee 23, 89081 Ulm, Germany; e-mail: hartmut.doehner@uniklinik-ulm.de.