Abstract
Modulating aberrant transcription of oncogenes is a relatively unexplored opportunity in cancer therapeutics. In approximately 10% of multiple myelomas, the initiating oncogenic event is translocation of musculoaponeurotic fibrosarcoma oncogene homolog (MAF), a transcriptional activator of key target genes, including cyclinD2. Our prior work showed that MAF is up-regulated in an additional 30% of multiple myeloma cases. The present study describes a common mechanism inducing MAF transcription in both instances. The second mode of MAF transcription occurred in myelomas with multiple myeloma SET domain (MMSET) translocation. MMSET knockdown decreased MAF transcription and cell viability. A small-molecule screen found an inhibitor of mitogen-activated protein kinase kinase (MEK), which activates extracellular signal-regulated kinase (ERK)-MAP kinases, reduced MAF mRNA in cells representing MMSET or MAF subgroups. ERK activates transcription of FOS, part of the AP-1 transcription factor. By chromatin immunoprecipitation, FOS bound the MAF promoter, and MEK inhibition decreased this interaction. MEK inhibition selectively induced apoptosis in MAF-expressing myelomas, and FOS inactivation was similarly toxic. Reexpression of MAF rescued cells from death induced by MMSET depletion, MEK inhibition, or FOS inactivation. The data presented herein demonstrate that the MEK-ERK pathway regulates MAF transcription, providing molecular rationale for clinical evaluation of MEK inhibitors in MAF-expressing myeloma.
Introduction
Multiple myeloma (MM) is a malignancy of plasma cells that is diagnosed in 20 000 people annually in the United States.1 Current therapy includes high-dose chemotherapy in conjunction with bone marrow transplantation as well as newer agents, such as bor-tezomib and lenalidomide.2 Although these treatments extend survival, they often do not achieve lasting cures, providing an impetus to search for novel therapeutic modalities.
MM is molecularly heterogeneous, with 7 subgroups defined by gene expression profiling.3 Four of these subgroups were associated with recurrent translocations in which an oncogene (musculoaponeurotic fibrosarcoma oncogene homolog [MAF]/MAFB, multiple myeloma SET domain [MMSET]/FGFR3, Cyclin D1 or Cyclin D3) is juxtaposed to immunoglobulin heavy chain (IgH) enhancer elements, causing aberrant oncogene expression. Of the 7 subgroups, the MMSET/FGFR3 and MAF subgroups have been associated with inferior overall survival.3
MAF is a B-ZIP transcription factor that is aberrantly expressed in myeloma by 2 distinct molecular mechanisms.4 Approximately 10% of primary myelomas bear a MAF translocation that is associated with very high MAF expression. Typically, the MAF translocation breakpoint occurs many kilobases upstream of the MAF transcriptional start site, suggesting that the IgH enhancer elements drive transcription by regulatory elements in the normal MAF promoter. Other myeloma cases lack an MAF translocation but nonetheless express MAF at levels above those in normal plasma cells.5 In myeloma cell lines overexpressing MAF by either mechanism, interference with MAF function blocked cell proliferation.5 This essential function of MAF is probably the result, in part, of its transactivation of cyclin D2, a key regulator of the G1-S phase transition of the cell cycle. In addition, MAF expression in myeloma cells causes them to adhere more avidly to bone marrow stromal cells, leading to greater vascular endothelial growth factor secretion by stromal cells; transactivation of integrin β7 by MAF contributes to these microenvironmental phenotypes.5 Furthermore, MAF may regulate invasion and metastasis of cancer cells through its activation of the AKT pathway.4
These studies established MAF as a target for therapeutic intervention in MM but did not provide a ready means to achieve this clinically. In the present study, we hypothesized that MAF transcription might be driven by intracellular signal transduction pathways, downstream of MMSET, that could be inhibited by clinically available small molecules. A key role for the mitogen-activated protein kinase kinase (MEK)–extracellular signal-regulated kinase (ERK) pathway in MAF transcription emerged from our studies. A large variety of growth factor receptors engage RAS, which activates a kinase cascade in which RAF phosphorylates MEK1, which in turn phosphorylates the ERK family kinase, ERK1 and ERK2.6,7 The ERK kinases activate the transcription of FOS, which encodes one subunit of the AP-1 transcription factors that regulate diverse genes involved in cellular and proliferation and stress responses. FOS heterodimerizes with JUN family members, which are controlled transcriptionally and posttranscriptionally by Jun N-terminal kinase kinase-mediated phosphorylation. FOS/JUN heterodimers transactivate diverse downstream target genes involved in cellular proliferation and stress responses.
Previous preclinical studies have documented activity of MEK inhibitors against myeloma cell lines and osteoclasts.8-10 A novel, potent inhibitor of MEK1/2 affected MAF protein levels in the MM cell line H929.11 We now extend these findings to a larger panel of MM cell lines to investigate whether particular myeloma subtypes would be especially sensitive to MEK inhibition. The current study dissects the contribution of MEK/ERK/AP-1 signaling to the transcription of MAF in MM. Our findings provide a sound mechanistic basis for rationally developing inhibitors of this signaling axis for the therapy of MM using MAF expression as a biomarker for response.
Methods
Retrovirally mediated transduction
For efficient retroviral transductions, cell lines were engineered to express the murine ecotropic retroviral receptor.12 The MAF expression construct was stably expressed in cell lines as described.5 Certain cell lines were also engineered to express the bacterial tetracycline repressor.13 All inducible constructs in tetracycline repressor lines were activated by doxycycline (20 ng/mL). To assess toxicity of an shRNA, retroviruses that coexpressed green fluorescent protein (GFP) were used as described.13 In brief, flow cytometry was performed 2 days after retroviral infection to determine the initial GFP+ proportion of live cells for each shRNA; then cells were subsequently cultured with doxycycline to induce shRNA and sampled over time. The GFP+ proportion at each time was normalized to that for the control shRNA and further normalized to the initial value.
Cell survival and cell cycle analysis
Cells were treated in culture with U0126 (Calbiochem). Assays for cell viability and number in uniform culture were performed with calcein, ethidium homodimer, and beads as previously described.14 Assays for DNA content and cell cycle analysis were performed with propidium iodide.12 VAD-FMK-FITC (Promega) was added directly to cell cultures and analyzed by flow cytometric quantitation, according to the manufacturer's instructions.
Quantitative RT-PCR
For quantitation of mRNA expression, total RNA was extracted from myeloma cell lines using Trizol (Invitrogen). cDNA was generated from 1 μg RNA using SuperScript II enzyme (Invitrogen). Primer/probe sets were purchased from Applied Biosystems. The level of MAF expression was calculated by normalization to the level of the control gene, B2M. Primer/probe sets used were MAF (Hs00170630_m1), MMSET (Hs00370212_m1), CCND2 (Hs00153380_m1), ITGB7 (Hs01565750_m1), CCR1 (Hs00174298_m1), B2M (4333766F), and FOS (Hs00170630_m1).
Primary MM patient samples
Microarray data from primary MM samples were obtained from accession #GSE2658 (http://www.ncbi.nlm.nih.gov/geo).3 Probe sets for the MAF signature were 200952_s_at (CCND2), 205098_at (CCR1), 205718_at (ITGB7), and 206363_at (MAF). Probe sets for the MAF average were 206363_at, 209347_s_at, 209348_s_at, and 229327_s_at.
Western blot
Protein was harvested from MM cell lines and fractionated using a Nuclear/cytosol fractionation kit (BioVision). Protein was quantified using the BCA method (Pierce Chemical) and separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis on a 4% to 12% acrylamide gradient. The following antibodies were used: c-fos, p42/44, phospho-p42/44 (Cell Signaling Technology), cyclinD2 (BD Biosciences PharMingen), B-tubulin (Sigma-Aldrich), and histone H2B (Imgenex).
Chromatin immunoprecipitation
A total of 5 million cells were treated in culture with formaldehyde to a final concentration of 1% for 10 minutes at 37°C. Cells were harvested by centrifugation, washed with 2 × 10 mL phosphate-buffered saline, and chromatin was immunoprecipitated as described.15 Acetylated histone H3 antibody (Upstate Biotechnology) or c-Fos antibody (Cell Signaling Technology) was used. After precipitation, 2 ng DNA was used for each real-time polymerase chain reaction (PCR). Quantitation was performed using SYBR Green on an ABI7500 TaqMan machine (40 cycles) using self-designed primers that had been tested for lack of primer-dimer artifacts and for single-species amplification. Values for immunoprecipitations were normalized to input DNA values, giving data in arbitrary units.
Results
MAF and its target genes are expressed in myeloma cases with an MMSET translocation
We examined the expression of MAF mRNA using gene expression profiling data from 47 MM cell lines and 451 patient samples taken at initial diagnosis. Primary myeloma samples were assigned to gene expression subgroups as previously described,3 and the myeloma cell lines were classified according to their IgH translocations. Not surprisingly, the “MF” myeloma subgroup, associated with MAF translocations, had the highest MAF expression (Figure 1A,C). Notably, the “MS” subgroup, associated with MMSET translocations, had the second highest MAF mRNA levels, which were significantly higher than those in the other 5 myeloma subgroups. Likewise, the cell line subsets bearing an MAF or MMSET translocation had the highest expression of MAF mRNA (Figure 1E). As a surrogate for MAF transcription factor activity, we developed an MAF signature by averaging the expression levels of MAF and 3 known MAF target genes, cyclin D2 (CCND2), integrin-β7 (ITGB7), and CCR1. These are previously discovered and validated MAF target genes.5 Although this MAF signature was highest in cell lines and patient samples in the MF subgroup (Figure 1B,D), myeloma patient samples and cell lines with the MMSET translocation had the next highest levels (Figure 1B,D,F). We conclude that the presence of an MMSET translocation in myeloma is associated with MAF overexpression and activity.
MAF and target genes are highly expressed in myelomas with MAF or MMSET translocations. Expression of MAF and target genes in primary MM patient samples were derived from Affymetrix U133plus2.0 gene expression profiling data from 451 purified bone marrow plasma cell populations collected from untreated patients with MM (A-D) or 47 MM cell lines (E-F). Samples are ranked according to the expression of MAF itself (A,C,E, average of 4 probe sets) or the average expression of MAF and 3 target genes (B,D,F). Expression was centered based on the mean value in normal plasma cells (GEO dataset GSE5900). Myeloma subgroups are designated according to Zhan et al.3
MAF and target genes are highly expressed in myelomas with MAF or MMSET translocations. Expression of MAF and target genes in primary MM patient samples were derived from Affymetrix U133plus2.0 gene expression profiling data from 451 purified bone marrow plasma cell populations collected from untreated patients with MM (A-D) or 47 MM cell lines (E-F). Samples are ranked according to the expression of MAF itself (A,C,E, average of 4 probe sets) or the average expression of MAF and 3 target genes (B,D,F). Expression was centered based on the mean value in normal plasma cells (GEO dataset GSE5900). Myeloma subgroups are designated according to Zhan et al.3
MMSET regulates MAF transcription
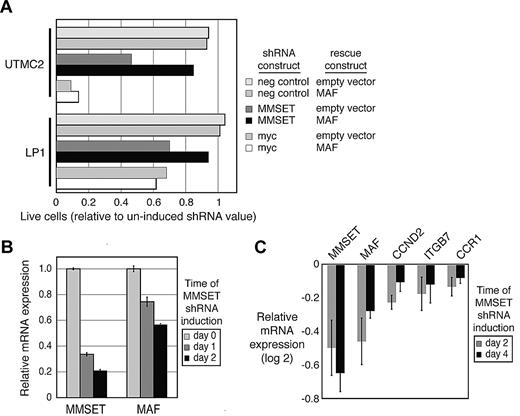
The close association of the MAF signature expression with the MMSET translocation in both cell lines and patient samples led us to speculate that MMSET regulates transcription of MAF itself in MM. Knockdown of MAF has been shown previously to decrease viability in cells of the MS subgroup.5 We therefore depleted MMSET and found decreased survival in 2 cell lines of the MS subgroup (Figure 2A). The MMSET shRNA targets both the short and long isoforms of MMSET (MMSET I and MMSET II16 ), but not REIIBP, another transcript from this genomic locus. This MMSET shRNA construct has been further validated by others. The quantitative PCR probe spans exons 6 to 7 in these isoforms and also does not detect REIIBP. To test the contribution of MAF loss to the decreased viability of these cells, we engineered these 2 cell lines of the MMSET translocation subgroup to stably express MAF using an exogenous promoter. In both cell lines, constitutive reexpression of MAF rescued the cells from toxicity of MMSET depletion (Figure 2A). MMSET depletion lowered MAF mRNA levels as determined by quantitative PCR during induction of the MMSET shRNA (Figure 2B). MYC was used as a positive control and was not rescued by MAF reexpression. Gene expression profiling after MMSET knockdown confirmed decrease in MAF target genes CCND2, CCR1, and ITGB7, as well as MMSET and MAF itself (Figure 2C).
MMSET regulates MAF transcription. (A) Myeloma cell lines UTMC2 and LP-1, both harboring the MMSET translocation, either were engineered to express MAF cDNA under a constitutive LTR promoter or contained an empty construct. These cells were secondarily transduced with a retrovirus expressing MMSET shRNA and coexpressing GFP (“Retrovirally mediated transduction”). Live, GFP-expressing cells were quantified by flow cytometry and normalized to day 2 after retroviral transduction. (B) MM cells were transduced with a TET-inducible MMSET shRNA construct and selected. MMSET shRNA was induced for the indicated times, and RNA was quantified by real-time PCR. (C) Gene expression was profiled during inducible MMSET knockdown. Experiments were repeated in triplicate. Shown are average values of target genes compared with control shRNA targeting luciferase (± SEM).
MMSET regulates MAF transcription. (A) Myeloma cell lines UTMC2 and LP-1, both harboring the MMSET translocation, either were engineered to express MAF cDNA under a constitutive LTR promoter or contained an empty construct. These cells were secondarily transduced with a retrovirus expressing MMSET shRNA and coexpressing GFP (“Retrovirally mediated transduction”). Live, GFP-expressing cells were quantified by flow cytometry and normalized to day 2 after retroviral transduction. (B) MM cells were transduced with a TET-inducible MMSET shRNA construct and selected. MMSET shRNA was induced for the indicated times, and RNA was quantified by real-time PCR. (C) Gene expression was profiled during inducible MMSET knockdown. Experiments were repeated in triplicate. Shown are average values of target genes compared with control shRNA targeting luciferase (± SEM).
Transcriptional activity at the MAF locus is decreased by MEK inhibition
To further dissect cellular pathways involved in the regulation of MAF transcription, we screened a panel of 6 small-molecule inhibitors of kinases known to trigger distinct signaling cascades (Figure 3A). Inhibition of ERK1/2 signaling using the MEK inhibitor U0126 decreased MAF expression in a dose-dependent fashion. By contrast, treatment with inhibitors of protein kinase C, PI3 kinase, p38 MAP kinase, IκB kinase β, or tyrosine kinases had little, if any, effect on MAF mRNA levels. Given the similar effect of MMSET depletion and MEK inhibition on decreasing MAF transcription, we next asked whether MMSET functioned in the MEK-ERK pathway. Interestingly, MMSET depletion decreased ERK phosphorylation and FOS expression (Figure 3B), suggesting that MMSET functions upstream to ERK activation to up-regulate MAF transcription in the MS myeloma subgroup.
MEK signaling regulates MAF transcription. (A) Myeloma cell line LP1 was treated with small-molecule kinase inhibitors targeting major cell signaling cascades at the indicated concentrations. MAF mRNA was measured by quantitative RT-PCR. (B) LP1 MM cells were transduced with a TET-inducible MMSET shRNA construct, selected, and MMSET shRNA was induced for the indicated times. Protein lysates were analyzed by Western blot. ERK phosphorylation and FOS expression were decreased in these cells. (C) MAF-expressing myeloma cell lines were treated with MEK inhibitor U0126 (10μM), and MAF mRNA was measured by real-time quantitative PCR. (D) MAF and target gene mRNA were measured by quantitative RT-PCR after treatment with a second MEK inhibitor, AZD6244 100nM, for the indicated times. (E) MM cells transduced with 2 individual TET-inducible MEK1 shRNA construct, selected, and MEK1 shRNA was induced for the indicated times. Protein lysates were analyzed by Western blot to confirm MEK1 protein decrease after shRNA-mediated knockdown. (F) MAF expression was quantified by quantitative RT-PCR after MEK1 depletion with each shRNA construct in 2 MM cell lines. (G) MAF and target gene mRNA expression were measured by quantitative RT-PCR after MEK1 depletion. Error bars represent mean plus or minus SD.
MEK signaling regulates MAF transcription. (A) Myeloma cell line LP1 was treated with small-molecule kinase inhibitors targeting major cell signaling cascades at the indicated concentrations. MAF mRNA was measured by quantitative RT-PCR. (B) LP1 MM cells were transduced with a TET-inducible MMSET shRNA construct, selected, and MMSET shRNA was induced for the indicated times. Protein lysates were analyzed by Western blot. ERK phosphorylation and FOS expression were decreased in these cells. (C) MAF-expressing myeloma cell lines were treated with MEK inhibitor U0126 (10μM), and MAF mRNA was measured by real-time quantitative PCR. (D) MAF and target gene mRNA were measured by quantitative RT-PCR after treatment with a second MEK inhibitor, AZD6244 100nM, for the indicated times. (E) MM cells transduced with 2 individual TET-inducible MEK1 shRNA construct, selected, and MEK1 shRNA was induced for the indicated times. Protein lysates were analyzed by Western blot to confirm MEK1 protein decrease after shRNA-mediated knockdown. (F) MAF expression was quantified by quantitative RT-PCR after MEK1 depletion with each shRNA construct in 2 MM cell lines. (G) MAF and target gene mRNA expression were measured by quantitative RT-PCR after MEK1 depletion. Error bars represent mean plus or minus SD.
The MEK inhibitor down-regulated MAF mRNA levels regardless of whether MAF overexpression was associated with an MAF translocation or an MMSET translocation (Figure 3C). A second small-molecule MEK inhibitor, AZD6244, confirmed the decrease in MAF and target gene expression (Figure 3D). To further pinpoint MEK signaling in the regulation of MAF transcription, we specifically knocked down MEK1 using 2 individual TET-inducible shRNA constructs (Figure 3E). Specific depletion of MEK1 also decreased MAF expression in MM cell lines (Figure 3F) as well as MAF target gene expression (Figure 3G).
These data suggested that regulatory elements within the MAF promoter may be activated by signaling through MEK. In cell lines with MAF overexpression, the MAF promoter region and intron had histone H3 acetylation at lysines 9 and 14, characteristic of active chromatin (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Notably, inhibition of MEK lowered the degree of histone acetylation in the MAF promoter in cell lines bearing either MAF or MMSET translocations (supplemental Figure 1B), suggesting that the MEK pathway modulates transcriptional activity in the MAF promoter.
MAF-expressing myeloma cells are dependent on MEK signaling
To examine the dependence of myeloma cell lines on MEK signaling, we inhibited MEK in 16 myeloma cell lines, representing the different translocation-associated subgroups, and 10 cell lines were killed in a dose-dependent manner (Figure 4A). Interestingly, all but one of the sensitive cell lines had an MAF or MMSET translocation and MAF overexpression. The exception was XG2, a cell line with an activating mutation in the K-RAS oncogene, which is able to stimulate MEK signaling.1,18 The MEK inhibitor-induced toxicity occurred in the absence of RAS pathway mutations in 5 of the other 9 cell lines (ANBL6, JJN3, OCI-My5, LP1, and UTMC2), whereas 2 had K-RAS mutations (RPMI8226 and KMS28BM), one had an N-RAS mutation (H929), and one had an FGFR3 mutation (OPM1). The levels of ERK or p-ERK levels were not predictive of sensitivity to MEK inhibition (supplemental Figure 2C). This is consistent with other cell types in the literature.19 MEK inhibition up-regulated the proapoptotic protein BIM20 (Figure 4B) and induced apoptosis, as measured by caspase 3 activation (Figure 4C), but not cell-cycle arrest (supplemental Figure 2A). The toxic effect of MEK inhibition was not abrogated in the presence of bone marrow stromal cells (Figure 4D), whereas sensitivity to dexamethasone was markedly diminished. This suggests that MEK inhibition, but not corticosteroid admnistration, blocks survival signals provided by the microenvironment.
MEK inhibitor toxicity is higher in MAF-expressing MM cell lines and rescued by MAF reexpression. (A) MM cell lines were incubated with indicated concentrations of MEK inhibitor U0126, and cell number was enumerated by flow cytometry after 8 days. Live cells were normalized to solvent (dimethyl sulfoxide)–treated control. (B) Proapoptotic BIM is up-regulated after MEK inhibition. Western blot also shows decreased ERK phosphorylation and FOS expression. (C) MEK inhibitor-induced apoptosis was detected by VAD-FITC labeling of cleaved caspase 3 in MM cell lines treated with U0126 (10μM) at the indicated time points. (D) MM cell lines were incubated with either dexamethasone (24nM) or U0126 (10μM) for 7 days in the presence or absence of HS-5 bone marrow stromal cell line. Live cells were quantified by sodium 3-[1-[(phenylamino) carbonyl]-2H-tetrazolium hydroxide assay and normalized to untreated control cells (± SEM). (E) MAF transgene was stably and constitutively expressed in MM cell lines using an exogenous LTR promoter. Live cells were enumerated by flow cytometry for up to 11 days of MEK inhibition (U0126, 10μM). Shown are representative growth curves from at least 3 independent experiments.
MEK inhibitor toxicity is higher in MAF-expressing MM cell lines and rescued by MAF reexpression. (A) MM cell lines were incubated with indicated concentrations of MEK inhibitor U0126, and cell number was enumerated by flow cytometry after 8 days. Live cells were normalized to solvent (dimethyl sulfoxide)–treated control. (B) Proapoptotic BIM is up-regulated after MEK inhibition. Western blot also shows decreased ERK phosphorylation and FOS expression. (C) MEK inhibitor-induced apoptosis was detected by VAD-FITC labeling of cleaved caspase 3 in MM cell lines treated with U0126 (10μM) at the indicated time points. (D) MM cell lines were incubated with either dexamethasone (24nM) or U0126 (10μM) for 7 days in the presence or absence of HS-5 bone marrow stromal cell line. Live cells were quantified by sodium 3-[1-[(phenylamino) carbonyl]-2H-tetrazolium hydroxide assay and normalized to untreated control cells (± SEM). (E) MAF transgene was stably and constitutively expressed in MM cell lines using an exogenous LTR promoter. Live cells were enumerated by flow cytometry for up to 11 days of MEK inhibition (U0126, 10μM). Shown are representative growth curves from at least 3 independent experiments.
MEK activity is dependent on MAF and regulated by MMSET
Given the strong correlation between MAF expression and sensitivity to MEK inhibition, we suspected that toxicity of MEK inhibitors was a result of MAF down-regulation. Therefore, we tested whether ectopic expression of MAF could rescue myeloma cell lines from MEK inhibitor toxicity, as it had rescued after MMSET depletion. We used a retrovirus to stably express MAF in 3 sensitive cell lines (H929, LP1, and UTMC2) and one insensitive cell line (KMS12).5 Enforced MAF expression significantly rescued myeloma cells from the toxicity of MEK inhibition (Figure 4E).
A large variety of growth factor receptors engage RAS, activating a kinase cascade whereby RAF phosphorylates MEK1, which in turn phosphorylates the ERK family kinases.6,7 The ERK kinases activate the transcription of FOS, which encodes one subunit of the AP-1 transcription factors that regulate diverse genes involved in cellular proliferation and stress responses. MEK inhibition decreased ERK phosphorylation and FOS expression, as expected (Figure 4B; supplemental Figure 2B). Whereas MEK inhibition down-regulated the expression of the MAF target gene cyclin D2 in control cells, ectopic MAF maintained cyclin D2 expression, consistent with a loss in MAF transactivation potential (supplemental Figure 2B). Exogenous expression of MAF did not alter the ability of the MEK inhibitor to decrease FOS levels.
FOS, downstream of ERK, directly regulates MAF transcription
We next sought to determine whether FOS played a direct role in the transcriptional regulation of MAF. Chromatin immunoprecipitation identified FOS bound to the MAF promoter in myeloma cells expressing MAF (Figure 5A), and inhibition of MEK decreased the binding of FOS (Figure 5B).
FOS regulates MAF transcription. (A) Chromatin immunoprecipitation of FOS demonstrates binding to MAF promoter in MM cells that is decreased with MEK inhibition (U0126, 10μM). (B) MEK inhibition (U0126, 10μM) decreased FOS binding to MAF locus by chromatin immunoprecipitation in MAF-expressing cell lines. (C) Knockdown of FOS is toxic to MAF-overexpressing myeloma cell lines. The indicated myeloma cell lines were transduced with a retrovirus expressing FOS shRNA and coexpressing GFP (“Retrovirally mediated transduction”). Live GFP+ cells were enumerated by fluorescence-activated cell sorter and normalized to the value at day 2 after retroviral infection. Reexpression of MAF rescues myeloma cells transduced with an shRNA targeting FOS. (D) Quantitative RT-PCR measurement of FOS mRNA levels after induction of the FOS shRNA in H929 cells. (E) Western blot of FOS protein levels after introduction of the FOS shRNA in H929 cells.
FOS regulates MAF transcription. (A) Chromatin immunoprecipitation of FOS demonstrates binding to MAF promoter in MM cells that is decreased with MEK inhibition (U0126, 10μM). (B) MEK inhibition (U0126, 10μM) decreased FOS binding to MAF locus by chromatin immunoprecipitation in MAF-expressing cell lines. (C) Knockdown of FOS is toxic to MAF-overexpressing myeloma cell lines. The indicated myeloma cell lines were transduced with a retrovirus expressing FOS shRNA and coexpressing GFP (“Retrovirally mediated transduction”). Live GFP+ cells were enumerated by fluorescence-activated cell sorter and normalized to the value at day 2 after retroviral infection. Reexpression of MAF rescues myeloma cells transduced with an shRNA targeting FOS. (D) Quantitative RT-PCR measurement of FOS mRNA levels after induction of the FOS shRNA in H929 cells. (E) Western blot of FOS protein levels after introduction of the FOS shRNA in H929 cells.
FOS activity can be decreased experimentally using the dominant negative protein A-FOS21 or by knocking down FOS expression using RNA interference. We used 2 different shRNA constructs to deplete FOS expression, confirmed at the transcriptional (Figure 5D; supplemental Figure 3C) and protein (Figure 5E; supplemental Figure 3D) levels. Attenuation of FOS expression was toxic to the MAF-expressing myeloma cells, but retroviral expression of MAF abrogated the toxicity (Figure 5C; supplemental Figure 3A). Blockade of FOS activity using the dominant negative A-FOS resulted in a similar decrease in myeloma cell viability, but once again, retroviral expression of MAF rescued the cells from toxicity because of loss of active FOS (supplemental Figure 3E).
Discussion
The present study provides genetic and functional data implicating the MEK-ERK pathway in MAF transcription in MM, providing a sound rationale for the development of MEK inhibitors for the treatment of myeloma cases that overexpress MAF. In approximately 10% of myeloma cases, MAF expression is achieved by a t(14;16) translocation that brings IgH enhancers in proximity to the MAF promoter. Perhaps more importantly, MAF and its downstream target genes were expressed in a much larger subset of cases in the absence of an MAF translocation. Many of these cases instead harbored the t(4;14) translocation involving MMSET, and we experimentally validated the requirement for MMSET in activating ERK phosphorylation, triggering MAF transcription and maintaining cell viability.
In myeloma cell lines that overexpress MAF by either mechanism, the MEK inhibitor U0126 lowered MAF mRNA levels and decreased the active chromatin surrounding the MAF promoter. In myeloma cell lines expressing MAF, MEK inhibition initiated apoptosis, whereas this treatment showed low toxicity for most other myeloma lines lacking MAF expression. Remarkably, toxicity from either MMSET depletion or MEK inhibition in cells harboring the MMSET translocation was abrogated by ectopic retroviral expression of MAF, demonstrating that MAF is a critical target gene regulated by the MEK pathway and MMSET. The AP-1 transcription factor subunit FOS was bound to the MAF promoter in cell lines with either an MAF or MMSET translocation, and MEK inhibition reduced this localization. Blockade of FOS transactivation using the dominant negative A-FOS or knockdown of FOS by RNA interference was toxic to the MMSET-translocated myeloma cells expressing MAF, and retroviral expression of MAF rescued cells from this toxicity. Interestingly, the decreased enrichment of FOS on the MAF regulatory element was greater in the MS cells than the MF group. It is possible that the MAF translocation to the immunoglobulin locus supplies additional stimuli to the MAF promoter via the immunoglobulin enhancer element that supersede the activity of FOS alone. In the case of the MMSET translocation, the FOS activity might play a more prominent role. These genetic and functional data demonstrate that the MEK-ERK pathway regulates MAF transcription via FOS in the MS subtype of MM.
Inhibition of MAF transcription thus emerges as a viable therapeutic strategy in myeloma cases that aberrantly express this oncogene. More generally, our study suggests that therapies targeting the aberrant transcription of oncogenes in cancer may complement approaches that target oncoproteins directly.
Various mechanisms may account for the activity of the MEK-ERK pathway in MM. Dependence on MEK was a characteristic feature of cell lines bearing a t(4;14) that causes abnormal expression of both MMSET and FGFR3. Conceivably, signals transmitted from FGFR3 could activate ERK in some of these cell lines, especially the subset that has mutations that activate the FGFR3 kinase constitutively.22 However, in approximately 30% of the t(4;14) myelomas, FGFR3 is not expressed, suggesting that the aberrant expression of MMSET drives the pathogenesis these myelomas.23 Indeed, MM patient samples that express MMSET but not FGFR3 nonetheless can have high expression of MAF and its target genes. RAS mutations occur in approximately 20% of primary myelomas and are not associated with any of the identified myeloma subgroups.24 Less than half of the MEK-dependent myeloma cell lines in the current study contain a mutation in RAS pathway (data not shown), but other as yet unknown lesions in the RAS pathway may be present in some cases. In addition, a multitude of cell surface receptors may activate MEK-ERK signaling via interactions with the bone marrow microenvironment, including the IL-6, insulin-like growth factor-1, vascular endothelial growth factor, and B cell–activating factor of tumor necrosis factor family receptors.10,25,26
MEK inhibition decreased MAF transcription, MAF promoter histone acetylation, and FOS binding to the MAF promoter in myeloma cell lines with and without an MAF translocation. Thus, even though IgH enhancer elements are probably responsible for the high level of MAF transcription in cases with an MAF translocation, MAF transcription nonetheless relied on FOS binding to the MAF proximal promoter. Inhibition of transcriptional enhancers themselves might be difficult to achieve because of the redundant action of multiple transcription factors binding to enhancers. On the other hand, our study indicates that therapies targeting activation of proximal promoter elements of the target gene may be effective.
At least 5 MEK inhibitors are currently in phase 1 or phase 2 clinical trials treating both solid tumors and lymphoid malignancies and have been well tolerated.27 In addition, sorafenib, an approved agent in renal cell and hepatocellular carcinoma, inhibits RAF and thus should also inhibit its downstream kinases, MEK and ERK. Previous preclinical work with the MEK inhibitor AZD6244 demonstrated that it induces apoptosis in myeloma cell lines and also inhibits osteoclastogenesis.28 Although that study did not investigate differential sensitivity of myeloma cell lines based on MAF expression, it is notable that the MEK inhibitor AZD6244 blocked tumor growth in vivo in a xenograft model based on the MAF-expressing myeloma cell line OPM1.28
Our findings provide a mechanistic basis for the development of MEK inhibitors in the treatment of the MM subset with functional overexpression of the MAF oncogene. Importantly, the MEK inhibitor U0126 induced apoptosis of MAF-expressing myelomas, suggesting that it could have cytotoxic rather than cytostatic clinical activity. Prior studies have shown cell-cycle arrest with MEK inhibition.29,30 We also expected to see similar results, especially after the effects of MAF down-regulation on Cyclin D2 levels. On the contrary, our data showed a greater effect in promoting apoptosis than in G1 arrest. Other studies with MM cells have also found caspase 3-dependent apoptosis in MM cells after MEK inhibition.11,28 It is possible that the G1 arrest was seen predominantly in the K-RAS mutant cell lines that are dependent on MEK signaling, whereas the MAF-expressing cells are more driven to apoptosis. In our hands and others, MEK inhibition induced myeloma cell apoptosis despite the presence of supportive contact with bone marrow stromal cells.10 Patients with MAF-expressing myeloma subtypes have the poorest outcome when treated with high-dose cytotoxic chemotherapy and tandem autologous stem cell transplantation.2 The mechanism responsible for this poor prognosis is unknown but could derive from the transcriptional network regulated by MAF. One MAF target gene, ITGB7, might promote resistance of myelomas to chemotherapy by enhancing myeloma interaction with bone marrow stromal cells, which survive the high-dose chemotherapy regimens involved in autologous bone marrow transplantation.31 We previously explored the role of ITGB7 downstream of MAF, using both RNAi and a dominant negative MAF construct that inhibited its ability to act as a homodimerized transcription factor. Expression of ITGB7 was decreased, as was cell viability. Interruption of the ITGB7/E-cadherin interaction decreased the ability of the MM cells to adhere to the bone marrow stromal cells, and decreased their secretion of stimulatory cytokines.5 Signals delivered through this integrin-cadherin interaction could sustain the myeloma cell despite systemic chemotherapy.32 Increased presence of the chemokine receptor CCR1 on the MAF-expressing myeloma cell may also confer a survival advantage because of amplification of signals from prosurvival chemokines in the bone marrow microenvironment, allowing the repopulation of the malignant clone.33 Down-modulation of MAF and its target genes by MEK inhibition could, in theory, overcome these mechanisms of chemotherapy resistance.
The design of clinical trials of MEK inhibitors should include molecular profiling of tumor cells before treatment given the likelihood that MEK inhibition will be preferentially toxic for MAF-expressing myeloma cells. The presence of MAF activity in a person's tumor could be detected by quantitative PCR for MAF itself and for the MAF target genes CCND2, ITGB7, and CCR1. Moreover, the achievement of an effective in vivo concentration of the MEK inhibitor could be judged by the down-regulation of MAF and its target genes in myeloma cells obtained during treatment. This latter requirement would suggest that clinical trials should be designed in which the MEK inhibitor is given first as a single agent, allowing the assessment of tumor response, followed by combination treatment with conventional chemotherapeutic agents.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research (L.M.S., C.M.A.).
National Institutes of Health
Authorship
Contribution: C.M.A. designed and performed experiments, analyzed data, and wrote the manuscript; R.E.D. designed and performed experiments; L.H. and A.Z. performed experiments; L.L. performed experiments and analyzed data; L.T.L., E.M.H., and A.L.S. designed experiments and analyzed data; and W.M.K. and L.M.S. designed experiments, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Louis M. Staudt, Bldg 10, Rm 4N115, 10 Center Dr, Bethesda, MD 20892-1374; e-mail: lstaudt@mail.nih.gov.



![Figure 4. MEK inhibitor toxicity is higher in MAF-expressing MM cell lines and rescued by MAF reexpression. (A) MM cell lines were incubated with indicated concentrations of MEK inhibitor U0126, and cell number was enumerated by flow cytometry after 8 days. Live cells were normalized to solvent (dimethyl sulfoxide)–treated control. (B) Proapoptotic BIM is up-regulated after MEK inhibition. Western blot also shows decreased ERK phosphorylation and FOS expression. (C) MEK inhibitor-induced apoptosis was detected by VAD-FITC labeling of cleaved caspase 3 in MM cell lines treated with U0126 (10μM) at the indicated time points. (D) MM cell lines were incubated with either dexamethasone (24nM) or U0126 (10μM) for 7 days in the presence or absence of HS-5 bone marrow stromal cell line. Live cells were quantified by sodium 3-[1-[(phenylamino) carbonyl]-2H-tetrazolium hydroxide assay and normalized to untreated control cells (± SEM). (E) MAF transgene was stably and constitutively expressed in MM cell lines using an exogenous LTR promoter. Live cells were enumerated by flow cytometry for up to 11 days of MEK inhibition (U0126, 10μM). Shown are representative growth curves from at least 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/117/8/10.1182_blood-2010-04-278788/5/m_zh89991166510004.jpeg?Expires=1769145047&Signature=TXG09yX2lFwk-GuxBa7H8hzfSG61Qdaq1VgY~irrfq~nW6iAz0JMXVrDCtZPBqzGYvXTjZ3m7oKTNB5Y7rKz4t8YgZoPYEKM3KwtBqe2zzhAo2pLhclPmL4~VNsaFbj0m-fpS7rzaMs6L8J8s9b9knQuoyOAZcngdwzFuXm76ygXnXqUGT05r2l3OFy2pdLUVHg6EkxutnnRFj05TIZ7RHNTKRD8as7OkoKYavTdBnbPMeU746idBFiENaHpQBfAwVKuSRo9VqRI7cBGYcewC5k6CUTiOpeALE56sqxrBvq0eB~IbEi0Wb8ODIsSd-IrEBfgXQaNLXRpQiD5TrST4Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal