Abstract
Fusion protein AML1-ETO, resulting from t(8;21) translocation, is highly related to leukemia development. It has been reported that full-length AML1-ETO blocks AML1 function and requires additional mutagenic events to promote leukemia. We have previously shown that the expression of AE9a, a splice isoform of AML1-ETO, can rapidly cause leukemia in mice. To understand how AML1-ETO is involved in leukemia development, we took advantage of our AE9a leukemia model and sought to identify its interacting proteins from primary leukemic cells. Here, we report the discovery of a novel AE9a binding partner PRMT1 (protein arginine methyltransferase 1). PRMT1 not only interacts with but also weakly methylates arginine 142 of AE9a. Knockdown of PRMT1 affects expression of a specific group of AE9a-activated genes. We also show that AE9a recruits PRMT1 to promoters of AE9a-activated genes, resulting in enrichment of H4 arginine 3 methylation, H3 Lys9/14 acetylation, and transcription activation. More importantly, knockdown of PRMT1 suppresses the self-renewal capability of AE9a, suggesting a potential role of PRMT1 in regulating leukemia development.
Introduction
Acute myeloid leukemia (AML) is closely associated with chromosomal abnormalities.1 The AML1 gene (also known as CBFA2, PEBP2αB, and RUNX1) was initially identified as a target of chromosomal translocation in t(8;21), which is associated with 15% of de novo AML cases and ≤ 40% in the AML subtype M2 of the French-American-British classification.2,3 This specific translocation at t(8;21) involves the AML1 gene on chromosome 21 and the ETO (also known as MTG8) gene on chromosome 8 and generates an AML1-ETO fusion transcription factor.4
AML1-ETO inherits the DNA binding RUNT domain from AML1 and is functionally characterized as a transcription factor for both gene repression and activation.3,5-7 It has been shown that AML1-ETO negatively regulates AML1 target genes, possibly through interaction with corepressor proteins such as mSin3A, N-CoR/SMRT (nuclear receptor corepressor/silencing mediator for retinoic acid receptor and thyroid hormone receptor), and HDACs (histone deacetylases).8-10 AML1-ETO could also act as a transactivator on certain genes. One of the possible mechanisms is by recruiting histone modifiers to make chromatin structure more accessible to the transcription activation machinery, resulting in gene activation. A recent finding shows that p300 binds to NHR1 domain of AML1-ETO to facilitate transcription.11 A variety of posttranslational modifications, including acetylation, methylation, and phosphorylation, on specific residues of histones and their corresponding enzymes has been discovered.12 It is well documented that a specific histone modification on a promoter could determine the state of transcription. Specifically, methylation on histone H4 arginine 3 (Arg3) by PRMT1 (protein arginine methyltransferase 1) generally correlates with transcription activation.13 PRMT1 is the most predominant arginine methyltransferase in mammalian cells and functions as a coactivator for transcription activators, including p53 and Yin Yang 1,14,15 and nuclear receptors, including farnesoid X receptor and androgen receptor.16,17 In addition to histone methylation and transcription regulation, PRMT1 could also exert its function by methylating a number of substrates and thus regulating multiple cellular processes, including RNA processing, transcription, protein–protein interactions, nuclear trafficking, transcriptional elongation, DNA damage response, and cell cycle checkpoints.18,19
Studies from our group and others have reported the potential role of AML1-ETO in the development of leukemia. However, expression of AML1-ETO itself fails to cause leukemia development in mice.20-22 AML1-ETO becomes leukemogenic when accompanied with additional mutagenic events.23,24 We have previously reported that the C-terminally truncated form of AML1-ETO, termed AML1-ETOtr (AEtr), is strongly leukemogenic.25 More importantly, we subsequently identified an alternatively spliced isoform of AML1-ETO, named AML1-ETO9a (AE9a), directly from leukemia patient samples.26 This novel isoform AE9a is structurally similar to AEtr, and its expression leads to rapid onset of leukemia as shown in a mouse retroviral transduction-transplantation model.
To further examine the molecular mechanism underlying AE9a-induced leukemia development, we seek to identify AE9a-associated factors directly from primary leukemic cells that express AE9a/AEtr and investigate their roles in AE9a function. In the present study, we uncovered the specific interaction between AE9a and PRMT1. Immunoprecipitated AE9a possesses histone H4 methyltransferase activity possibly via PRMT1 association. AE9a can be weakly methylated at Arg142 by PRMT1. PRMT1 is required for AE9a-driven transcription activation on specific genes. Results from ChIP assay suggest that AE9a recruits PRMT1 to and directs histone H4 methylation at these AE9a-activated promoters. Furthermore, we found a crucial role of PRMT1 in maintaining the self-renewal capability of AE9a.
Methods
Cell culture
293T cells were grown in DMEM supplemented with 10% bovine calf serum. K562 and Kasumi-1 cells were grown in RPMI 1640 supplemented with 10% FBS. FLAG-AE9a–expressing K562 cells were previously described.27
Plasmid construction
MigR1-HA-AE9a and MigR1-FLAG-AEtr vectors were previously described.26 The cDNA of human AE9a was subcloned into p3xFLAG-CMV-7.1 via EcoRI and XbaI sites to generate p3xFLAG-AE9a. This vector was used to generate p3xFLAG-AE9a R142A with the use of QuikChange Site-Directed Mutagenesis Kit (QIAGEN). Vectors for expressing HA-PRMT1 in mammalian cells and GST-PRMT1 in bacteria are precious gifts from Dr Michael R. Stallcup (University of Southern California).28 An ∼ 1.9-kb fragment upstream of MT2A gene (−22 to −1916 from transcription initiation site) was PCR amplified and subcloned into pGLX2 vector,29 a modified version of pGL2-Basic (Promega), via KpnI and XhoI sites to generate pGLX2-MT2A-Luc for reporter assay. Vector for expressing shPrmt1 in murine cells was made by subcloning 19-bp Prmt1 target oligonucleotide (#1: 5′-GACATGACATCCAAAGACT-3′; #2: 5′-CAAGTGAAGAGGAACGACT-3′) into pSUPER.retro.puro vector.30
Transfection and nucleofection
Transfection of 293T cells was performed with polyethylenimine. For small interfering RNA (siRNA) transfection, 1 × 106 K562 cells were nucleofected with 3 μg of siRNA according to the manufacturer's protocol (Lonza). Negative control siRNA was purchased from Ambion (no. AM4636). siRNA for human PRMT1 (5′-CGTCAAAGCCAACAAGTTA-3′) was synthesized by Dharmacon. For luciferase reporter assay, 1 × 106 K562 cells were nucleofected with 1 μg of pGLX2-MT2A-Luc, 0.2 μg of pRL-null, 2 μg of p3xFLAG-AE9a, and 3 μg of siRNA and cultured for 24 hours. Approximately 40% of cell lysates were assayed for transcription activity by Dual-Luciferase Reporter Assay System (Promega) and Monolight 3010 Luminometer (BD Biosciences). Approximately 10% of cell lysates were analyzed for protein expression by Western blot analysis.
Whole-cell extracts and nuclear extracts preparation
For primary leukemic cells from mouse spleen, whole-cell lysates were prepared with lysis buffer (50mM This-HCl, pH 7.5, 150mM NaCl, 1% Triton X-100) supplemented with protease inhibitor tablets (Roche), 1mM PMSF, and phosphatase inhibitors (1mM NaF and 1mM Na3VO4). For all other cells, whole-cell lysates were prepared by lysing in modified RIPA buffer (50mM This-HCl, pH 7.6, 150mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate) supplemented with protease inhibitor tablets (Roche), 1mM PMSF and phosphatase inhibitors (1mM NaF and 1mM Na3VO4). Nuclear extracts were prepared with high salt as described31 with supplement of benzonase nuclease (Sigma-Aldrich). All immunoprecipitation experiments were performed with respective extraction buffer.
Antibodies
The rabbit polyclonal anti-PRMT1 (07-404), antiasymmetric dimethylarginine (aDMA; ASYM24, 07-414), and anti-H3K9/14Ac (06-599) Abs were purchased from Upstate Biotechnology. The rabbit IgG (I5006), mouse IgG (I5381), mouse monoclonal anti–α-tubulin (T9026), mouse monoclonal anti–FLAG-HRP (A8592) Abs, and anti-FLAG M2 Affinity Gel (A2220) were purchased from Sigma-Aldrich. The rabbit polyclonal anti-H4R3Me2a (39705) Ab was purchased from Active Motif. The mouse monoclonal anti-HA (HA.11, MMs-101P) Ab was purchased from Covance. The rabbit polyclonal anti–AML1-ETO Ab was generated with a peptide (DGPREPRNRTEKHS) that covers the conjunction of AML1 and ETO to specifically detect human AML1-ETO fusion protein. Method for generating Methyl RUNT (R142) Ab is essentially the same as described previously, with the exception that methyl R142 Runt peptide was used.32
Mass spectrometry analyses
After FLAG-IP from primary leukemic cells, AE9a and its associated proteins were eluted with FLAG peptides and subjected to mass spectrometry analyses. Multidimensional Protein Identification Technology and tandem mass spectra analyses were performed as described previously.33
In vitro methyltransferase assay
FLAG-tagged AE9a was expressed in 293T cells that were cultured with 20μM Adenosine-2′,3′-dialdehyde (Adox; Sigma-Aldrich) for 48 hours to minimize endogenous methylation. It was then immunoprecipitated by FLAG M2 agarose from nuclear extracts to enrich AE9a. The in vitro methylation assay was performed as described previously.34 For histone H4 methylation shown in Figure 2A, the immune complex was incubated with 3H-S-adenosylmethionine (SAM; PerkinElmer) and 1 μg of recombinant histone H4 (Upstate Biotechnology). For AE9a methylation, the immune complex was incubated with recombinant GST-PRMT1 and either 3H-SAM or unlabeled SAM (Sigma-Aldrich). For Figure 3D, GST-PRMT1 and 3H-SAM were incubated with various amounts of recombinant histone H4 or FLAG-IP–enriched AE9a. Band intensity from fluorography was quantified by ImageJ Version 1.43 software (National Institutes of Health). Signals from histone H4 were used to generate a standard curve. Methylation for each molecule of AE9a (67.1 kDa) was then compared with histone H4 (11.4 kDa).
RNA preparation and microarray analysis
Total RNA was isolated from K562 cells by QIAGEN RNeasy Mini Kit. Human HT-12 v4 Expression BeadChip (BD-103-0204) Kit from Illumina) was used for microarray analysis. Standard procedure was performed according to the manufacture's protocol at BIOGEM core at University of California, San Diego. Raw and normalized data have been deposited in Gene Expression Omnibus as the series accession number GSE28135. Significance of all data was further analyzed with BRB-ArrayTools, developed by Dr Richard Simon and BRB-ArrayTools Development Team (http://linus.nci.nih.gov/BRB-ArrayTools.html). A P value < .001 was considered statistically significant.
RT and real-time qPCR
Total RNA was isolated from K562 cells by QIAGEN RNeasy Mini Kit. Approximately 5 μg of total RNA was reverse transcribed by GeneAmp RNA PCR Kit from Applied Biosystems. Quantitative PCR (qPCR) was performed with 1/100 of RT product, primers, and Power SYBR Green PCR Master Mix on Applied Biosystems 7300 Real-Time PCR System. See supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article) for all primers used. Transcript levels of the genes of interest were quantitated by Δ-Δ threshold cycle (Ct) method, using the house keeping gene GAPDH for normalization.
ChIP assay
Chromatin preparation and immunoprecipitation was performed as described previously.35 Approximately 2 × 107 cells and 10 μg of Ab were used per ChIP reaction. Immunoprecipitated DNA (1/100) was amplified by qPCR with primers specific for promoters of interest and Power SYBR Green PCR Master Mix on Applied Biosystems 7300 Real-Time PCR System. See supplemental Table 2 for all primers used. The DNA relative enrichment was calculated by Δ-Δ Ct method. The PCR signals obtained for each promoter region were normalized to input DNA (total chromatin fraction) and compared with a control region on ACTB (β-actin) promoter.
Retroviral transduction and replating assay
For the production of retrovirus, 293T cells were cotransfected with MigR1 or MigR1-AE9a, pSUPER.retro.puro vector containing control shRNA or shPrmt1, and Ecopac (pIK6.1MCV.ecopac.UTd) by the polyethylenimine method. The viral supernatant fluid was harvested after 48 hours and filtered through 0.45-μm filters. BM from C57B6/J animals was collected by flushing femurs and tibias and cultured in BM medium (20 ng/mL SCF, 10 ng/mL IL-3, 10 ng/mL IL-6, 15% FBS, 1% penicillin and streptomycin, and 1% Glu). On the second day of culture, viral supernatant fluids were added at 20% of the final volume with 4 μg/mL polybrene. Cells were spinoculated at 3000 rpm for 3 hours at 32°C in an Allegrea-12R centrifuge with a SX4750 rotor (Beckman Coulter). The procedure was repeated on the next day. Cells were then plated in 35-mm plates in 1 mL of M3434 medium (StemCell Technologies) in the presence of 1 μg/mL puromycin. After 7 days, enhanced green fluorescent protein–positive (EGFP+) cells were sorted by FACS. The same number of cells from each transduction was replated, and colony counting was repeated every 7 days. For AE9a-leukemic cell replating, cells were transduced by retrovirus containing either pSUPER.retro.puro-control shRNA or pSUPER.retro.puro-shPrmt1. Cells were subjected to aforementioned selection and replating assay.
Results
Identification of PRMT1 as a novel AE9a-interacting protein
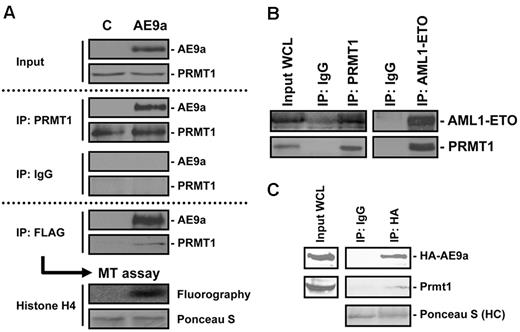
The fusion protein AML1-ETO has long been associated with the development of AML. To find proteins that participate in AML1-ETO–mediated leukemogenesis through protein–protein interactions, we took advantage of our AE9a/AEtr-induced leukemia mouse model and purified AE9a/AEtr and its associated proteins directly from primary leukemic cells. As shown in Figure 1, we made cell extracts from spleen cells of leukemic mice that express either FLAG-tagged AEtr or HA-tagged AE9a and performed immunoaffinity purification of AE9a/AEtr protein complex by anti-FLAG M2 agarose beads. HA-AE9a–containing cell lysate served as a negative control. Mass spectrometry analysis found that, in addition to several previously known AML1-ETO–interacting proteins, peptides from PRMT1 were abundantly recovered exclusively from immunoprecipitation of FLAG-tagged AEtr (supplemental Table 3). The finding of PRMT1 as a potential AE9a-interacting protein was further assured when we transiently expressed and analyzed the immunoprecipitated AE9a from 293T cells. Mass spectrometry analysis on this immunoprecipitated sample confirmed the presence of PRMT1 (data not shown). The interaction between FLAG-AE9a and endogenous PRMT1 was subsequently confirmed by both FLAG-IP and PRMT1-IP from 293T cells transiently expressing FLAG-AE9a (Figure 2A). With the use of bead-bound FLAG-AE9a complex as enzyme, we performed in vitro methyltransferase assay on histone H4 to show that the immunoprecipitated AE9a complex possesses histone methyltransferase activity (Figure 2A bottom panel). Taken together, our results clearly imply the interaction between AE9a and endogenous PRMT1.
Purification strategy for AE9a/AEtr and its associated factors. (A) Schematic representation of AE9a/AEtr fusion protein and the retroviral construct MigR1. Two constructs were used in the retroviral infection and transplantation experiments: MigR1-HA-AE9a and MigR1-FLAG-AEtr. (B) Purification scheme for AE9a/AEtr from leukemic mice.
Purification strategy for AE9a/AEtr and its associated factors. (A) Schematic representation of AE9a/AEtr fusion protein and the retroviral construct MigR1. Two constructs were used in the retroviral infection and transplantation experiments: MigR1-HA-AE9a and MigR1-FLAG-AEtr. (B) Purification scheme for AE9a/AEtr from leukemic mice.
Identification of PRMT1 as an AE9a-interacting protein. (A) AE9a-PRMT1 interaction is confirmed by both FLAG-IP and PRMT1-IP in 293T cells transiently expressing FLAG-tagged AE9a. Control lane (C) represents 293T cells transfected with empty vector. Endogenous PRMT1 associated with AE9a is enzymatically active as shown by in vitro methyltransferase assay on recombinant histone H4. (B) Interaction of endogenous AML1-ETO and PRMT1 in leukemic cells positive for t(8;21), Kasumi-1. Endogenous AML1-ETO is immunoprecipitated by PRMT1 Ab in Kasumi-1 cells. Reciprocal immunoprecipitation of AML1-ETO pulls down endogenous PRMT1 in Kasumi-1 cells. (C) Interaction of AE9a and Prmt1 in leukemic cells expressing HA-AE9a. Cell lysates from leukemic cells expressing HA-AE9a were subjected to HA-IP (human influenza hemagglutinin–immunoprecipitation) to pull down endogenous Prmt1.
Identification of PRMT1 as an AE9a-interacting protein. (A) AE9a-PRMT1 interaction is confirmed by both FLAG-IP and PRMT1-IP in 293T cells transiently expressing FLAG-tagged AE9a. Control lane (C) represents 293T cells transfected with empty vector. Endogenous PRMT1 associated with AE9a is enzymatically active as shown by in vitro methyltransferase assay on recombinant histone H4. (B) Interaction of endogenous AML1-ETO and PRMT1 in leukemic cells positive for t(8;21), Kasumi-1. Endogenous AML1-ETO is immunoprecipitated by PRMT1 Ab in Kasumi-1 cells. Reciprocal immunoprecipitation of AML1-ETO pulls down endogenous PRMT1 in Kasumi-1 cells. (C) Interaction of AE9a and Prmt1 in leukemic cells expressing HA-AE9a. Cell lysates from leukemic cells expressing HA-AE9a were subjected to HA-IP (human influenza hemagglutinin–immunoprecipitation) to pull down endogenous Prmt1.
Next, we examined the interaction between endogenous AML1-ETO and PRMT1 in a t(8;21) leukemia patient-derived cell line, Kasumi-1. Immunoprecipitation by anti-PRMT1 Ab pulled down AML1-ETO from Kasumi-1 cell lysates and reciprocal immunoprecipitation by anti–AML1-ETO Ab pulled down PRMT1 (Figure 2B). Furthermore, we also confirmed AE9a-PRMT1 interaction with the use of lysates from primary leukemic splenocytes expressing HA-AE9a (Figure 2C).
To test whether AE9a directly binds PRMT1, we conducted a protein interaction study with the use of recombinant GST-PRMT1 (supplemental Figure 1A) and in vitro transcribed/translated AE9a (supplemental Figure 1B lane 1). After GST pull-down, AE9a was detected in the presence of GST-PRMT1 (supplemental Figure 1B lane 3), suggesting that AE9a directly binds PRMT1. Finally, we used a series of AE9a deletion mutants to find the critical domain for PRMT1 interaction (supplemental Figure 2). Lysates from 293T cells coexpressing FLAG-PRMT1 and HA-AE9a (or its deletion mutants) were subjected to FLAG-IP. Both AML1 and ETO domains of the fusion protein are important for the interaction with PRMT1. Although the C-terminus (last 36 aa, downstream of NHR2 domain) of AE9a was dispensable for PRMT1 binding, further deletion toward N-terminus rendered AE9a incapable to bind PRMT1. Taken together, our data suggest that > 1 region, including both AML1 and ETO parts of AE9a, is involved in PRMT1 binding.
AE9a is methylated at Arg142 by PRMT1
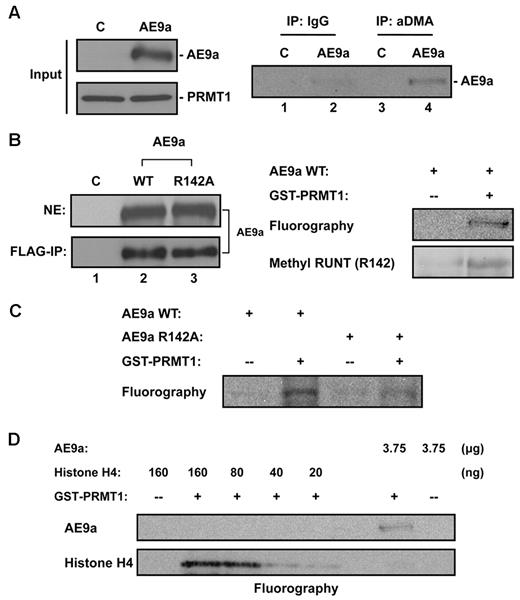
Considerable evidence from previous studies has shown that PRMT1-interacting proteins are often also its methylation targets. AE9a contains several Arg-X-Arg sequences, which is considered a consensus methylation site by PRMT1.36 To determine whether AE9a is indeed arginine methylated in vivo, we transiently expressed HA-tagged PRMT1, to enhance bulk protein methylation, and FLAG-tagged AE9a in 293T cells. Derived cell lysates were subjected to immunoprecipitation with the use of a polyclonal Ab against aDMA. The isolated pool of arginine methylated proteins was then analyzed for the presence of AE9a by Western blotting. AE9a is specifically recognized and immunoprecipitated by aDMA Ab (Figure 3A lanes 2,4). This experiment showed that transiently expressed AE9a is possibly asymmetrically dimethylated on arginine and is probably a PRMT1 target. We next determined whether AE9a is a direct methylation target of PRMT1 with the use of the in vitro methyltransferase assay. 293T cells were transiently transfected with FLAG-tagged AE9a-expressing construct and cultured with Adox to minimize endogenous arginine methylation. Nuclear extracts were prepared and subjected to immunoprecipitation by anti-FLAG M2 affinity gel. Enriched bead-bound AE9a was confirmed by Western blot analysis with anti-FLAG Ab (Figure 3B left panel lanes 1-2), visualized by Coomassie blue staining for quantification, and used as substrates for the in vitro methyltransferase assay. Reaction mixtures containing AE9a, recombinant GST-PRMT1, and methyl donor 3H-SAM were incubated and analyzed by SDS-PAGE and fluorography (Figure 3B right panel). The result of this in vitro assay indicates that AE9a is methylated in the presence of GST-PRMT1.
Methylation of AE9a in vivo and in vitro. (A) In vivo methylation of AE9a. Lysates of 293T cells coexpressing HA-PRMT1 and FLAG-AE9a was subjected to immunoprecipitation by anti-aDMA followed by Western blot analysis with FLAG Ab. Control lane (C) represents 293T cells transfected with empty vector. (B) PRMT1 can methylate AE9a in vitro at R142 residue. FLAG-tagged AE9a, WT or R142A, was transiently transfected and expressed in 293T cells. Control lane (C) represents 293T cells transfected with empty vector. After transfection, cells were treated with 20μM Adox. Nuclear extracts (NEs) were made and subjected to FLAG immunoprecipitation to enrich AE9a. Bead-bound AE9a WT was incubated with recombinant GST-PRMT1 and 3H-SAM (for fluorography) or unlabeled SAM (for Western blot analysis) for in vitro methylation. Results were analyzed by SDS-PAGE, and methylated FLAG-AE9a WT was either visualized by fluorography or detected by Western blot analysis with methyl RUNT (R142) Ab. (C) Arg142 residue of AE9a is the main methylation site for PRMT1. After FLAG-IP enrichment, equal amount of WT or R142A AE9a was used to incubate with recombinant GST-PRMT1 and 3H-SAM for in vitro methylation. Results were analyzed by SDS-PAGE, and methylated FLAG-AE9a was visualized by fluorography. (D) Methylation on AE9a by PRMT1 is much weaker than histone H4. AE9a and various amounts of histone H4 were used in methyltransferase assay. Protein methylation was analyzed by SDS-PAGE and visualized by fluorography. Methylation on AE9a was compared with histone H4 to obtain relative methylation status.
Methylation of AE9a in vivo and in vitro. (A) In vivo methylation of AE9a. Lysates of 293T cells coexpressing HA-PRMT1 and FLAG-AE9a was subjected to immunoprecipitation by anti-aDMA followed by Western blot analysis with FLAG Ab. Control lane (C) represents 293T cells transfected with empty vector. (B) PRMT1 can methylate AE9a in vitro at R142 residue. FLAG-tagged AE9a, WT or R142A, was transiently transfected and expressed in 293T cells. Control lane (C) represents 293T cells transfected with empty vector. After transfection, cells were treated with 20μM Adox. Nuclear extracts (NEs) were made and subjected to FLAG immunoprecipitation to enrich AE9a. Bead-bound AE9a WT was incubated with recombinant GST-PRMT1 and 3H-SAM (for fluorography) or unlabeled SAM (for Western blot analysis) for in vitro methylation. Results were analyzed by SDS-PAGE, and methylated FLAG-AE9a WT was either visualized by fluorography or detected by Western blot analysis with methyl RUNT (R142) Ab. (C) Arg142 residue of AE9a is the main methylation site for PRMT1. After FLAG-IP enrichment, equal amount of WT or R142A AE9a was used to incubate with recombinant GST-PRMT1 and 3H-SAM for in vitro methylation. Results were analyzed by SDS-PAGE, and methylated FLAG-AE9a was visualized by fluorography. (D) Methylation on AE9a by PRMT1 is much weaker than histone H4. AE9a and various amounts of histone H4 were used in methyltransferase assay. Protein methylation was analyzed by SDS-PAGE and visualized by fluorography. Methylation on AE9a was compared with histone H4 to obtain relative methylation status.
PRMT1 is known to methylate histone H4 at Arg3. In addition to Arg-X-Arg motifs, AE9a also contains a histone H4 N-terminal tail-like motif (SGRGK) at amino acids 140-144. In AML1, arginine methylation in this exact motif has been reported previously.32 We therefore speculated that AE9a could also be methylated by PRMT1 at Arg142. To address this hypothesis, in vitro methyltransferase assay with the use of unlabeled SAM as methyl donor was performed. Methylated AE9a was detected in Western blot analysis with the use of Ab specific for methylated R142 residue in RUNT domain (Figure 3B right panel). To further validate that Arg142 of AE9a is the methylation target of PRMT1, we next repeated in vitro methyltransferase assay with the use of wild-type (WT) or R142A-mutant AE9a as substrates. Bead-bound AE9a was prepared as described earlier, and the same amount of AE9a (WT or R142A) was used to compare the methylation status (Figure 3B left panel lanes 2-3). Unlike WT AE9a, only marginal methylation is detected when R142A-mutant AE9a is present (Figure 3C). On the basis of our results, we conclude that AE9a not only associates with, but also is a direct methylation target of, PRMT1. Arg142 of AE9a, which mimics the histone H4 N-terminal Arg3, is probably the major methylation site for PRMT1. Notably, methylation of AE9a by PRMT1 is ∼ 15 times weaker than with histone H4 methylation (Figure 3D).
PRMT1 is critical for AE9a-mediated transcription activation
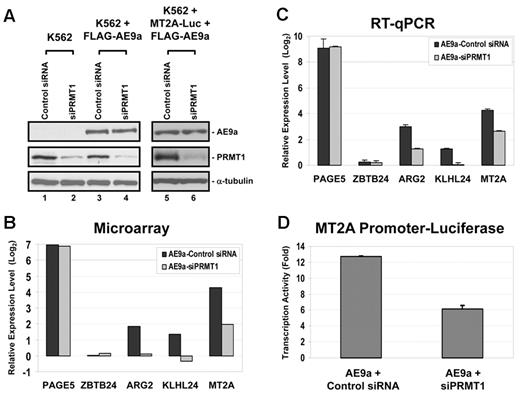
The fact that AE9a is a relatively weak PRMT1 methylation target suggests that PRMT1 is more probable to participate in regulating AE9a function as a cofactor. The role of PRMT1 as a transcription coactivator in gene regulation has been extensively studied. We therefore hypothesized that transcription factor AE9a could bind to DNA and recruit PRMT1 to promoters for histone modification to facilitate the transcription process. To test this hypothesis, we took advantage of siRNA and microarray analysis to examine the effect of PRMT1 on genome-wide AE9a transcription profile in the hematopoietic cell line K562. Parental K562 cells or a pool of FLAG-AE9a–expressing K562 cells were transfected with either control or PRMT1 siRNA. Knockdown efficiency of PRMT1 was determined by Western blot analysis that displayed a reduced level of PRMT1 by ∼ 75% after normalizing to α-tubulin (Figure 4A lanes 1-4). We also prepared total RNA from 3 independent sets of siRNA-treated cells and performed microarray gene expression profiling analysis to examine the influence of siPRMT1 on AE9a-regulated genes. As expected, expression of AE9a resulted in significant gene expression changes, both positively and negatively. siPRMT1 alone did not seem to have much influence on genome-wide transcription in parental K562 cells, which was also observed previously in HeLa cells.37 However, siPRMT1 did change the expression of a number of genes when AE9a was present. Considering the high correlation between PRMT1 and transcription activation, we analyzed the effect of siPRMT1 from 914 AE9a-activated genes. Among these genes, PRMT1 negatively regulated only 12 of them, but positively regulated 51 genes (supplemental Table 4). From these 51 AE9a/PRMT1-dependent genes, ARG2 (arginase, type II), KLHL24 (kelch-like 24), and MT2A (metallothionein 2A) were picked (Figure 4B) to confirm their expression in K562-AE9a cells by RT-qPCR under control siRNA or siPRMT1 knockdown conditions. Results on these genes from RT-qPCR correlated well with microarray data not only in AE9a-dependent gene activation but also in sensitivity to siPRMT1 knockdown (Figure 4C). We also included PAGE5 (P Ag family, member 5; AE9a-activated, PRMT1-independent), and ZBTB24 (zinc finger and BTB domain containing 24; AE9a- and PRMT1-independent) as control genes which did not respond to PRMT1 siRNA. In addition, the role of AE9a in activating transcription of MT2A gene in K562 cells was confirmed by luciferase reporter assay (Figure 4D). More importantly, AE9a-dependent transactivation of MT2A is diminished when PRMT1 is knocked down, as shown in reporter assay (Figure 4A lanes 5-6,D).
Knockdown of PRMT1 affects AE9a-driven transcription activation. (A) Knockdown of PRMT1 by siRNA. Knockdown efficiency of endogenous PRMT1 was examined by Western blot analysis. In lanes 1 through 4, WT K562 cells or AE9a-expressing K562 cells were transfected with either control siRNA or siPRMT1. These cells were used in microarray analysis. In lanes 5 and 6, WT K562 cells were cotransfected with pGLX2-MT2A-Luc, p3xFLAG-AE9a, and either control siRNA or siPRMT1. These cells were used in luciferase reporter assay. α-tubulin is used as a loading control. (B) Bar chart represents relative expression level in log2 scale of selected genes in microarray analysis. Results from AE9a-Control siRNA are normalized to K562-Control siRNA cells. Results from AE9a-siPRMT1 are normalized to K562-siPRMT1 cells. (C) Bar chart represents relative expression level in log2 scale of selected genes by RT-qPCR to validate results from microarray analysis. GAPDH is used as an internal control. Results from AE9a-Control siRNA are normalized to K562-Control siRNA cells. Results from AE9a-siPRMT1 are normalized to K562-siPRMT1 cells. Error bars represent SDs of 3 independent experiments. (D) PRMT1 is important for AE9a-dependent transcription of MT2A. WT K562 cells were cotransfected with pGLX2-MT2A-Luc, p3xFLAG-AE9a, and either control siRNA or siPRMT1. Error bars represent SDs of 3 independent experiments.
Knockdown of PRMT1 affects AE9a-driven transcription activation. (A) Knockdown of PRMT1 by siRNA. Knockdown efficiency of endogenous PRMT1 was examined by Western blot analysis. In lanes 1 through 4, WT K562 cells or AE9a-expressing K562 cells were transfected with either control siRNA or siPRMT1. These cells were used in microarray analysis. In lanes 5 and 6, WT K562 cells were cotransfected with pGLX2-MT2A-Luc, p3xFLAG-AE9a, and either control siRNA or siPRMT1. These cells were used in luciferase reporter assay. α-tubulin is used as a loading control. (B) Bar chart represents relative expression level in log2 scale of selected genes in microarray analysis. Results from AE9a-Control siRNA are normalized to K562-Control siRNA cells. Results from AE9a-siPRMT1 are normalized to K562-siPRMT1 cells. (C) Bar chart represents relative expression level in log2 scale of selected genes by RT-qPCR to validate results from microarray analysis. GAPDH is used as an internal control. Results from AE9a-Control siRNA are normalized to K562-Control siRNA cells. Results from AE9a-siPRMT1 are normalized to K562-siPRMT1 cells. Error bars represent SDs of 3 independent experiments. (D) PRMT1 is important for AE9a-dependent transcription of MT2A. WT K562 cells were cotransfected with pGLX2-MT2A-Luc, p3xFLAG-AE9a, and either control siRNA or siPRMT1. Error bars represent SDs of 3 independent experiments.
To further strengthen our findings, we next examined whether PRMT1 is important for AML1-ETO–mediated transcriptional activation in Kasumi-1 cells. We treated Kasumi-1 cells with either siPRMT1 or PRMT1 inhibitory compound RM65. Knockdown or inhibition of PRMT1 was confirmed by PRMT1 (supplemental Figure 3A lane 2) or H4R3Me2a (supplemental Figure 3A lane 4) Western blot analysis. In the presence of RM65, cell growth rate was reduced (supplemental Figure 3B). On the basis of previous reports on AML1-ETO–activated genes in Kasumi-1 cells,38 we picked 9 of them to study whether knockdown or inhibition of PRMT1 affect their expression. Among these 9 examined genes, expression of EDIL3, MTSS1, and SLC2A3 was down-regulated in response to siPRMT1 or RM65 treatment (supplemental Figure 3C-D), suggesting that PRMT1 is involved in AML1-ETO–mediated transcription activation in Kasumi-1 cells.
AE9a recruits PRMT1 to promoters for histone modifications that favor transcription
To further establish the role of PRMT1 in AE9a-driven transcription, we sought to determine whether AE9a could function as a PRMT1 recruiter for promoter binding on selected genes (Figure 5A). ChIP assays from parental or FLAG-AE9a–expressing K562 cells were performed with Abs against FLAG epitope or PRMT1, followed by qPCR amplification of the promoter region that contained ≥ 1 consensus AML1 site. We observed promoter binding of AE9a on all genes checked (Figure 5B). More importantly, binding of AE9a resulted in enrichment of PRMT1 only on promoters of those 3 PRMT1-dependent genes (Figure 5C). Next, we investigated whether the recruitment of PRMT1 altered methylation status of H4 Arg3 on promoters. Results from histone H4R3Me2a ChIP analysis clearly showed increased histone H4 Arg3 methylation on AE9a-activated ARG2, KLHL24, and MT2A promoters, but not on control PAGE5 and ZBTB24 promoters (Figure 5D). Histone H4 Arg3 methylation and histone H3 Lys9/Lys14 acetylation are both considered transcriptional activation markers. It has been reported that histone H4 Arg3 methylation catalyzed by PRMT1 facilitates histone H3 Lys9/Lys14 acetylation on transcriptionally active promoters.39,40 We observed similar histone modification changes as shown by histone H3K9/14Ac ChIP analysis (Figure 5E). In addition, we also performed ChIP analysis for PRMT1 and histone H4R3Me2a in siPRMT1-treated K562-AE9a or Kasumi-1 cells to confirm the direct role of PRMT1 as shown in Figure 5. As expected, we observed significant decreases of both PRMT1 and H4R3Me2a on promoters of either AE9a/PRMT1 target genes in K562-AE9a cells (supplemental Figure 4) or AML1-ETO/PRMT1 target genes in Kasumi-1 cells (supplemental Figure 3E-F) after siPRMT1 treatment. Taken together, our findings provide strong supporting evidence that AE9a recruits PRMT1 to activate transcription.
ChIP analyses of AE9a, PRMT1, histone H4R3Me2a, and histone H3K9/14Ac on promoters of interest. (A) Schematic diagrams show promoters analyzed by ChIP. Positions of the predicted transcription initiation sites (solid arrows) are indicated. Relative positions of consensus AML1 sites (white boxes) and primers (dotted arrows) in each promoter are shown. (B) ChIP results for AE9a. (C) ChIP results for PRMT1. (D) ChIP results for H4R3Me2a. (E) ChIP results for H3K9/14Ac. Promoter of ACTB is used as an internal control. Data represented are relative enrichment of specific ChIP target in K562-AE9a cells over WT K562 cells. Error bars represent SDs of 3 independent experiments.
ChIP analyses of AE9a, PRMT1, histone H4R3Me2a, and histone H3K9/14Ac on promoters of interest. (A) Schematic diagrams show promoters analyzed by ChIP. Positions of the predicted transcription initiation sites (solid arrows) are indicated. Relative positions of consensus AML1 sites (white boxes) and primers (dotted arrows) in each promoter are shown. (B) ChIP results for AE9a. (C) ChIP results for PRMT1. (D) ChIP results for H4R3Me2a. (E) ChIP results for H3K9/14Ac. Promoter of ACTB is used as an internal control. Data represented are relative enrichment of specific ChIP target in K562-AE9a cells over WT K562 cells. Error bars represent SDs of 3 independent experiments.
Knockdown of PRMT1 suppresses the proliferative potential of AE9a in progenitor cells
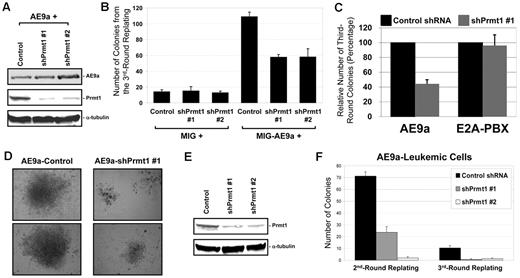
We have previously reported the identification of AE9a as a potent inducer of leukemia development in mice. To study the biologic effects of PRMT1 on AE9a, we used in vitro retroviral transduction/transformation assay.20,41,42 To this aim, we transduced AE9a- and shPrmt1-expressing vectors into murine BM cells that were subjected to serial replating and CFU assay to assess the clonogenic potential of AE9a and the influence of Prmt1 knockdown. The efficiency of transduced shPrmt1 was confirmed by Western blot analysis, which exhibited reduction in Prmt1 protein expression (Figure 6A). Compared with vector control, AE9a-transduced cells dramatically increased the number of colonies formed, indicating its self-renewal capability. Interestingly, colony number was significantly reduced in the presence of shPrmt1 (using 2 independent shPrmt1 sequences), which suggested a critical function for the specific recruitment of Prmt1 in AE9a leukemogenesis (Figure 6B). The specificity was further supported when we included E2A-PBX, a previously reported Prmt1-independent oncoprotein, in our assay.30 Unlike AE9a-transduced cells, knockdown of Prmt1 did not affect colony-forming ability of E2A-PBX–transduced cells (Figure 6C). We also noticed a phenotypic difference in colonies formed. In general, colonies from shPrmt1-transduced cells are slightly smaller in size than from control shRNA-transduced cells (data not shown). However, AE9a-shPrmt1 cotransduced cells produced much smaller colonies in all replating rounds (Figure 6D shows typical image of the third-round colonies). We did not observe any obvious difference in morphology of AE9a-control shRNA and AE9a-shPrmt1 cells (data not shown). However, we noticed that there is an increase of Gr-1+/Cd11b+ cells and a decrease of c-Kit+/Sca-1+ cells in AE9a-shPrmt1 cells (supplemental Figure 5), suggesting that Prmt1 knockdown reduces AE9a-involved differentiation block. In addition, Prmt1 knockdown (Figure 6E) reduced the growth and survival of primary AE9a leukemic cells (Figure 6F), which provides additional support for the role of PRMT1 in leukemia development.
The role of Prmt1 in AE9a-mediated self-renewal in murine BM cells. (A) Western blot analysis for AE9a and Prmt1 expression from BM cells cotransduced with AE9a, control shRNA, or shPrmt1 retroviruses after puromycin selection. α-tubulin is used as a loading control. (B) Bar chart represents the number of colonies from the third-round replating of BM cells. Error bars represent SDs of 3 independent experiments. (C) BM cells were cotransduced with AE9a or E2A-PBX in combination with control shRNA or shPrmt1 #1. Bar chart represents the relative number of colonies from the third-round replating of BM cells. Error bars represent SDs of 2 independent experiments. (D) Typical image of the third-round colonies generated from BM cells cotransduced with AE9a, control shRNA, or shPrmt1 retroviruses. (E) Western blot analysis for Prmt1 expression from AE9a-leukemic cells transduced with control shRNA or shPrmt1 retroviruses after puromycin selection. α-tubulin is used as a loading control. Images were taken using Nikon Eclipse TS100 microscope with 10×/0.25 objective lens and Nikon DS Camera Control Unit DS-U2 system. (F) Bar chart represents the number of colonies from the second- and third-round replating of primary AE9a leukemic cells. Error bars represent SDs of 3 independent experiments.
The role of Prmt1 in AE9a-mediated self-renewal in murine BM cells. (A) Western blot analysis for AE9a and Prmt1 expression from BM cells cotransduced with AE9a, control shRNA, or shPrmt1 retroviruses after puromycin selection. α-tubulin is used as a loading control. (B) Bar chart represents the number of colonies from the third-round replating of BM cells. Error bars represent SDs of 3 independent experiments. (C) BM cells were cotransduced with AE9a or E2A-PBX in combination with control shRNA or shPrmt1 #1. Bar chart represents the relative number of colonies from the third-round replating of BM cells. Error bars represent SDs of 2 independent experiments. (D) Typical image of the third-round colonies generated from BM cells cotransduced with AE9a, control shRNA, or shPrmt1 retroviruses. (E) Western blot analysis for Prmt1 expression from AE9a-leukemic cells transduced with control shRNA or shPrmt1 retroviruses after puromycin selection. α-tubulin is used as a loading control. Images were taken using Nikon Eclipse TS100 microscope with 10×/0.25 objective lens and Nikon DS Camera Control Unit DS-U2 system. (F) Bar chart represents the number of colonies from the second- and third-round replating of primary AE9a leukemic cells. Error bars represent SDs of 3 independent experiments.
Discussion
PRMT1 is the predominant arginine methyltransferase in cells that regulates a variety of cellular processes by methylating protein factors or histones. Here, we reported the association of PRMT1 with AE9a and its crucial role on AE9a function, including transcription regulation and self-renewal capability.
AE9a harbors several consensus methylation motifs that make it a potential PRMT1 target, but it is only found to be weakly methylated by PRMT1. We have shown that PRMT1 could methylate AE9a at R142 residue that is in the histone H4 N-terminal tail-like motif (SGRGK). The arginine within SGRGK of histone H4 is methylated by PRMT1.16 Compared with histone H4, arginine methylation within SGRGK of AE9a is much weaker, which may be because of the imperfect positioning of the PRMT1 enzyme active site to reach R142 when bound to AE9a. Mutation of R142 significantly decreases the transactivation ability of AE9a in a reporter assay (supplemental Figure 6A). However, it has been reported that R142A mutation weakens DNA-binding ability of the RUNT domain without disturbing its overall structure and affinity to CBFβ.43,44 In fact, we also have data to support the negative effect of R142A mutation on DNA binding of AE9a (supplemental Figure 6B) as well as cell replating capability (supplemental Figure 6C). The negative effect of R142A mutation on AE9a-driven transactivation or transformation may be contributed by methylation deficiency and/or weaker DNA affinity. At this point, whether methylation on AE9a R142 itself is significant enough to be involved in regulating any cellular functions remains unclear. Importantly, AE9a-associated PRMT1 is active in methylating histone H4 which suggests that it may regulate AE9a function by altering chromatin dynamics. In this case, PRMT1 functions as a cofactor and is recruited by transcription factor AE9a to specific promoters for histone modifications that favor transcription activation.
Full-length AML1-ETO has been reported to interact with several transcription repressors, including N-CoR/SMRT, mSin3A and HDACs, through various ETO regions. Although missing the C-terminal NHR3 and NHR4 domains of ETO, AE9a is still capable of interacting with those corepressors presumably via NHR2 domain.45,46 The fact that AE9a could interact with PRMT1 for transcription activation and corepressors for transcription repression is uncommon, but not unprecedented. Sequence-specific DNA-binding transcription factor Yin Yang 1 could recruit PRMT1 and HDACs to either positively or negatively regulate transcription.47 Transcription factor AML1 is found to interact with and be methylated by PRMT1.32 AML1 is methylated at its mSin3A-interacting region, and the PRMT1-dependent methylation of AML1 abrogates mSin3A binding, thus potentiating its transcription activity. It should be noted that the reported methylation site of AML1 is not on the N-terminus of AML1 that fuses to ETO to generate AML1-ETO. Given that AE9a could regulate gene expression both positively and negatively, it will be of great importance to discover the biologic signal or switch that could determine the transcription outcome driven by AE9a. It is worth mentioning that AE9a also positively regulated gene expression in a PRMT1-independent manner. It remains to be determined whether and how other cofactors are involved in transcription regulation by AE9a.
Posttranslational modifications on histone tails play important roles in chromatin function. Crosstalk between these covalent modifications often occurs to achieve specific regulatory activities. Histone H4 Arg3 methylation by PRMT1 has been reported to facilitate subsequent acetylation of H4 tails that leads to transcription activation mediated by nuclear receptor and p53.15,16 The critical role of PRMT1 in transcription activation cascade is further defined in vivo on β-globin transcription whereby H4 Arg3 methylation is essential for histone acetyltransferase binding, histone H3 and H4 acetylation, and promoter/enhancer communications that lead to efficient recruitment of preinitiation complexes to activate transcription.39,40 Here, we report another example of PRMT1 recruitment that alters both histone methylation (H4 Arg3) and acetylation (H3 Lys9/14), thus positively modulating a specific set of AE9a-regulated genes. Our findings provide a potential molecular mechanism for AE9a function on transcription level that could ultimately play a key role in leukemia development. Interestingly, AML1-ETO has recently been reported to interact with p300, which was essential for AML1-ETO–mediated gene activation, self-renewal, and leukemogenesis.11 Given the previously reported cooperative functions of PRMT1 and p300,15,16 it is probable that PRMT1 functions as another transcription coactivator to facilitate leukemogenicity of AML1-ETO.
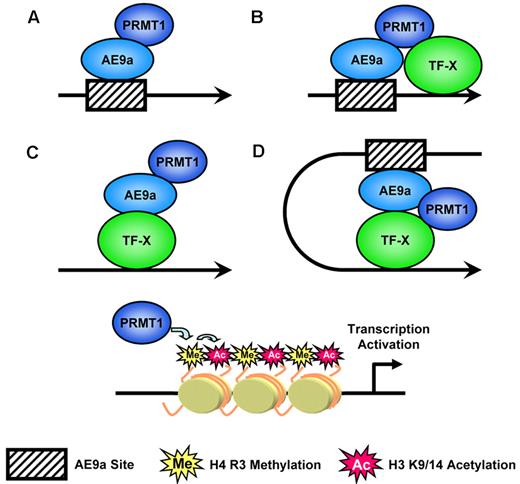
We provide strong evidence that AE9a functions as a PRMT1 recruiter to facilitate transcription. AE9a could act as a sole transcription factor that brings PRMT1 to promoters by binding to consensus sites. However, we could not rule out the possibility that other transcription factors were also involved in PRMT1 recruitment (Figure 7). The interaction of PRMT1 with a number of transcription factors has been reported. It is possible that AE9a cooperates with another transcription factor that binds to DNA near AE9a binding site to synergistically recruit PRMT1. We also observed AE9a and PRMT1 binding at promoters that lacked consensus AE9a binding sites. In this case, AE9a might be indirectly recruited to promoters by unknown transcription factors which also resulted in PRMT1 enrichment. On the basis of the reported role of PRMT1 in enhancer-promoter communication mentioned earlier, it is certainly possible that AE9a could bind DNA in upstream regulatory elements and bring in PRMT1 to participate in gene activation over long distances. Ultimately, all these possible mechanisms alter histone modifications on promoters that lead to transcription activation.
Models for AE9a-mediated PRMT1 recruitment that leads to transcription activation. AE9a could recruit PRMT1 to promoter directly, indirectly, or in cooperation with other transcription factors to alter histone modifications and to facilitate transcription.
Models for AE9a-mediated PRMT1 recruitment that leads to transcription activation. AE9a could recruit PRMT1 to promoter directly, indirectly, or in cooperation with other transcription factors to alter histone modifications and to facilitate transcription.
Dysregulation of transcription and aberrant chromatin modifications are often found in human cancer. A number of proteins responsible for a specific histone modification have been identified and extensively studied for their involvement in cancer and disease development.12 Abnormal PRMT1 expression is found in colon and bladder cancers,48,49 and its correspondent H4 Arg3 methylation could be used to predict recurrence of prostate cancer.50 The specific role of PRMT1 in hematopoiesis has also been investigated. In human leukemia cells, expression of PRMT1 suppresses hematopoietic differentiation via modulating the p38 MAPK pathway.51 B-cell translocation 1 is reported to regulate glucocorticoid receptor autoinduction in acute lymphoblastic leukemia through cooperative interaction with PRMT1.52 AML1 is methylated by PRMT1 in a lineage-dependent manner which may dictate the pattern of hematopoietic cell differentiation.32 More importantly, as an essential component of a mixed lineage leukemia transcriptional complex that modifies histones, the direct role of PRMT1 in transcriptional up-regulation during cancer progression has been clearly shown.30 In support of this discovery, we also provide strong evidence to show the importance of PRMT1 in AE9a-mediated transcription and self-renewal. Further studies on elucidating the biologic role of PRMT1 in the development of leukemia and human malignancies are particularly imperative. Given that there are several PRMT1-specific inhibitory compounds identified, it will be of interest to determine whether these inhibitors exert any therapeutic effect on leukemia development in the future.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Russell DeKelver for editing the manuscript, all other members of D.-E.Z.'s laboratory for valuable discussions, Dr Michael R. Stallcup for providing PRMT1-expressing vectors, Dr Eric So and Dr Suming Huang for providing shPrmt1 vectors, and Dr Mark Kamps for providing E2A-PBX vector.
This work was supported by the Leukemia & Lymphoma Society (fellowship 5140-08, W.-J.S.) and the National Institutes of Health (grant P41RR011823, J.Y.; and grant CA104509, D.-E.Z.).
National Institutes of Health
Authorship
Contribution: W.-J.S. designed and performed the research, analyzed the data, and wrote the manuscript; A.J.O. designed and performed experiments and analyzed the data; M.Y. designed and performed experiments; M.-C.L., S.M., and Y.K. performed experiments; A.S. and J.R.Y. provided experimental assistance and data analysis; X.Z. and S.D.N. provided critical Abs and experimental advice; and D.-E.Z. supervised experimental design, data analysis, and manuscript preparation.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dong-Er Zhang, Mail Drop 0815, Rm 5328, Moores Cancer Center, University of California, San Diego, 3855 Health Sciences Dr, La Jolla, CA 92093; e-mail: d7zhang@ucsd.edu.