Despite increased understanding of molecular pathogenesis of multiple myeloma and implementation of therapies such as bortezomib and thalidomide, only 10% of patients survive more than 10 years after diagnosis. Until recently, new therapies for myeloma have not been developed based on a detailed understanding of the molecular pathology of the disease. In this issue of Blood, Annunziata et al report a rationale for the use of mitogen-activated or extracellular signal-regulated protein kinase (MEK) inhibitors in the subset of myeloma patients expressing high levels of the MAF oncogene.1
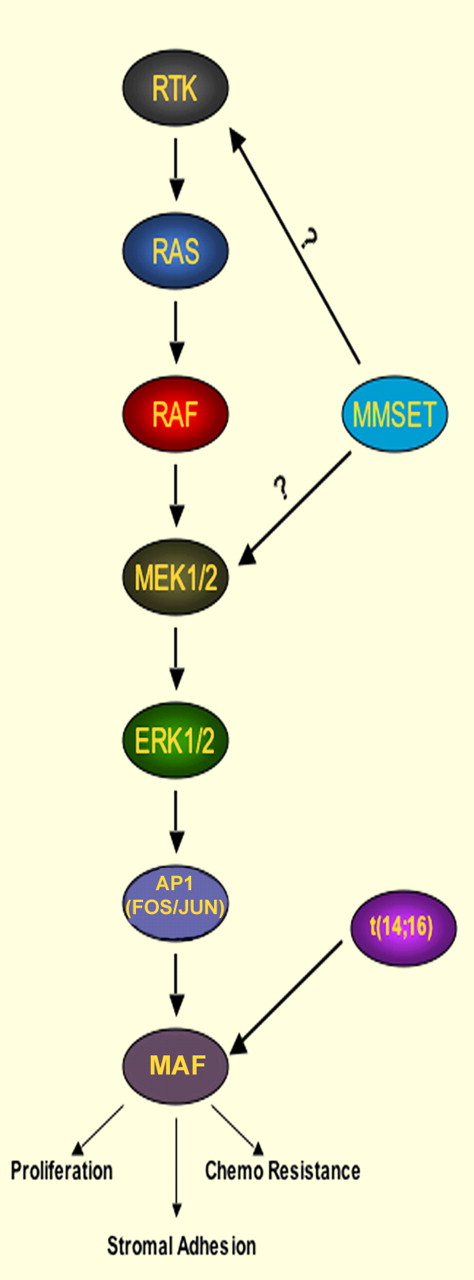
The molecular pathogenesis of multiple myeloma has become clearer with the identification of recurrent chromosomal translocations in approximately 40% of cases. These rearrangements represent aberrant class switching resulting in the linkage of the immunoglobulin promoter/enhancer to the Cyclin D1, Cyclin D3, MMSET and FGFR3, c-MAF or MAF-B genes. Gene expression profiling found that myeloma could be distinguished into 7 distinct subsets, 4 of which are defined by the translocations above.2 Translocation t(14;16), leading to elevated expression of MAF, occurs in approximately 10% of cases. Annunziata et al found that high levels of MAF were also observed in samples with t(4;14) translocation associated with overexpression of MMSET and FGFR3. Both t(14;16) and t(4;14) myeloma subgroups are associated with poorer progression-free and overall survival.
Signaling cascades controlling MAF expression in multiple myeloma.
Signaling cascades controlling MAF expression in multiple myeloma.
A previous study suggested that increased levels of MAF enhance myeloma cell proliferation and adhesion to the bone marrow stroma. In addition, myeloma cells with t(14;16) required continued MAF expression to engraft and form tumors in nonobese diabetic–severe combined immunodeficient mice.3 To determine the pathways maintaining MAF expression, the authors treated a panel of myeloma cell lines, including those with the t(14;16), with inhibitors targeting pathways such as Protein kinase C, PI3 kinase, and Iκβ kinase and found that only inhibition of mitogen-activated protein (MAP) kinase signaling using MEK inhibitors decreased expression of MAF and its target genes. Down-regulation of the MEK1 or MMSET proteins using shRNA had similar effects on MAF expression. Furthermore, the group showed that knockdown of MMSET in t(4;14)+ cells led to decreased levels of activated phospho–extracellular signal-regulated kinase (ERK) and decreased levels of the c-FOS transcription factor, which stimulates the transcription of MAF. Curiously, MEK inhibitors depleted MAF levels both in patients with MMSET overexpression or MAF gene rearrangement. In addition, loss of MEK1 expression led to decrease of histone H3 acetylation, an activating epigenetic mark in the MAF promoter, suggesting that while the immunogloblulin promoter/enhancer may help drive MAF overexpression in t(14;16)+ cells, regulatory elements within MAF promoter still play a role in controlling its expression.
MMSET is a histone methyltransferase (HMT) and transcriptional cofactor whose overexpression in myeloma is associated with global changes in chromatin methylation and shifts in gene expression.4-6 How MMSET affects MAP kinase activity in t(4;14) myeloma has yet to be defined (see figure). Nevertheless, MAF is clearly an important target of MMSET, as the loss of cell survival noted after shRNA-mediated depletion of MMSET was rescued by constitutive expression of MAF. While Inhibitors of the HMT activity of MMSET remains the most targeted therapy for t(4;14)-associated myeloma, MEK inhibitor therapy appears more immediately testable.
The tranlational importance of the work of Annuziata et al became apparent when the group showed that MEK inhibitors specifically halted proliferation and induced apoptosis in myeloma cell lines with t(14;16) and t(4;14) translocations and high MAF expression. The lethality can be significantly rescued with exogenous MAF expression. Importantly, given the tendency of many antimyeloma agents to be less effective when cells are grown on stroma and the importance of MAF in myeloma/stromal interactions, the authors showed that the effectiveness of MEK inhibitors persisted under coculture conditions with bone marrow stromal cells.
MEK inhibitors, representing the first selective inhibitors of MAP kinase signaling, are being studied in early-phase clinical trials. In preclinical studies, MEK inhibitors were particularly effective in killing melanoma and colon tumor cells with RAF mutations. In a xenograft model, MEK inhibitor completely abrogated the growth of RAF-mutant tumors, but only had a partial effect on RAS-mutant cells,7 possibly because constitutive signaling by rat sarcoma (RAS) can affect the PI3K as well as the MAP kinase pathway. In the setting of myeloma, MEK inhibitors could kill cell lines resistant to conventional chemotherapy, including some expressing high levels of MAF. MEK inhibitors prolonged overall survival and decreased phosphorylation of ERK in a xenograft mouse model based on MAF overexpressing t(4;14)+ cell line OPM1.8 In addition, inhibition of MEK enhanced myeloma cell killing in the presence of dexamethasone, bortezomib, or lenolidomide, further suggesting that MEK inhibitors could enhance the efficacy of current myeloma therapy. In light of the current work, future clinical trials of MEK inhibitors in myeloma must be sure to include subsets of patients with MMSET and MAF rearrangements that collectively represent only one-third of all patients, lest a positive result be lost among the other two-thirds of patients who might not respond.
While gene expression profiling identifies 7 types of myeloma, currently therapy is based on empirical studies that predate the molecular classification of disease. Although it was clear that chromosomal anomalies and gene expression patterns have prognostic importance, the current study is one of the first to elucidate a therapy specific to molecularly defined subsets. Whether MEK inhibitors will prove effective in the clinic may also depend on the nature of other genetic changes found in myeloma. Besides the recurrent chromosomal translocations that represent initial genetic events, there are additional genetic events commonly found in myeloma, including activating mutations in RAS and NFKB pathways, rearrangements leading to MYC deregulation, and inactivating mutations in important cellular and epigenetic regulators such as p53 and UTX.9 With the cost of whole genome and exome sequencing rapidly declining, myeloma therapy in the next decade might involve genotyping of individual tumors and the use of rational therapy combinations to intervene with multiple affected pathways.
Conflict-of-interest disclosure: The authors declare no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal