Abstract
In a zebrafish mutagenesis screen to identify genes essential for myelopoiesis, we identified an insertional allele hi1727, which disrupts the gene encoding RNA helicase dead-box 18 (Ddx18). Homozygous Ddx18 mutant embryos exhibit a profound loss of myeloid and erythroid cells along with cardiovascular abnormalities and reduced size. These mutants also display prominent apoptosis and a G1 cell-cycle arrest. Loss of p53, but not Bcl-xl overexpression, rescues myeloid cells to normal levels, suggesting that the hematopoietic defect is because of p53-dependent G1 cell-cycle arrest. We then sequenced primary samples from 262 patients with myeloid malignancies because genes essential for myelopoiesis are often mutated in human leukemias. We identified 4 nonsynonymous sequence variants (NSVs) of DDX18 in acute myeloid leukemia (AML) patient samples. RNA encoding wild-type DDX18 and 3 NSVs rescued the hematopoietic defect, indicating normal DDX18 activity. RNA encoding one mutation, DDX18-E76del, was unable to rescue hematopoiesis, and resulted in reduced myeloid cell numbers in ddx18hi1727/+ embryos, indicating this NSV likely functions as a dominant-negative allele. These studies demonstrate the use of the zebrafish as a robust in vivo system for assessing the function of genes mutated in AML, which will become increasingly important as more sequence variants are identified by next-generation resequencing technologies.
Introduction
Genes lost or mutated in human leukemias are often found to be essential for normal developmental hematopoiesis in animal models. Accordingly, developmental hematopoiesis is frequently disrupted in murine knockout models of tumor suppressor genes implicated in the pathogenesis of human leukemias. Using an animal model to identify and functionally assess genes necessary for developmental hematopoiesis could facilitate the characterization of new genes mutated in human leukemias.1-5 Importantly, as the zebrafish is highly genetically tractable, the functions of genes whose loss or mutation results in abnormal hematopoiesis can be analyzed in vivo.
DEAD-box 18 (DDX18) encodes a DEAD-box protein (DBP), which are members of an evolutionarily conserved family of energy-dependent RNA helicases involved a broad range of functions associated with RNA (including ribosome biogenesis, microRNA processing, and mRNA transport). DBPs contain a central core required for their enzymatic helicase activity and diverse carboxy- and N-termini presumed to confer substrate specificity. DBPs are members of superfamily II (SFII) of helicases/translocases. Members of this large superfamily all possess the capacity for ATP-dependent translocation on single- or double-stranded nucleic acid.6 There are 37 characterized DEAD-box helicases in eukaryotes, showing minimal redundancy in function. Even the highly homologous proteins DDX5 and DDX17 (which are 90% identical to one another and 80% and 72% identical to their single yeast orthologue) are both essential genes in murine development indicating nonoverlapping functions.7 DDX18, also known as Myc-related DEAD-box 18 (MrDB; because its identification as a direct target of Myc-Max heterodimers8 ), is a highly conserved protein (72% identical amino acid sequence between zebrafish and human) whose orthologue in yeast, HAS1 (exhibiting 60% amino acid identity with human DDX18) is essential for ribosomal biogenesis. Human DDX18 is located on chromosome band 2q14.1, which has not been associated with nonrandom cytogenetic abnormalities in acute myeloid leukemia (AML) or myelodysplastic syndrome (MDS). However, low levels of DDX18 expression form part of a gene expression signature in normal karyotype human AML associated with low ERG or BAALC expression.9 In this study, patients over 60 with normal karyotype AML and low expression levels of ERG and/or BAALC were identified as having a benefit in overall survival, independent of any other variables. Gene expression profiles of patients with either low ERG or low BAALC showed down-regulation of a subset of genes (962 genes and 127 genes, respectively) which, in both datasets, included down-regulation of DDX18 expression to ∼ 75% of normal.9 Specific functions of DDX18 in mammals have not been studied, and a role for DDX18 in hematopoiesis has not previously been described. Several other genes involved in ribosome biogenesis have been implicated in the pathogenesis of hematopoietic disorders. Heterozygous mutations in ribosomal protein (RP) genes of both the small and the large ribosomal subunits (RPS19, RPS24, RPS17, RPL35A, RPL5, RPL11, RPS7, RPS10, and RPS26) have recently been identified as the causative genetic lesions accounting for 50% of patients with Diamond-Blackfan anemia (DBA).10-15 In addition, several other genes essential for ribosome biogenesis have been implicated in congenital syndromes, associated with abnormal hematopoiesis including cartilage hair hypoplasia, and Shwachman-Bodian-Diamond syndrome.16,17 Somatic loss of another RP gene, RPS14 has been implicated in the pathogenesis of anemia in myelodysplastic syndromes with loss of chromosome 5q.18,19
As development of new technology has permitted the sequencing of the entire genome in individual patients with leukemia, the number of genes known to be mutated in such patients is growing exponentially. However, many of the newly identified mutations affect relatively small proportions of AML patients. Although such resequencing technology has tremendous power, there are considerable challenges associated with the large volumes of data which are becoming available. Furthermore, new genes found to be mutated in human leukemias cannot be considered to implicate novel pathways in molecular pathogenesis unless their functional significance has been determined.20,21 Thus, there is great need for in vivo models to test the biologic significance of novel, low frequency genetic mutations found in human leukemias. The use of such models will greatly complement the current bioinformatics approaches used to predict the functional significance of novel nonsynonymous sequence variants (NSVs).
Here we report that a genetic screen in the zebrafish identified ddx18 as a novel gene essential for primitive hematopoiesis. Loss of Ddx18 caused p53-dependent apoptosis and cell-cycle arrest, which was most severe in primitive myeloid cells. Rescue of myelopoiesis in the Ddx18 mutant zebrafish model proved an ideal method to interrogate DDX18 sequence variants identified in human patients with AML and MDS. Thus, the zebrafish system represents a powerful animal model for in vivo analysis of the functional significance of genetic mutations found in humans with hematologic malignancies.
Methods
Patients
Patient BM samples were collected from the Dana-Farber Cancer Institute and Memorial Sloan-Kettering after obtaining their informed consent and with institutional review board (IRB) approval of the relevant institution. All patients had a confirmed diagnosis of MDS or AML. DBA patient samples were obtained after obtaining informed consent in accordance with the Declaration of Helsinki and with IRB approval at Children's Hospital Boston.
Purification and in vitro culture of human CD34+ HSPCs from CB
Mononuclear cells were isolated from cord blood (CB; obtained from the New York Blood Center on a contractual basis) by Ficoll-Hypaque Plus density centrifugation. CD34+ hematopoietic stem and progenitor cells (HSPCs) were purified by positive selection using the Midi-magnetic-activated cell sorting LS-separation columns and isolation kit (Miltenyi Biotec). CD34+ cells were cultured in IMDM (Cellgro) containing 20% BIT 9500 medium (StemCell Technologies) supplemented with Epo (6 IU/mL) and SCF (100 ng/mL) to support erythroid differentiation or SCF (100 ng/mL), FLT-3 (10 ng/mL), IL-3 (20 ng/mL), IL-6 (20 ng/mL), GM-CSF (20 ng/mL), and G-CSF (20 ng/mL) to support myeloid differentiation.
RNA was extracted on the same day of CD34 cell isolation and on day 2 and day 7 of liquid culture using the RNeasy mini kit (QIAGEN).
Zebrafish maintenance
Wild-type stocks of AB fish, transgenics, and mutant lines were maintained as previously described.24 The ddx18 mutant allele ddx18hi1727/+, Tg(pu.1:EGFP), Tg(gata1:dsRed), Tg(cmyb:EGFP), and tp53zdf1/zdf1 (tp53m214k/m215k referred to in the text as p53e7/e7) have been previously reported and described elsewhere.22-24 Staging was performed as described.25
Microinjection
Morpholinos (MO) directed against ddx18 exon 2–intron 2 junction (ddx18E2I2 MO) and ATG/5′ UTR were designed by Genetools LLC. p53 MO was used as previously described.26 Morpholino sequences are shown in Table 1. RNAs were in vitro transcribed from pCS2+ using the Ambion mMessage Sp6 kit (Ambion). MO and RNAs were injected at the 1- to 4-cell stage. Doses were titrated to the working concentrations shown in Table 1.
Morpholino sequences
| Morpholino . | Sequence . | Dose . |
|---|---|---|
| ddx18 E2-I2 | AGCATCATTAAACACTCACTGGATG | 0.8–4 pg |
| ddx18 ATG/5′ UTR | GAATCTTCATCTGCATGTCCGCCAT | 0.4 pg |
| p53 ATG/5′ UTR | GCGCCATTGCTTTGCAAGAATTG | 1.6 pg |
| Genetools standard control | CCTCTTACCTCAGTTACAATTTATA | Variable (max dose of other MO(s) in experiment) |
| Morpholino . | Sequence . | Dose . |
|---|---|---|
| ddx18 E2-I2 | AGCATCATTAAACACTCACTGGATG | 0.8–4 pg |
| ddx18 ATG/5′ UTR | GAATCTTCATCTGCATGTCCGCCAT | 0.4 pg |
| p53 ATG/5′ UTR | GCGCCATTGCTTTGCAAGAATTG | 1.6 pg |
| Genetools standard control | CCTCTTACCTCAGTTACAATTTATA | Variable (max dose of other MO(s) in experiment) |
Bold regions indicate ATG start sites within the respective genes for ATG/5[prime ] UTR MOs.
Whole-mount in situ hybridization
Embryos at the desired time points were fixed overnight in 4% paraformaldehyde (PFA) at 4°C. Whole-mount in situ hybridization (WISH) was performed as previously described.15
Cloning and subcloning
To generate a WISH probe for ddx18 in zebrafish, a 450-bp fragment of zebrafish ddx18 was PCR amplified from RNA derived from pooled 24 hours postfertilization (hpf) wild-type (WT) embryos using the QIAGEN one-step RT-PCR kit. The resulting PCR product was cloned into the pCRII-Topo Dual Promoter vector (Invitrogen). Primers used for the amplification are shown in Table 2. The direction of the clone was confirmed using direct sequencing from M13 forward primers and an antisense digoxigenin-labeled probe was synthesized using NotI and Sp6.
Primer sequences
| . | Sequences . |
|---|---|
| ddx18hi1727 genotyping | ddx18 forward: 5′-GCGGACATGCAGATGAAGAT-3′ |
| Reverse viral (nltr3): 5′-CTGTTCCATCTGTTCCTGAC-3′ | |
| ddx18 reverse wt: 5′-CATTAAATCACACATAGCCACGA-3′ | |
| Zebrafish ddx18 short probe | Forward: 5′-ggctattcatgggaagcaga-3′ |
| Reverse: 5′-TGTAGGCCTCCTGAGCTGAT-3′ | |
| Zebrafish ddx18 qPCR | Forward: 5′-CGGCTGAGGAGGATAGTGATG-3′ |
| Reverse: 5′-ACGCACCTGTCAGACCAGAAG-3′ | |
| Zebrafish β-actin qPCR | Forward: 5′-CTGGTCGTTGACAACGGCT-3′ |
| Reverse: 5′-TCCATCACAATACCAGTAGT-3′ | |
| Human DDX18 qPCR | Forward: 5′-GGGACTGACAGGAGCTTTTGAG-3′ |
| Reverse: 5′-GCCTTCAGAGTGTTTTCATTGACA-3′ | |
| Site-directed mutagenesis primers | hsDDX18_G548A: 5′-GGATACTTCGTTTGCTTCTCTATATAATCTTGTCAATGAAAACACTC-3′ |
| hsDDX18_G548A-r: 5′-GAGTGTTTTCATTGACAAGATTATATAGAGAAGCAAACGAAGTATCC-3′ | |
| hsDDX18_G1111A: 5′-GCCACCCAAACTCGAAAAATTGAAGACCTGGCAA-3′ | |
| hsDDX18_G1111A-r: 5′-TTGCCAGGTCTTCAATTTTTCGAGTTTGGGTGGC-3′ | |
| hsDDX18_G1861A: 5′-TCAAGGTGCCTCCCTTCATTGATCTGAACGTCAAC-3′ | |
| hsDDX18_G1861A-r: 5′-GTTGACGTTCAGATCAATGAAGGGAGGCACCTTGA-3′ | |
| Human DDX18 sequencing primers | Forward: 5′-CGGGATCCTGTTGGGCAGAATGTCACAC-3′ |
| Reverse: 5′-ACTCGAGAAGGAAGGCATGTGTTCAGTG-3′ | |
| middle: 5′-TCCTCTTCAAACCCCACATC-3′ | |
| middle2: 5′-GCTTGATAATGGGTGGCAGT-3′ |
| . | Sequences . |
|---|---|
| ddx18hi1727 genotyping | ddx18 forward: 5′-GCGGACATGCAGATGAAGAT-3′ |
| Reverse viral (nltr3): 5′-CTGTTCCATCTGTTCCTGAC-3′ | |
| ddx18 reverse wt: 5′-CATTAAATCACACATAGCCACGA-3′ | |
| Zebrafish ddx18 short probe | Forward: 5′-ggctattcatgggaagcaga-3′ |
| Reverse: 5′-TGTAGGCCTCCTGAGCTGAT-3′ | |
| Zebrafish ddx18 qPCR | Forward: 5′-CGGCTGAGGAGGATAGTGATG-3′ |
| Reverse: 5′-ACGCACCTGTCAGACCAGAAG-3′ | |
| Zebrafish β-actin qPCR | Forward: 5′-CTGGTCGTTGACAACGGCT-3′ |
| Reverse: 5′-TCCATCACAATACCAGTAGT-3′ | |
| Human DDX18 qPCR | Forward: 5′-GGGACTGACAGGAGCTTTTGAG-3′ |
| Reverse: 5′-GCCTTCAGAGTGTTTTCATTGACA-3′ | |
| Site-directed mutagenesis primers | hsDDX18_G548A: 5′-GGATACTTCGTTTGCTTCTCTATATAATCTTGTCAATGAAAACACTC-3′ |
| hsDDX18_G548A-r: 5′-GAGTGTTTTCATTGACAAGATTATATAGAGAAGCAAACGAAGTATCC-3′ | |
| hsDDX18_G1111A: 5′-GCCACCCAAACTCGAAAAATTGAAGACCTGGCAA-3′ | |
| hsDDX18_G1111A-r: 5′-TTGCCAGGTCTTCAATTTTTCGAGTTTGGGTGGC-3′ | |
| hsDDX18_G1861A: 5′-TCAAGGTGCCTCCCTTCATTGATCTGAACGTCAAC-3′ | |
| hsDDX18_G1861A-r: 5′-GTTGACGTTCAGATCAATGAAGGGAGGCACCTTGA-3′ | |
| Human DDX18 sequencing primers | Forward: 5′-CGGGATCCTGTTGGGCAGAATGTCACAC-3′ |
| Reverse: 5′-ACTCGAGAAGGAAGGCATGTGTTCAGTG-3′ | |
| middle: 5′-TCCTCTTCAAACCCCACATC-3′ | |
| middle2: 5′-GCTTGATAATGGGTGGCAGT-3′ |
Bold, underlined text indicates restriction enzyme linkers.
DDX18 full-length cDNA clone ID 3452935 was purchased from Open Biosystems. Direct sequencing confirmed the full-length open reading frame was present (GenBank no. BC001238.1). The WT DDX18 clone was excised from pCMV-SPORT6 by restriction enzyme digest with EcoRI and XbaI and ligated using T4 ligase (Roche) into pCS2+ using the same restriction enzyme sites (pCS2+DDX18-WT).
Genotyping
For embryo stages < 24 hpf and rescue experiments, ddx18hi1727 embryos were individually genotyped using 1 forward and 2 reverse primers (Table 2). The forward primer was located in the first exon of ddx18, with one reverse primer located in the viral genomic sequence and one in the ddx18 first intron after the viral insertion. Bands at 306 and 360 bp represented the WT and ddx18hi1727 insertional allele, respectively.
Cell-cycle analysis
Tg(pu.1:EGFP) fish were bred to ddx18hi1727/+ to generate Tg(pu.1:EGFP);ddx18hi1727/+ zebrafish. Tg(pu.1:EGFP)/ddx18hi1727/+ were then bred with ddx18hi1727/+ and at 27-hpf embryos were sorted into ddx18hi1727 mutant or siblings based on the developmental morphology. Embryos (30 per condition) were dissociated in liberase blendzyme III (Roche) at a dilution of 1:200 at 32°C for 30 minutes. Cell suspensions were washed twice in PBS containing 5% FBS. Green fluorescent protein (GFP)–expressing cells were sorted on a FACSAria flow cytometer. Sorted cells were lysed in hypotonic PI solution (containing 50μM propidium iodide, 0.1% sodium citrate, and 0.1% Triton X-100) and incubated for 1 hour before analysis. Cell-cycle analysis was carried out on a FACSCanto and analyzed using Modfit software.
Imaging
Embryos were imaged on Leica M420 microscope with a Nikon camera, using ACT2U or NIS Elements software (both Nikon Instruments Inc). For live bright field images, embryos were treated with tricaine 0.4% and mounted in 1.5% methylcellulose. Fixed embryos were mounted in 90% glycerol.
Quantitative PCR analysis
Tg(cmyb:EGFP);ddx18hi1727/+, Tg(gata1:dsRed);ddx18hi1727/+, and Tg(pu.1:EGFP);ddx18hi1727/+ were incrossed and sorted at 32-hpf (cmyb) and 24-hpf (gata1 and pu.1) into ddx18hi1727 mutants and siblings. Embryos were dissociated as described in “Cell-cycle analysis” and sorted on a FACSAria. For Tg(cmyb:EGFP) only cells with high forward scatter/side scatter were sorted as previously described.16 RNA from sorted cells was extracted using TRIzol reagent (Invitrogen) with GlycoBlue (Ambion) to facilitate RNA precipitation and visualization. cDNA was synthesized using Superscript III (Invitrogen) for both zebrafish and in vitro–differentiated primary human cells. Quantitative PCR (qPCR) was carried on a Bio-Rad i-Cycler using SYBR Green. Fold change was calculated using the δ-δ CT method, normalized to β actin. Primer sequences are shown in Table 2.
Western blotting
Embryos for protein extraction were sorted into ddx18hi1727 mutant and siblings where appropriate and manually de-yolked with forceps. Extracts from 10-20 embryos per condition were lysed in radio-immunoprecipitate assay (RIPA) buffer containing protease inhibitor cocktail (Sigma-Aldrich). Fifteen- to 50 μg of protein lysate was separated using 4%-12% NuPage Bis-Tris Gel (Invitrogen) and then transferred onto a nitrocellulose membrane using standard protocols. Rabbit anti-DDX18 (Bethyl Laboratories), mouse anti–human β-actin, and anti–rabbit-HRP and anti–mouse-HRP secondary conjugates were used.
Sequencing of human AML and MDS BM samples
Sanger sequencing was performed exon by exon using gDNA from BM samples from adults with AML or MDS obtained from the Dana-Farber Cancer Institute by Agencourt Bioscience Corporation. Traces were analyzed using Mutation Surveyor software (Softgenetics LLC). Further AML and MDS patient BM samples were obtained from Memorial Sloan-Kettering and sequenced by Genewiz Inc. DBA patient samples were sequenced in collaboration with H.T.G.
Site-directed mutagenesis
Site-directed mutagenesis was carried out using the Stratagene quikchange PCR mutagenesis kit (Agilent Technologies Inc). Primers were designed using the QuikChange Primer Design Program (sequences can be found Table 2; https://www.genomics.agilent.com/CollectionSubpage.aspx?Page-Type = Tool&SubPageType = ToolQCPD&PageID = 15).
Using the mutation-specific primers, the pCS2+hsDDX18-WT was PCR amplified using PFU ultra and digested with DpnI to remove parent plasmid.
The entire coding sequence of resultant clones was determined using direct sequencing to confirm the presence of the mutation and no additional PCR-induced errors.
Results
Loss of ddx18 results in reduced myeloid and erythroid cells
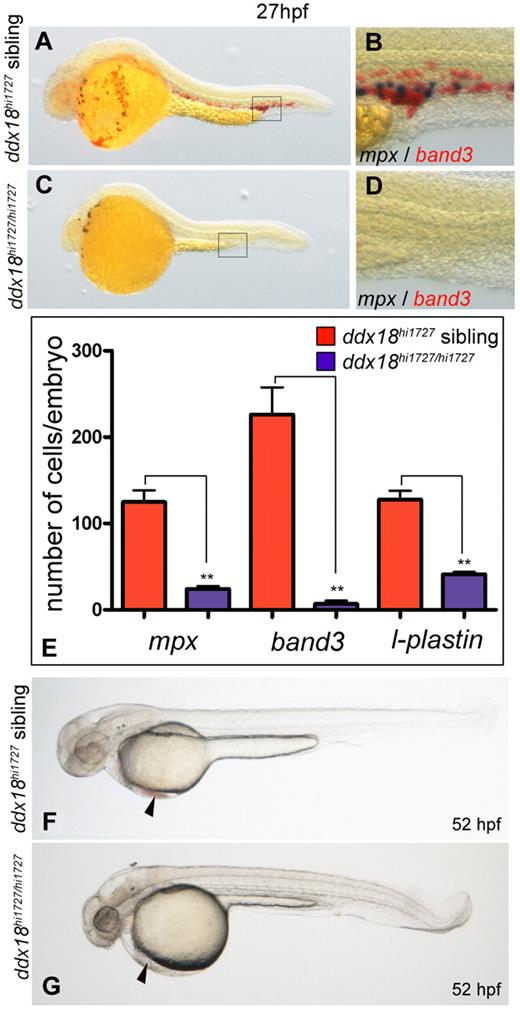
To identify novel genes involved in myelopoiesis, a library of 276 insertionally mutated zebrafish lines were analyzed at 2 dpf for deficient or abnormal expression of the pan-granulocyte peroxidase, myeloid peroxidase (mpx) using WISH.23,27-29 Zebrafish embryos homozygous for the ddx18hi1727 allele, which results from a viral insertion within the first intron of the ddx18 gene were identified as having a profound reduction in the number of cells expressing mpx at 27 hpf (Figure 1A-E). Homozygous ddx18hi1727/hi1727 embryos also showed reduced expression of band3 (slc4a1), a marker of erythroid cells, and the pan-leukocyte marker l-plastin (Figure 1E, supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).30
Loss of Ddx18 results in reduced number of primitive myeloid and erythroid cells. (A-D) Embryos (27 hpf) showing WISH for mpx (black) and band3 (slc4a1; red) to distinguish myeloid and erythroid lineage cells, respectively. Lateral view with head to the left and dorsal upward. ddx18hi1727 mutants have markedly reduced myeloid cells and erythroid cells (C) and close-up view of the ICM (D), compared with siblings (A) and close-up of ICM (B). (E) Bar chart quantifying numbers of mpx, band3, and l-plastin cells per embryo at 27 hpf. **P < .002, Student t test. (F-G) Brightfield images of 52-hpf embryos showing reduced number of erythroid cells in the ducts of Cuvier (arrowhead) in ddx18hi1727 mutants (G) compared with sibling controls (F).
Loss of Ddx18 results in reduced number of primitive myeloid and erythroid cells. (A-D) Embryos (27 hpf) showing WISH for mpx (black) and band3 (slc4a1; red) to distinguish myeloid and erythroid lineage cells, respectively. Lateral view with head to the left and dorsal upward. ddx18hi1727 mutants have markedly reduced myeloid cells and erythroid cells (C) and close-up view of the ICM (D), compared with siblings (A) and close-up of ICM (B). (E) Bar chart quantifying numbers of mpx, band3, and l-plastin cells per embryo at 27 hpf. **P < .002, Student t test. (F-G) Brightfield images of 52-hpf embryos showing reduced number of erythroid cells in the ducts of Cuvier (arrowhead) in ddx18hi1727 mutants (G) compared with sibling controls (F).
ddx18hi1727/hi1727 mutants at 27 hpf also showed several developmental defects visible by brightfield microscopy. These included a small head with cerebral edema, and later, brain necrosis, a narrowed, shortened yolk extension and small size (Figure 1G). In addition, there was a reduction in RBCs visible by brightfield microscopy (Figure 1G arrowhead). Based on such developmental abnormalities, ddx18hi1727 homozygous mutants could be reliably distinguished morphologically from WT or heterozygous embryos within the same clutch from 24 hpf, compared with assignment based on genotyping for WT and mutant alleles.
To confirm that loss of Ddx18 was responsible for the observed phenotypes, we used a morpholino targeting the splice donor sites of exon 2–intron 2 (ddx18E2I2 MO) of the ddx18 gene. Embryos injected with this morpholino at the 1-cell stage phenocopied the developmental and hematopoietic defects observed in ddx18hi1727 mutants, thus implicating loss of Ddx18 as the cause of all the features of ddx18hi1727 mutants (supplemental Figure 2).
To determine the earliest time point at which Ddx18 affects the development of blood in ddx18hi1727/hi1727 mutants, in situ hybridization was performed with a range of markers delineating erythroid (gata-1), myeloid (pu.1) and hemangioblast (lmo2) precursors (supplemental Figure 3). Early in development (14-20 somites) the distribution and quantity of cells recognized by these markers was similar in ddx18hi1727/hi1727 mutant embryos compared with siblings. Thus, primitive hematopoietic cells are developmentally specified, but they are unable to expand and differentiate into mature forms in the ddx18hi1727/hi1727 mutant animals.
ddx18hi1727/hi1727 mutants and WT siblings were also assessed for defects specific to definitive HSCs. WISH for c-myb showed that the number of HSCs was markedly reduced in the aorta and caudal hematopoietic tissue of ddx18hi1727/hi1727 mutants compared with WT siblings indicating that mutants possessed reduced numbers of HSCs (supplemental Figure 4A-B). However, ddx18hi1727/hi1727 mutants also show abnormal intersegmental vessel formation (supplemental Figure 4C-D) as marked by expression of the vascular marker flk1 and aberrant cardiac contractility resulting in reduced blood flow (supplemental Videos 1-2). Reduced blood flow has been shown to contribute to normal HSC development, and thus these defects may contribute to the low levels of c-myb staining in the ventral wall of the aorta.31
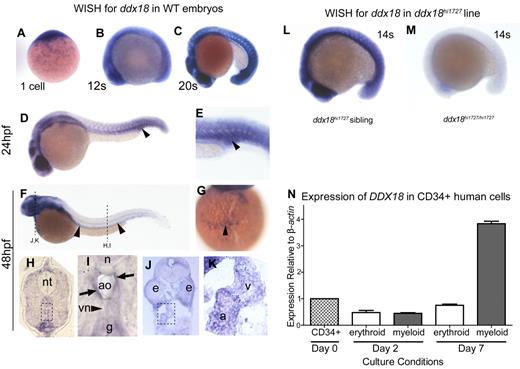
Expression analysis of endogenous ddx18
Expression of ddx18 in WT embryos was determined by in situ hybridization (Figure 2A-K). Expression of ddx18 was observed at the 1-cell stage indicating maternal gene expression (Figure 2A) and was ubiquitous through gastrulation. By 12 somites (15 hpf), a more distinct expression pattern was observed in the developing brain, eye, and somites of WT animals (Figure 2B). From 20 somites to 24 hpf, this pattern of expression continued and became more pronounced (Figure 2C-E). ddx18 was strongly expressed in the posterior blood island and intermediate cell mass (ICM; where erythroid cells reside before the onset of circulation) at 24 hpf, as shown in Figure 2D and E (arrowheads). At 48 hpf, ddx18 expression appeared to be present along the length of the aorta (Figure 2F arrowheads) and in circulating blood cells over the yolk, in the ducts of Cuvier (Figure 2G arrowhead). To more specifically delineate the location of ddx18 expression in these regions, cryosections of 48-hpf embryos were analyzed. Individual cells showed expression of ddx18 in the wall of the aorta (Figure 2H boxed and I) suggestive of expression in HSC and progenitor cells. Expression of ddx18 was also observed in the heart (Figure 2J boxed, arrow and K). In contrast to WT embryos, ddx18 was not expressed by homozygous ddx18hi1727 mutant animals, indicating that the retroviral insertion within the first intron of the gene disrupts normal ddx18 RNA expression (Figure 2L-M shown at 14 somites).23
Expression pattern of zebrafish and human ddx18. (A-K) WISH for ddx18 in WT embryos, from 1 cell to 48 hpf. Lateral view, head to the left (B-K). (A) ddx18 is expressed at the 1-cell stage indicating it is maternally supplied. (B-C) Ubiquitous expression is observed during early development. (D) Expression in the ICM region of the 24-hpf embryo is indicated by the arrowhead. Close-up view is shown in panel E. (F) Expression of ddx18 becomes more localized to the head and along the trunk in the region of the aorta (arrowheads) at 48 hpf. (G) Ventral view of expression in blood cells over the yolk. (H-K) transverse cryosections from WISH embryos at 48 hpf, imaged on a Zeiss Axio imager Z1 microscope using a Zeiss 63×/1.4 NA Apochromat oil lens (Carl Zeiss) and Openlab software (PerkinElmer). The levels of the sections are indicated in panel F by dashed lines. (H) ddx18-expressing cells are found in the wall of the aorta, which are likely to be HSCs, the neural tube is denoted in this view (nt). Close-up view of the boxed region is shown in panel I, indicating expression of ddx18 in cells within the wall of the aorta (ao; arrows), compared with the cardinal vein (vn;, arrowhead). ddx18 expression is also noted in the gut (g) and notochord (n). (J) ddx18 is expressed in cardiac muscle. Close-up view of boxed regions in J shown in K identifies the atrium (a) and ventricle (v). (L-M) WISH for ddx18 in ddx18hi1727 siblings (L) and homozygous mutants (M) show the viral insertion results in loss of ddx18 transcript. (N) Quantitative PCR for DDX18 in primary human CB selected for CD34+ on day 0 (D0) and at 2 days and 7 days following liquid culture in myeloid or erythroid media. DDX18 is expressed in all cell subsets is highest in myeloid culture after 7 days. Expression is shown relative to β actin and normalized to expression in CD34+ cells at day 0. e indicates eye; a, atrium; v, ventricle; nt, neural tube; n, notochord; g, gut; ao, aorta; and vn, vein.
Expression pattern of zebrafish and human ddx18. (A-K) WISH for ddx18 in WT embryos, from 1 cell to 48 hpf. Lateral view, head to the left (B-K). (A) ddx18 is expressed at the 1-cell stage indicating it is maternally supplied. (B-C) Ubiquitous expression is observed during early development. (D) Expression in the ICM region of the 24-hpf embryo is indicated by the arrowhead. Close-up view is shown in panel E. (F) Expression of ddx18 becomes more localized to the head and along the trunk in the region of the aorta (arrowheads) at 48 hpf. (G) Ventral view of expression in blood cells over the yolk. (H-K) transverse cryosections from WISH embryos at 48 hpf, imaged on a Zeiss Axio imager Z1 microscope using a Zeiss 63×/1.4 NA Apochromat oil lens (Carl Zeiss) and Openlab software (PerkinElmer). The levels of the sections are indicated in panel F by dashed lines. (H) ddx18-expressing cells are found in the wall of the aorta, which are likely to be HSCs, the neural tube is denoted in this view (nt). Close-up view of the boxed region is shown in panel I, indicating expression of ddx18 in cells within the wall of the aorta (ao; arrows), compared with the cardinal vein (vn;, arrowhead). ddx18 expression is also noted in the gut (g) and notochord (n). (J) ddx18 is expressed in cardiac muscle. Close-up view of boxed regions in J shown in K identifies the atrium (a) and ventricle (v). (L-M) WISH for ddx18 in ddx18hi1727 siblings (L) and homozygous mutants (M) show the viral insertion results in loss of ddx18 transcript. (N) Quantitative PCR for DDX18 in primary human CB selected for CD34+ on day 0 (D0) and at 2 days and 7 days following liquid culture in myeloid or erythroid media. DDX18 is expressed in all cell subsets is highest in myeloid culture after 7 days. Expression is shown relative to β actin and normalized to expression in CD34+ cells at day 0. e indicates eye; a, atrium; v, ventricle; nt, neural tube; n, notochord; g, gut; ao, aorta; and vn, vein.
The expression of ddx18 in hematopoietic cells was also demonstrated by analysis of FACS-sorted cells from Tg(pu.1:EGFP) (early myeloid), Tg(cmyb:EGFP) (HSC) and Tg(gata1:dsRed) (erythroid) lines. Quantitative RT-PCR for ddx18 showed that ddx18 was expressed in all 3 cell populations in WT or ddx18hi1727 heterozygous mutant animals, compared with the very low level of expression detected in ddx18hi1727 homozygous mutant animals (supplemental Figure 5). To show that DDX18 was also expressed in human hematopoietic cells, we determined its expression level in primary CD34+ cells isolated from CB and after liquid culture for 2 days and 7 days in media supporting either erythroid or myeloid differentiation. DDX18 was expressed at all time points. After 2 days of culture in erythroid or myeloid differentiation media, the expression level of DDX18 fell to around half of that observed in day-0 CD34+ cells; however, after 7 days in culture, expression levels of DDX18 rose in both erythroid and myeloid differentiation media. This was most marked in myeloid conditions where the level of DDX18 reached 4 times that observed in erythroid culture conditions (Figure 2N). Thus, DDX18 is expressed in human hematopoietic cells and is up-regulated during myelopoiesis.
Human DDX18 rescues the effects of Ddx18 loss in ddx18hi1727 mutants
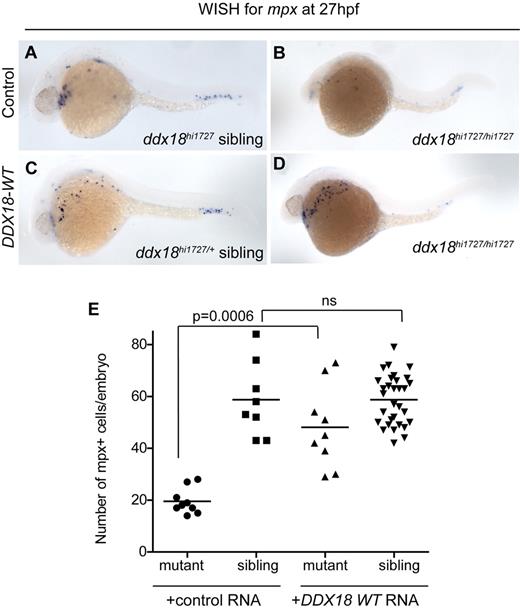
To establish that the defects observed in the ddx18hi1727 mutant animals resulted from loss of Ddx18, and to assess whether DDX18 overexpression affected normal hematopoiesis, in vitro–transcribed RNA encoding human DDX18 was injected into 1-cell stage embryos from clutches of incrossed ddx18hi1727 heterozygous fish. One hundred picograms of DDX18 RNA resulted in complete or partial rescue of the developmental and myeloid phenotype of 27-hpf homozygous ddx18hi1727/hi1727 mutant embryos (Figure 3).
ddx18hi1727/hi1727 myeloid and developmental defects can be rescued with human DDX18. (A-D) Lateral view, head to the left embryos at 27 hpf. WISH for mpx in ddx18hi1727 siblings (A,C) and mutants (B,D), after injection of control (mCherry encoding; A-B) or human DDX18-encoding (C-D) RNA. Expression of the human DDX18 results in rescue of the ddx18hi1727/hi1727 mutant phenotype from both developmental and myeloid defects. (E) Scatter plot showing number of mpx-expressing cells per embryo. Line denotes the mean value. Statistical analysis performed using the Student t test with Welch correction. ns indicates not significant.
ddx18hi1727/hi1727 myeloid and developmental defects can be rescued with human DDX18. (A-D) Lateral view, head to the left embryos at 27 hpf. WISH for mpx in ddx18hi1727 siblings (A,C) and mutants (B,D), after injection of control (mCherry encoding; A-B) or human DDX18-encoding (C-D) RNA. Expression of the human DDX18 results in rescue of the ddx18hi1727/hi1727 mutant phenotype from both developmental and myeloid defects. (E) Scatter plot showing number of mpx-expressing cells per embryo. Line denotes the mean value. Statistical analysis performed using the Student t test with Welch correction. ns indicates not significant.
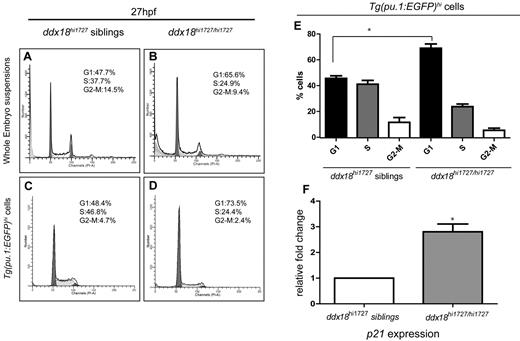
Loss of ddx18 results in G1 cell-cycle arrest
To determine the mechanism of loss of hematopoietic cells, DNA content analysis was carried out in both mutant and sibling embryos at 27 hpf using flow cytometry. PI-stained single-cell suspensions were analyzed from whole embryos, as well as FACS-sorted GFPhi-expressing cells from Tg(pu.1:EGFP) fish. Both whole embryo and early myeloid cells from homozygous ddx18hi1727/hi1727 mutant animals were blocked in the G1 phase of the cell cycle, with a corresponding reduction of cells in S phase and G2-M phases (Figure 4A-E). Cells from the whole embryo but not sorted myeloid progenitors showed sub-G1 cells, suggesting that Ddx18 loss could induce apoptosis in some cell populations but not more prominently in early myeloid progenitors. TUNEL and annexin V analysis supported this interpretation (supplemental Figure 5). To further investigate the block in cell-cycle progression in ddx18hi1727hi1727 mutant animals, we analyzed p21 expression levels in GFP-sorted cells from Tg(pu.1-EGFP);ddx18hi1727/hi1727 mutants and siblings. qPCR showed low levels of expression of p21 in the siblings with a marked increase in p21 expression in the mutant myeloid cells (Figure 4F), suggesting that the cell-cycle defect might result from p53 activation in hematopoietic cells from ddx18hi1727/hi1727 mutant animals.
Loss of Ddx18 results in G1 cell-cycle arrest and elevated p21 levels. (A-D) DNA histograms from whole embryo cell suspensions (A-B) and Tg(pu.1:EGFP)–expressing cells (C-D) stained with propidium iodide. ddx18hi1727/hi1727 mutants (B,D) show increased numbers of cells in G1 and fewer cells in S and G2 phase compared with sibling controls (A,C). (E) Bar chart showing the number of cells in each phase of the cell-cycle in pu.1:EGFP+ cells from 3 experiments. *P < .05 (Student t test). (F) qRT-PCR for p21 shows expression of p21 is up-regulated in sorted Tg(pu.1:EGFP)–expressing myeloid cells from ddx18hi1727/hi1727 compared with sibling controls. Results of 3 experiments normalized to β-actin. *P < .05 (Student t test).
Loss of Ddx18 results in G1 cell-cycle arrest and elevated p21 levels. (A-D) DNA histograms from whole embryo cell suspensions (A-B) and Tg(pu.1:EGFP)–expressing cells (C-D) stained with propidium iodide. ddx18hi1727/hi1727 mutants (B,D) show increased numbers of cells in G1 and fewer cells in S and G2 phase compared with sibling controls (A,C). (E) Bar chart showing the number of cells in each phase of the cell-cycle in pu.1:EGFP+ cells from 3 experiments. *P < .05 (Student t test). (F) qRT-PCR for p21 shows expression of p21 is up-regulated in sorted Tg(pu.1:EGFP)–expressing myeloid cells from ddx18hi1727/hi1727 compared with sibling controls. Results of 3 experiments normalized to β-actin. *P < .05 (Student t test).
p53 loss, but not Bclxl overexpression, rescues the developmental and hematopoietic defects in ddx18hi1727 mutants
The G1 cell-cycle arrest, accompanied with increased expression of p21 RNA levels, prompted us to test whether cell-cycle arrest observed in ddx18hi1727/hi1727 mutants was dependent on intact p53 function. We therefore used antisense morpholinos directed at the ATG/5′ UTR of the p53 gene in ddx18hi1727 clutches. Loss of p53 rescued the developmental defects associated with ddx18hi1727/hi1727 mutants until 48 hpf, such that mutant embryos could not be distinguished from their siblings (Figure 5C and D compared with A and B). Morpholinos decrease in concentration after 48 hours, so to examine the effects of loss of p53 on development after 48 hpf, we bred the ddx18hi1727 mutants with the p53e7/e7 mutants carrying a loss-of-function mutation in the p53 DNA-binding domain.22 Embryos homozygous for loss of both Ddx18 and functional p53 had less severe developmental defects than embryos with loss of Ddx18 and WT p53 (supplemental Figure 6, compare panels B and E to C and F) but the double mutant embryos could be identified by their smaller head and eyes and abnormal cardiac contractility after 48 hpf (supplemental Videos). Homozygous loss of ddx18hi1727 in the absence of functional p53 still resulted in embryologic lethality by 5 dpf. Thus, loss of p53 rescued some but not all of the overt developmental abnormalities that accompany homozygous loss of Ddx18. This indicates that Ddx18 has both p53-dependent and p53-independent functions, and suggests that Ddx18 exerts pleiotropic tissue-specific functions.
p53 loss but not overexpression of bcl-xl rescues development and myeloid defects in ddx18hi1727/hi1727 mutants. (A-F) Lateral views, head to the left, dorsal upwards of 27-hpf embryos. (A,C,E) ddx18hi1727 siblings and (B,D,F) ddx18hi1727 homozygous mutants injected with control (mcherry-encoding [100 pg] RNA; A-B), p53 morpholino (1.6 ng; C-D), or Bcl-xl–encoding RNA (100 pg; E-F). Embryos are stained using 2-color WISH for mpx in black and ddx18 in red. ddx18hi1727/hi1727 were identified by the absence of expression of ddx18. Arrowheads denote ddx18 expression in the eye and forebrain of sibling embryos which is absent in mutant embryos. Injection of p53 morpholino but not bcl-xl RNA was able to completely rescue both developmental and myeloid defects observed at 27 hpf. (G) Scatter plot quantifying number of mpx+ cells per embryo. ***P < .0005 unpaired Student t test. (H-I) mCherry(H) and bcl-xl (I) RNA-injected embryos were irradiated with 12 Gy at 24 hpf and stained using the supravital dye acridine orange 6 hours postirradiation (HPI) to detect cell death. bcl-xl–injected embryos were resistant to irradiation-induced cell death indicating functional Bcl-xl protein is translated in vivo. Epifluorescent images were acquired on a Nikon SMZ1500 zoom stereomicroscope (Nikon Instruments Inc) using a 488-nm filter. ns indicates not significant.
p53 loss but not overexpression of bcl-xl rescues development and myeloid defects in ddx18hi1727/hi1727 mutants. (A-F) Lateral views, head to the left, dorsal upwards of 27-hpf embryos. (A,C,E) ddx18hi1727 siblings and (B,D,F) ddx18hi1727 homozygous mutants injected with control (mcherry-encoding [100 pg] RNA; A-B), p53 morpholino (1.6 ng; C-D), or Bcl-xl–encoding RNA (100 pg; E-F). Embryos are stained using 2-color WISH for mpx in black and ddx18 in red. ddx18hi1727/hi1727 were identified by the absence of expression of ddx18. Arrowheads denote ddx18 expression in the eye and forebrain of sibling embryos which is absent in mutant embryos. Injection of p53 morpholino but not bcl-xl RNA was able to completely rescue both developmental and myeloid defects observed at 27 hpf. (G) Scatter plot quantifying number of mpx+ cells per embryo. ***P < .0005 unpaired Student t test. (H-I) mCherry(H) and bcl-xl (I) RNA-injected embryos were irradiated with 12 Gy at 24 hpf and stained using the supravital dye acridine orange 6 hours postirradiation (HPI) to detect cell death. bcl-xl–injected embryos were resistant to irradiation-induced cell death indicating functional Bcl-xl protein is translated in vivo. Epifluorescent images were acquired on a Nikon SMZ1500 zoom stereomicroscope (Nikon Instruments Inc) using a 488-nm filter. ns indicates not significant.
To determine whether loss of p53 rescued the number of myeloid cells in ddx18hi1727/hi1727 mutants, 2-color WISH was performed at 27 hpf with mpx (black) and ddx18 (red; to identify homozygous ddx18 mutants; Figure 5G). Surprisingly, knockdown of p53 in ddx18hi1727/hi1727 mutants resulted in a complete rescue of myeloid cell numbers to levels observed in siblings injected with control morpholino, which were the same as uninjected WT embryos (Figure 5A-D and G). Although ddx18hi1727 homozygous mutants showed only a modest increase in apoptosis of myeloid cells (supplemental Figure 5), we also injected these embryos with RNA encoding the antiapoptotic protein Bcl-xl. Expression of Bcl-xl blocked irradiation-induced cell death in control animals (Figure 5H and G), but was unable to rescue the developmental phenotype or myeloid cell numbers in ddx18hi1727/ hi1727 mutant animals (Figure 5E and F compared with 5A and B quantified in 5G). Thus, these results combined with measurements of p21 levels are highly suggestive that the loss of mpx-expressing hematopoietic cells observed in ddx18hi1727/ hi1727 mutant animals is because of p53-dependent cell-cycle arrest and not apoptosis.
Functional analysis of human sequence variants identified from AML/MDS patients
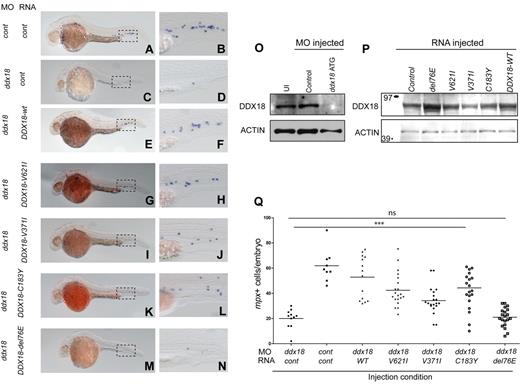
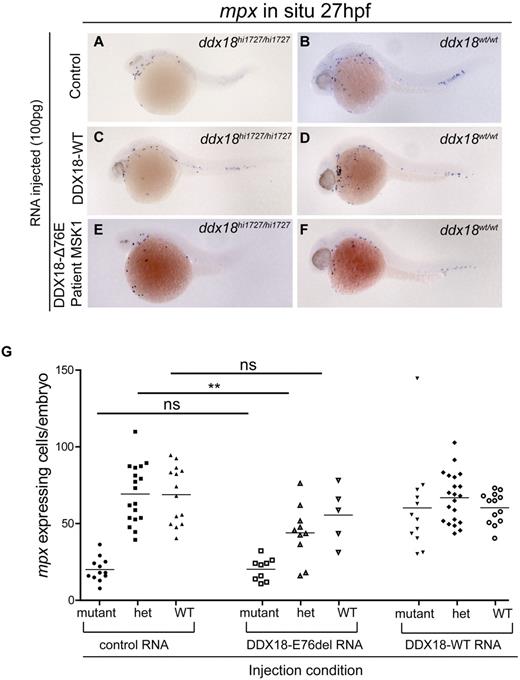
Given the essential role of ddx18 in hematopoiesis and its known function in rRNA processing, we hypothesized that DDX18 could be responsible for cases DBA that are not currently attributable to ribosomal protein deficiency. Thus, we sequenced the cohort of 96 samples from DBA probands negative for all known DBA gene mutations (RPS19, RPS24, RPS17, RPL35A, RPL5, RPL11, RPS7, RPS10, and RPS26) looking for mutations within the DDX18 coding exons. No sequence variants were identified in this cohort suggesting DDX18 is not a DBA-associated disease gene. We further hypothesized that DDX18 may act as a tumor suppressor in human myeloid malignancies. To this end, we sequenced gDNA derived from BM samples of 262 patients with either MDS or AML. Four heterozygous NSVs were identified in BM cell DNA of 3 patients (Table 3). The National Center for Biotechnology Information (NCBI) database for single nucleotide polymorphisms (dbSNP) and the International HapMap Project were searched to determine whether these NSVs represented previously described SNPs. None of the variants were identified in the databases searched. Unfortunately, no nonhematopoietic or remission gDNA was available from the 3 patients to determine whether these abnormalities were inherited or acquired. Because WT human DDX18 is able to rescue the mutant phenotype of ddx18hi1727/hi1727 zebrafish embryos, we were able to use this system to test the functional significance of DDX18 sequence variants. RNAs transcribed in vitro from DDX18 cDNA's harboring each of the sequence variants were injected into WT embryos, along with a specific morpholino targeting the ATG/5′ UTR of the ddx18 gene or control morpholino. This morpholino was validated using an Ab that cross-reacted with zebrafish Ddx18 (Figure 6O). WISH analysis to detect the numbers of cells expressing mpx was then performed at 27 hpf. RNAs encoding DDX18 sequence variants V621I, C183Y, and V371I were able to rescue the developmental and myeloid effects of Ddx18 loss (Figure 6A-B [control injected] and E-L compared with 6C and D, quantified in 6Q); however, the DDX18-E76del RNA was unable to rescue the effects of Ddx18 loss, indicating that this mutation disrupts the function of DDX18 (Figure 6M and N compared with A and B, quantified in 6Q). RNAs encoding the DDX18 sequence variants V621I, C183Y, and V371I did not induce any developmental alterations or altered myeloid cell numbers when injected into WT embryos, suggesting that these mutations did not result in dominant-negative or gain-of-function changes in DDX18. Western blotting on embryo lysates from 15 embryos per condition with a DDX18-specific Ab showed expression of protein from all conditions injected with RNAs encoding each of the variants, including the DDX18-E76del mutant (Figure 6P). To further address whether DDX18-E76del mutant might affect the function of Ddx18 in the heterozygous setting we injected RNA encoding this variant, control or DDX18-WT RNAs into clutches of embryos derived from ddx18hi1727/+ × ddx18hi1727/+ crosses. ISH for mpx was performed at 27 hpf, and embryos imaged, mpx cells per embryo quantified, and genotyped (Figure 7A-F). ddx18hi1727/+ embryos injected with control RNA or DDX18-WT did not differ in developmental morphology or myeloid cell number from WT (ddx18+/+) embryos. By contrast, ddx18hi1727/+ embryos injected with RNA encoding the DDX18-E76del mutant resulted in a significant reduction in myeloid cells compared with WT siblings and to ddx18hi1727/+ embryos injected with control or DDX18-WT RNA (Figure 7G). Conversely, their developmental morphology was indistinguishable from WT ddx18+/+ sibling embryos. These results indicate that the DDX18 sequence variant DDX18-E76del likely acts as a dominant-negative mutation that is antagonistic to the product of the remaining WT allele and results in a reduction in myeloid cell numbers in the absence of any other detectable developmental abnormalities. The V621I, C183Y, and V371I alterations appear to be inconsequential and most likely represent SNPs or passenger mutations.
Diagnostic details of AML patients with DDX18 nonsynonymous sequence variants
| . | Age, y . | Sequence variant . | M/F . | Diagnosis/WHO classification . | Cytogenetics . | Molecular characteristics . | BLAST cell immunophenotype . |
|---|---|---|---|---|---|---|---|
| PT33 | 81 | C183Y and C371I | M | Relapsed AML M6 | 46XY | Not available | CD45+, CD13+, CD33+, CD34+, CD117+ |
| PT40 | 20 | C671I | M | De novo AML | 46XY | Not available | CD34+, CD13+, CD33+, CD7+ |
| MSK1 | 72 | E76del | F | AML evolved from MDS | 46XX | Wild type (TET2, ASXL1, IDH2, FLT3, NPM1, CEBPA, p53) R132C IDH1 | Not available |
| . | Age, y . | Sequence variant . | M/F . | Diagnosis/WHO classification . | Cytogenetics . | Molecular characteristics . | BLAST cell immunophenotype . |
|---|---|---|---|---|---|---|---|
| PT33 | 81 | C183Y and C371I | M | Relapsed AML M6 | 46XY | Not available | CD45+, CD13+, CD33+, CD34+, CD117+ |
| PT40 | 20 | C671I | M | De novo AML | 46XY | Not available | CD34+, CD13+, CD33+, CD7+ |
| MSK1 | 72 | E76del | F | AML evolved from MDS | 46XX | Wild type (TET2, ASXL1, IDH2, FLT3, NPM1, CEBPA, p53) R132C IDH1 | Not available |
Assessment of developmental effects and myeloid cell numbers identifies DDX18-E76del a nonfunctional allele of DDX18 found in AML. (A-N) Lateral views, head to the left and dorsal upward. Boxed region shows area of corresponding close-up in the caudal hematopoietic tissue. In situ hybridization for mpx at 27 hpf in AB embryos injected with ddx18 morpholino (C-N) or control morpholino (A-B) along with control RNA (A-D) or DDX18 RNAs encoding WT (E-F) or sequence variant (G-N) proteins. Injection of DDX18 WT-, V621I-, V371-, or C183Y-encoding RNA was able to rescue the developmental and myeloid defects of ddx18 morphants (E-L compared with C and D). DDX18-E76del–encoding RNA was not able to rescue any of the ddx18 morphants (M-N) compared with panels C and D. (O) Western blot for Ddx18 showing the ddx18ATG/5′ UTR morpholino results in the loss of Ddx18 protein. (P) Western blot for DDX18 showing expression of sequence variant proteins. Control-injected embryos show expression because of cross-reactivity of the Ab with the zebrafish protein. (Q) Scatter plot showing number of mpx-expressing cells per embryo. ***P ≤ .0002. ns indicates not significant (Student t test).
Assessment of developmental effects and myeloid cell numbers identifies DDX18-E76del a nonfunctional allele of DDX18 found in AML. (A-N) Lateral views, head to the left and dorsal upward. Boxed region shows area of corresponding close-up in the caudal hematopoietic tissue. In situ hybridization for mpx at 27 hpf in AB embryos injected with ddx18 morpholino (C-N) or control morpholino (A-B) along with control RNA (A-D) or DDX18 RNAs encoding WT (E-F) or sequence variant (G-N) proteins. Injection of DDX18 WT-, V621I-, V371-, or C183Y-encoding RNA was able to rescue the developmental and myeloid defects of ddx18 morphants (E-L compared with C and D). DDX18-E76del–encoding RNA was not able to rescue any of the ddx18 morphants (M-N) compared with panels C and D. (O) Western blot for Ddx18 showing the ddx18ATG/5′ UTR morpholino results in the loss of Ddx18 protein. (P) Western blot for DDX18 showing expression of sequence variant proteins. Control-injected embryos show expression because of cross-reactivity of the Ab with the zebrafish protein. (Q) Scatter plot showing number of mpx-expressing cells per embryo. ***P ≤ .0002. ns indicates not significant (Student t test).
DDX18-E76del expression in ddx18hi1727/+ animals identifies a dominant-negative effect on myeloid cells. (A-F) Expression of control (mCherry encoding; A-B), DDX18-WT encoding (C-D), or DDX18-E76del (E-F) encoding RNA in ddx18hi1727/hi1727 mutant (A,C,E) or ddx18+/+ (B,D,F) embryos shows the DDX18-E76del variant cannot rescue the phenotype of ddx18hi1727/hi1727. Lateral views, head to the left, dorsal upward of 27-hpf embryos. (G) Quantitation of myeloid cell numbers from individual genotyped (imaged) embryos injected with control, DDX18-WT, and DDX18-E76del encoding mRNAs. Embryos injected with control or DDX18-WT encoding RNA show no difference in myeloid cell numbers between heterozygous ddx18hi1727/+ and ddx18+/+ siblings. Embryos injected with DDX18-E76del encoding mRNA, however, have a statistically significant reduction in myeloid cell numbers compared with control or DDX18-WT–injected ddx18hi1727/+ siblings. **P = .002 (unpaired Student t test).
DDX18-E76del expression in ddx18hi1727/+ animals identifies a dominant-negative effect on myeloid cells. (A-F) Expression of control (mCherry encoding; A-B), DDX18-WT encoding (C-D), or DDX18-E76del (E-F) encoding RNA in ddx18hi1727/hi1727 mutant (A,C,E) or ddx18+/+ (B,D,F) embryos shows the DDX18-E76del variant cannot rescue the phenotype of ddx18hi1727/hi1727. Lateral views, head to the left, dorsal upward of 27-hpf embryos. (G) Quantitation of myeloid cell numbers from individual genotyped (imaged) embryos injected with control, DDX18-WT, and DDX18-E76del encoding mRNAs. Embryos injected with control or DDX18-WT encoding RNA show no difference in myeloid cell numbers between heterozygous ddx18hi1727/+ and ddx18+/+ siblings. Embryos injected with DDX18-E76del encoding mRNA, however, have a statistically significant reduction in myeloid cell numbers compared with control or DDX18-WT–injected ddx18hi1727/+ siblings. **P = .002 (unpaired Student t test).
Discussion
In this report, we describe the identification of Ddx18 as a novel gene required for cell-cycle traverse by primitive hematopoietic cells during zebrafish development. We have shown that the mechanism by which Ddx18 loss results in disruption of hematopoiesis involves the induction of p53-dependent cell-cycle arrest. We then took advantage of the prominent in vivo hematopoietic phenotype to show by RNA rescue that 1 of the 4 sequence variants identified from BM samples of AML and MDS patients (DDX18-E76del) is unable to rescue Ddx18 loss, and that expression of the mutant protein results in a reduction in mpx expression in ddx18hi1727/+ heterozygous animals, suggesting that it antagonizes the activity of the normal Ddx18 that is required for myelopoiesis.
In our zebrafish studies, Ddx18 was not required for primitive myeloid and erythroid specification. However, further maturation of these lineages was impaired in ddx18hi1727/hi1727 mutants as evidenced by reduced mpx and band 3 expression from 27 hpf. The definitive wave of hematopoiesis, which is independent of the production of primitive myeloid and erythroid cells, was also affected by loss of Ddx18. However, the defect observed in HSCs may result from impaired vascular development and reduced cardiac contractility.31
Our results indicate that the failure of maturation of primitive hematopoietic cells in ddx18hi1727/hi1727 mutants was because of G1 cell-cycle arrest. This effect was associated with elevated p21 levels and was dependent on functional p53. A role for Ddx18 in vertebrate hematopoiesis has not previously been described. However, in yeast, there is evidence that Has1p (the Ddx18 yeast orthologue) plays an essential role in ribosomal RNA (rRNA) processing.32-34 Stabilization of p53 is a major cellular response to ribosomal stress resulting from disruption of rRNA processing,35 suggesting that loss of zebrafish Ddx18 results in p53 activation through this mechanism. In addition, several studies have determined that human DDX18 interacts with several RPs including RPS3, RPS3a, RPS6, RPS14, and RPS1936,37 and thus ddx18hi1727/hi1727 mutants may also have a relative RP-deficient state. Ribosomal stress induction in RP-deficient states, results from translational up-regulation RPL11,38 which then binds and sequesters MDM2. This in turn leads to stabilization of p53. Further studies will therefore be necessary to determine whether Mdm2 is sequestered in ddx18hi1727/hi1727 mutants, and whether relative RP deficiency contributes to the observed phenotype.39
Although the major determinant of the mutant phenotype of zebrafish ddx18hi1727/hi1727 mutants was G1 cell-cycle arrest, these animals also showed significantly increased apoptosis. The predominance of cell-cycle arrest over increased apoptosis may simply reflect the high proportion of actively cycling hematopoietic cells during normal embryogenesis. Another potential explanation for the predominance of cell-cycle effects in ddx18hi1727/hi1727 mutants may be a result of the role of Ddx18 in assembly of the large ribosomal subunit.40 P53-dependent G1 cell-cycle arrest has been shown to be the dominant effect of p53 stabilization when rRNA processing is disrupted early during the biogenesis of the large subunit.41
Although Ddx18 loss resulted in other developmental defects, these were less marked than the disruption of hematopoiesis. This is in keeping with the hypothesis that Ddx18 loss results in disruption of ribosome biogenesis, as the hematopoietic system appears to be especially susceptible to loss of RP genes.42 Our results also indicate that disruption of ribosome biogenesis is not sufficient to result in abnormal hematopoiesis because insertional mutants affecting 28 other essential genes involved in ribosome biogenesis were tested in our screen, and ddx18hi1727 was the only mutant line with a specific reduction in mpx expression at 2 dpf. Thus, while Ddx18 loss is incompatible with survival, even in the absence of functional p53, somatic loss of DDX18 in human HSCs could have a profound and selective effect on hematopoiesis, particularly if there is also a loss of functional p53, which is commonly seen in acquired hematopoietic disorders.
Our findings in zebrafish ddx18hi1727/hi1727 mutants along with data suggesting that DDX18 is able to bind NPM1 (data not shown and Sekiguchi et al36 ) RPS14 and HSPA9B,36 3 putative tumor suppressor genes in AML/MDS located on chromosome 5q,43-45 led us to further investigate whether human DDX18 plays a role in myeloid malignancies. We undertook sequencing studies in DNAs from BM samples from AML and MDS patients and identified 4 heterozygous sequence variants. Using our zebrafish embryonic model, we showed that 3 of the 4 variants identified had no apparent functional significance; however, using morpholinos we were able to identify DDX18-E76del as functionally unable to rescue ddx18 morphants. Importantly, because the deletion resulting in DDX18-E76del was heterozygous, we also determined using ddx18hi1727/+ embryos that DDX18-E76del likely exerts a dominant-negative effect on myeloid cells. The reason that this variant of DDX18 results in dominant-negative effects is not clear. One reason may be associated with the fact that the deleted glutamic acid residue in the DDX18-E76del variant resides within a conserved consensus SQE motif, a binding motif for PI3K-related kinases (PIKKs). Binding of PIKKs (such as DNA damage proteins ATM and ATR) to their target binding sites is thought to be required for kinase activation.46 Furthermore, a recent study of the Plasmodium falciparum orthologue of DDX18 showed that, in addition to the 9 conserved DEAD-box helicase motifs, the N-terminus of the protein is also required for its helicase activity.47 The patient with DDX18-E76del mutation had a normal karyotype AML which had evolved from prior MDS. This patient also had the IDH1-R132C mutation. IDH1 mutations, particularly those affecting the R132 residue, have been described in MDS patients early in the course of disease as well as secondary AML.48 Notably however, in JAK2V617F myeloproliferative neoplasms, IDH mutations are acquired during transformation, and can occur in JAK2 mutant or WT clones.49 Thus, mutation of DDX18 may occur as a primary event preceding IDH1-R132 mutation, or secondary event during disease evolution leading to deregulation of myeloid and erythroid proliferation and maturation. Our studies in zebrafish show complete loss of ddx18 results in p53-dependent cell-cycle arrest and apoptosis. By contrast, ddx18hi1727/+ heterozygotes have no increase in apoptosis, and no identifiable blood abnormalities. DDX18-E76del expression in ddx18hi1727/+ heterozygotes appears to result in a dominant-negative effect on the WT ddx18 allele sufficient to disrupt hematopoiesis while maintaining normal developmental patterning in zebrafish embryos. This may suggest that a threshold level of functional DDX18 is required for normal myeloid development, and that this level is sufficient to maintain ribosomal integrity in other tissues. Such dosage-mediated mechanisms have also been postulated for the anemia associated with DBA.42
Our sequencing studies also contained 96 probands with DBA without identified mutations in RPs. No DDX18 mutations were identified in this cohort indicating that congenital heterozygosity for DDX18 does not cause pure red cell aplasia.
In conclusion, we have identified Ddx18 as a novel gene involved in primitive hematopoiesis whose loss results in p53-dependent cell-cycle arrest in G1. We have successfully identified DDX18-E76del, present in the BM of a human MDS/AML patient, as a variant of DDX18 exerting a dominant-negative effect on myelopoiesis using an in vivo strategy in zebrafish embryos. In the era of whole-genome resequencing, an increasing number of NSVs are being identified in small proportions of patients with myeloid malignancies. There is currently an unmet need for a simple in vivo model in which to determine the functional significance of novel NSVs. This is necessary to identify and prioritize new oncogenic pathways that may be amenable to therapeutic interventions. Our work in this study demonstrates that the zebrafish is a powerful and robust in vivo model system, which could be widely used to assess the functional significance of NSVs of uncertain significance identified by the resequencing of DNA from hematopoietic malignancies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors gratefully acknowledge Adam Amsterdam for the provision of embryos for the original insertional screen and for supplying the ddx18hi1727/+ line. They also thank Jeff Davies for critical review of the manuscript, Michael Landowski for technical assistance with the DBA sample sequencing, and Feng Guo for technical assistance with cryosections.
N.B. has been supported by a “Princess Borghese and Anna Bulgari Fellowship” from The American-Italian Cancer Foundation, and is currently a Leukemia and Lymphoma Society Special Fellow. E.M.P. is the recipient of the GSK Greg Harper Clinical Research Training Fellowship from Leukemia and Lymphoma Research UK, and was formerly supported by the National Institutes of Health (NIH-5T32 CA009382-26). This work was supported by NIH grant CA93152 (A.T.L. and J.P.K.). H.T.G. is supported by the Diamond-Blackfan Anemia Foundation Research grant.
National Institutes of Health
Authorship
Contribution: E.M.P. designed, performed, and analyzed research and wrote the manuscript; N.B. performed and analyzed research; J.R. performed research; R.L., C.V.H., R.S., and A.H.B. supplied patient samples; O.I.A.-W. supplied patient samples and performed and analyzed research; A.K.-G. supplied key reagents; H.S. performed research; H.T.G. supplied and sequenced DBA patient samples; F.E.C. supplied key reagents; and J.P.K. and A.T.L. designed research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Elspeth M. Payne, University College London Cancer Institute, Paul O'Gorman bldg, 72 Huntley St, London, WC1E6BT. e-mail: e.payne@ucl.ac.uk; or A. Thomas Look, Dana-Farber Cancer Institute, Mayer Bldg, Rm M630, 450 Brookline Ave, Boston MA 02115; e-mail: Thomas_look@dfci.harvard.edu.

![Figure 5. p53 loss but not overexpression of bcl-xl rescues development and myeloid defects in ddx18hi1727/hi1727 mutants. (A-F) Lateral views, head to the left, dorsal upwards of 27-hpf embryos. (A,C,E) ddx18hi1727 siblings and (B,D,F) ddx18hi1727 homozygous mutants injected with control (mcherry-encoding [100 pg] RNA; A-B), p53 morpholino (1.6 ng; C-D), or Bcl-xl–encoding RNA (100 pg; E-F). Embryos are stained using 2-color WISH for mpx in black and ddx18 in red. ddx18hi1727/hi1727 were identified by the absence of expression of ddx18. Arrowheads denote ddx18 expression in the eye and forebrain of sibling embryos which is absent in mutant embryos. Injection of p53 morpholino but not bcl-xl RNA was able to completely rescue both developmental and myeloid defects observed at 27 hpf. (G) Scatter plot quantifying number of mpx+ cells per embryo. ***P < .0005 unpaired Student t test. (H-I) mCherry(H) and bcl-xl (I) RNA-injected embryos were irradiated with 12 Gy at 24 hpf and stained using the supravital dye acridine orange 6 hours postirradiation (HPI) to detect cell death. bcl-xl–injected embryos were resistant to irradiation-induced cell death indicating functional Bcl-xl protein is translated in vivo. Epifluorescent images were acquired on a Nikon SMZ1500 zoom stereomicroscope (Nikon Instruments Inc) using a 488-nm filter. ns indicates not significant.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/4/10.1182_blood-2010-11-318022/4/m_zh89991175390005.jpeg?Expires=1767697799&Signature=QqpmWJYxtPWhWaorFNSF9eIu6THfruRidU9GvfRXqR-QaljpkpNTpFdk-4~CNoJF51xVXhn8rbT1y7RzLbWQFNQjIYxUga8cvtfV8k785pPrKyDEOkApAs4h2k3U-Tp-ywdh4prlH6C0vsjE5Zu--bMF-HLKGXzRUbHej3BkD35pJUOb7ylOfIcrTIP-8xh~088GowRVrK~XavkgiP02fwW5OlUjkapFsIbb7tDLZcuFmWqnGuTZZzXaFU-uOFNMPwhvpvJLH3~00eK1qoD2SdlRjR5Neg0B-4AfQ1HCZm~tcrONyxl5HkEaIL2avFRZcFRK~FEM3uGmU6tSImdckg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal