Abstract
Mitoferrin1 is 1 of 2 homologous mitochondrial iron transporters and is required for mitochondrial iron delivery in developing erythroid cells. We show that total deletion of Mfrn1 in embryos leads to embryonic lethality. Selective deletion of Mfrn1 in adult hematopoietic tissues leads to severe anemia because of a deficit in erythroblast formation. Deletion of Mfrn1 in hepatocytes has no phenotype or biochemical effect under normal conditions. In the presence of increased porphyrin synthesis, however, deletion of Mfrn1 in hepatocytes results in a decreased ability to convert protoporphyrin IX into heme, leading to protoporphyria, cholestasis, and bridging cirrhosis. Our results show that the activity of mitoferrin1 is required to manage an increase in heme synthesis. The data also show that alterations in heme synthesis within hepatocytes can lead to protoporphyria and hepatotoxicity.
Introduction
The biosynthesis of heme and iron-sulfur clusters occurs within mitochondria, and both processes rely on iron as a substrate. Homologous members of the family of mitochondrial carrier proteins, Mrs3/Mrs4 in yeast and mitoferrin1 (Mfrn1) and mitoferrin2 (Mfrn2) in vertebrates, have been implicated in the acquisition of mitochondrial iron. Deletion of the yeast homologs MRS3 and MRS4 results in mitochondrial iron deficiency under conditions of limiting iron, suggesting that these transporters are high-affinity iron transporters.1-3 The loss of Mfrn1 in zebrafish and developing erythroid cells results in impaired iron delivery and defective heme synthesis.4 This deficiency cannot be suppressed by ectopic expression of the homologous Mfrn2. Mfrn2 is functionally homologous to Mfrn1, as shown by its expression in yeast4 and nonerythroid mammalian cells.5 The inability of Mfrn2 to suppress the loss of zebrafish Mfrn1 results from differences in the regulation of the 2 mitoferrins. Transcription of Mfrn1 is regulated by the GATA1 transcription factor and is increased during erythropoiesis.4 Further, the stability of Mfrn1 is increased during erythropoiesis by its binding to ABCB10, protein induction of which is also increased during erythropoiesis.5,6 Transcripts for Mfrn1 can be found in nonerythropoietic tissues but at levels lower than Mfrn2, which is ubiquitously expressed in all tissues.4
Support for the tissue-specific expression of Mfrn1 comes from studies of zebrafish mutants (frascati) and mouse embryos, which contain a gene-trap cassette in Mfrn1.4 These embryos die early in development at times that are consistent with the development of RBCs.4 The coincidence of embryonic lethality and RBC developmental defects suggests a casual relationship—that death is due to defective hemoglobinization. There are 2 caveats to the interpretation that Mfrn1 expression is specific for RBC development. First, the gene-trap disruption of Mfrn1 contains the neomycin-resistance gene and there have been cases where the inclusion of that gene affects viability. Second, a finding that Mfrn1 is required for viability because of its role in RBC hemoglobinization does not exclude its functioning in other tissues. To address the role of Mfrn1 in embryonic development and organ function, we generated Mfrn1 deletion constructs that lacked the neomycin gene and, using Cre/Lox technology, examined the effect of cell-type–specific deletions. We determined that the mortality of embryos homozygous for a Mfrn1 deletion was not due to the neomycin construct, but rather could be ascribed to defective hematopoiesis. Deletion of Mfrn1 in adult hematopoietic progenitors resulted in a severe anemia, suggesting that lethality was due to defective hematopoiesis. We determined that the loss of Mfrn1 in hepatocytes had no effect under basal conditions. In contrast, increased dietary delta aminolevulinic acid (δALA) in hepatocyte-specific Mfrn1-knockout mice resulted in high levels of protoporphyrin IX and liver pathology, indicating that in hepatocytes, Mfrn1 may be required under conditions of increased heme synthesis.
Methods
Targeting vector for disruption of the murine Mfrn1 (Slc25a37) gene
We used the bacterial artificial chromosome clone RPCI23-132H6 (Invitrogen) containing Mfrn1 (Slc25a37) from mouse strain C57Bl/6J. All PCRs were carried out with Expand High Fidelity PCR (Roche). The backbone of the targeting vector was the pTK1 vector provided by the University of Utah transgenic core facilities, which was slightly modified to contain compatible restriction enzyme sites. The following PCR products were cloned into the pTK1 vector in this order: the 5′ homologous region (5′ HR) consisting of a 2.1-kb fragment upstream the EcoRV restriction site located in the intron 1 adjacent to exon 2; a loxP-flippase recognition target site (FRT)–neomycin-FRT cassette containing a BamHI restriction site; the 820-bp fragment starting at the EcoRV site, spanning exon 2 and ending 305 bp after the end of exon 2 in intron 2; a loxP site; and finally the 3′ homologous region (3′ HR) consisting of a 8.4-kb fragment from 305 bp after the end of exon 2 and including exon 3, intron 3, and exon 4. Sequencing of both loxP sites, the FRT-neomycin-FRT cassette, and exons 2, 3, and 4 confirmed the absence of mutations. The targeting vector was linearized at the end of the 5′ HR by digestion with NotI, electroporated into G4 embryonic stem (ES) cells, and selected for G418 and ganciclovir resistance.
Screening of the ES cell clone and genotyping of animals
156 ES cell clones resistant to G418 and ganciclovir were analyzed for correct homologous recombination by Southern blot and PCR analyses. For Southern blot analysis, DNA was purified, digested by BamHI, and probed with a 5′-flanking probe generated as a PCR product using the forward primer 5′-TGGTTGCCCCAAGTTCTTAC-3′ and the reverse primer 5′-ATGACAACCAAAGGGACAGC-3′. PCR analyses confirmed the presence of the loxP-FRT-neomycin-FRT cassette, exon 2, and the second loxP site in the recombinant Mfrn1 locus. Cells from a single heterozygous clone were injected into C57BL/6J-derived blastocysts that were implanted into foster mothers. Chimeric animals were identified by coat color, and males were mated with C57BL/6J females to produce mice heterozygous at the Mfrn1 locus. The Mfrn1floxneo offspring were screened by PCR using a forward primer, Mfrn1-F (5′-CCACAACCCCTTTGTTTCAT-3′), and 2 reverse primers, Mfrn1-R1 (5′-GCACCGCCTGTGCTCTAGTA-3′), which hybridizes in the loxP-FRT-neomycin-FRT cassette and generates a 178-bp product, and Mfrn1-R2 (5′-ACAGCACTTGTGACCCATGC-3′), which hybridizes in intron 1 near exon 2 and generates a 62-bp wild-type product. The neomycin-resistance cassette was subsequently excised by recombination of FRT sites by breeding Mfrn1flox−NEO/+ mice with flp/flp mice that express Flp recombinase under the Rosa promoter to obtain heterozygous Mfrn1flox/+animals.
Disruption of the murine Mfrn1 gene
Mfrn1flox/+ male mice were bred with Hprt-Cre female mice expressing Cre recombinase under the Hprt promoter to excise exon 2 of Mfrn1 at the zygote stage and produce a global deletion of Mfrn1 in all tissues (Mfrn1+/− mice). These mice were bred with Tie2-Cre mice (provided by Ann Moon, University of Utah), alphaMHC-Cre mice (provided by Abel Dale, University of Utah), Alb-Cre mice (purchased from The Jackson Laboratory), and Mx-Cre mice (provided by Nancy Andrews, Duke University, Durham, NC). The background of the Cre mice was C57Bl. The background of the FLP-expressing mice was a mixture of C57Bl/Sj, but had been backcrossed to BL6 for at least 3 generations. Mfrn1flox/flox mice were bred with Mfrn1+/−;Tie2-Cre mice to selectively inactivate the Mfrn1 gene in the hematopoietic and endothelial cells, with Mfrn1+/−;αMHC-Cre mice to inactivate the Mfrn1 gene in the cardiac myocytes, with Mfrn1+/−;Alb-Cre mice to inactivate the Mfrn1 gene in the hepatocytes, or with Mfrn1+/−;Mx-Cre mice to inactivate the Mfrn1 gene mainly in the liver, spleen, and bone marrow and partially in other tissues.7 Mfrn1+/−;Mx-Cre mice received 3 IP injections of 250 μg each of poly(I:C) for 5 days every 2 days to activate the Mx promoter, and were studied at least 3 days after the last injection. Tail DNA was subjected to PCR for genotyping. We used primers Mfrn1-F (as in the preceding paragraph), reverse primer Mfrn1-R3 (5′-AACCCCTCCTTAACCTTGGA-3′), which hybridizes in intron 1 near exon 2 and gives a 173-bp product with a wild-type allele and a 273-bp product with the floxed allele, and primer Mfrn1-R4 (5′-GTCTCCGGCAGCTTAAAGTG-3′), which hybridizes in intron 2 near exon 3 and gives a 428-bp product for the negative allele.
Animals
All procedures involving animals were approved by the University of Utah Animal Care Committee. All mice were born and housed in the University of Utah transgenic facility or mouse house facility. Mice were maintained on a standard rodent diet that included 350 parts per million of iron. Mice were given drinking water supplemented with 2 mg/mL of δ-ALA to bypass the rate-limiting step in hepatic porphyrin synthesis (ALA-synthase 1), as described previously.8 In all experiments, littermates from the same breeding pair were used as controls.
TAT-Cre transduction
Skin fibroblasts were isolated from Mfrn1flox/− and Mfrn1+/− mice and transduced with recombinant TAT-Cre proteins (kindly provided by Dr Matt Hockin, University of Utah) for 2 hours. Sixteen hours later, the cells were exposed to 2 mg/mL of ALA in culture medium for 8 hours, and analyzed for protoporphyrin IX content and Cre-mediated recombination.
Protoporphyrin IX and Zn-protoporphyrin measurements
Liver was homogenized at 100 mg/mL in 10mM Tris-HCl, pH 7.0. For total protoporphyrin IX measurement, lysates were extracted with ethyl acetate and acetic acid (4:1) and centrifuged for 15 minutes at 1500 rpm to remove precipitated proteins. Protoporphyrin IX was extracted in 1.5N HCl and measured in a spectrofluorometer at a 405-nm excitation wavelength and a 604-nm emission wavelength. Total serum protoporphyrin IX and washed erythrocyte protoporphyrin IX analyses were performed as described for liver total protoporphyrin IX analysis without the homogenizing step. Erythrocyte protoporphyrin was expressed as concentration per liter of packed cells. For Zn-protoporphyrin IX measurement, liver lysates were extracted with ethanol. The extracts were measured in a spectrofluorometer at a 415-nm excitation wavelength and a 589-nm (Zn-protoporphyrin) or 632-nm (protoporphyrin IX) emission wavelength. Protoporphyrin standards were purchased from Porphyrin Products.
Other procedures
Blood was collected from anesthetized animals by heart puncture into heparinized tubes. Hematocrit, hemoglobin, RBC numbers, and mean corpuscular volume (MCV) were measured. Hematoxylin-eosin and trichrome staining were performed by ALM Laboratories. Western blots were developed using Western Lightning reagent (Perkin-Elmer). Aconitase was assayed as described previously.2 A xanthine oxidase assay was carried out on liver lysates using the Amplex Red Xanthine Oxidase Assay Kit (Invitrogen) following the manufacturer's instructions. Ferrochelatase was assayed by a modification of the method used by Sellers and Dailey.9 Briefly, enzyme activity was determined by measuring the absorbance at 550 nm over time from a reaction mixture containing 1mM deuteroporphyrin, 50μM NiCl2, and the lysate of freshly isolated mitochondria. Respiratory complex activity was assayed as described by Barrientos,10 and the assay for malate dehydrogenase can be found at: https://www.mnstate.edu/provost/MDHAssayProtocol.pdf.
RT-PCR
For gene amplification, 50 ng of mRNA was used for quantitative RT-PCR using the One-Step qRT-PCR kit according to the manufacturer's instructions (Invitrogen). The primers used amplified a sequence in exon 1 (F-ACGAGAACCTACCGACCAGC) and exon 4 (R-GGCACGCAGGGTAACTGTAT). A wild-type mRNA gives rise to a transcript amplification of 443 bp; a deletion of exon 2 at the genomic level gives rise to a transcript amplification of 214 bp.
Results
Targeted disruption of Mfrn1
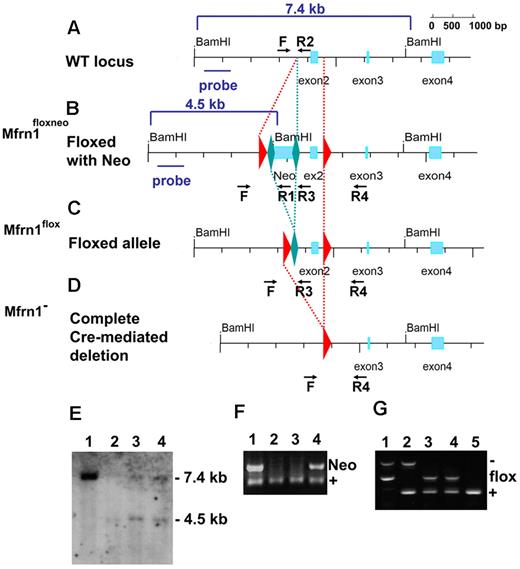
The murine Mfrn1 (Slc25a37) gene consists of 4 exons spread over more than 43 kb on mouse chromosome 14 on the reverse strand, with exon 1 far from the other 3 (Figure 1A). We designed a targeting construct in which coding exon 2 was flanked with loxP sites. The neomycin-resistance gene, flanked with FRT sites to permit its excision, was inserted in the first intron (Figure 1B). This construct was introduced by electroporation into mouse G4 ES cells (C57BL6/129 hybrid). Homologous recombination was confirmed by Southern blot analysis (Figure 1E) and PCR analysis. ES cells carrying the Mfrn1floxneo allele were injected into blastocysts to generate germline chimeric animals (Figure 1F). The neomycin cassette in the targeting construct, which was flanked with FRT sites, was removed by crossing heterozygous Mfrn1+/neo with flp recombinase mice to produce mice carrying the Mfrn1flox allele (Figure 1C,G). Deletion of exon 2 was produced by a cross of Mfrn1flox/+ mice with zygote-expressing Cre recombinase mice to produce Mfrn1+/− mice (Figure 1D,G). Mice were maintained on a C57BL/6 background and Mfrn1+/− mice appeared normal. Mice homozygous for Mfrn1flox/− showed no anatomical abnormalities (eg, in size or weight), hematologic parameters, or plasma and liver porphyrin concentration abnormalities compared with wild-type littermates (data not shown).
Generation of floxed Mfrn1 alleles. Schematic description of the wild-type Mfrn1 locus (A) and the locus after introduction of a loxP-FRT-neomycin resistance-FRT cassette in intron 1 and an additional loxP site between exons 2 and 3 (B). The position of the 5′-flanking probe used for Southern blot analysis is shown as a blue bar in panels A and B. (C) Flp recombinase–mediated excision in which the neomycin cassette was removed by FRT recombination but the coding sequence was left intact. (D) Complete Cre recombinase–mediated excision of exon 2 to inactivate the Mfrn1 gene. For panels A through D, LoxP sites are shown as red triangles and FRT sites are shown as green diamonds. (E) Southern blot analysis of the 5′ end of the locus in ES cells after BamHI digestion. Lane 1 shows the wild-type allele at 7.4 kb, and lanes 2-4 targeted recombination with a wild-type allele at 7.4 kb and a recombinant allele at 4.5 kb, as indicated in panels A and B. (F) PCR analysis of the offspring of Mfrn1floxneo/+ and Mfrn1+/+ chimeric mice. PCR was done using primers F, R1, and R2, as indicated in panels A and B. Lane 1 and lane 4 show the Mfrn1floxneo/+ genotype and lanes 2 and 3 the Mfrn1+/+ genotype. The presence of Neo (primers F × R1) gives a product of 120 bp; the absence of Neo (primers F × R2) gives a product of 60 bp. (G) PCR analysis of Cre recombinase–mediated excision of exon 2. PCR was done using primers F, R3, and R4, as indicated in panels C and D. The wild-type allele gives a product of 173 bp (primers F × R3), the flox allele gives a product of 273 bp (primers F × R3), the negative allele gives a product of 428 bp (primers F × R4), and R3 does not hybridize anymore. Lane 1 shows the Mfrn1flox/− genotype, lane 2 shows the Mfrn1+/− genotype, lanes 3 and 4 show the Mfrn1flox/+ genotype, and lane 5 shows the Mfrn1+/+ genotype.
Generation of floxed Mfrn1 alleles. Schematic description of the wild-type Mfrn1 locus (A) and the locus after introduction of a loxP-FRT-neomycin resistance-FRT cassette in intron 1 and an additional loxP site between exons 2 and 3 (B). The position of the 5′-flanking probe used for Southern blot analysis is shown as a blue bar in panels A and B. (C) Flp recombinase–mediated excision in which the neomycin cassette was removed by FRT recombination but the coding sequence was left intact. (D) Complete Cre recombinase–mediated excision of exon 2 to inactivate the Mfrn1 gene. For panels A through D, LoxP sites are shown as red triangles and FRT sites are shown as green diamonds. (E) Southern blot analysis of the 5′ end of the locus in ES cells after BamHI digestion. Lane 1 shows the wild-type allele at 7.4 kb, and lanes 2-4 targeted recombination with a wild-type allele at 7.4 kb and a recombinant allele at 4.5 kb, as indicated in panels A and B. (F) PCR analysis of the offspring of Mfrn1floxneo/+ and Mfrn1+/+ chimeric mice. PCR was done using primers F, R1, and R2, as indicated in panels A and B. Lane 1 and lane 4 show the Mfrn1floxneo/+ genotype and lanes 2 and 3 the Mfrn1+/+ genotype. The presence of Neo (primers F × R1) gives a product of 120 bp; the absence of Neo (primers F × R2) gives a product of 60 bp. (G) PCR analysis of Cre recombinase–mediated excision of exon 2. PCR was done using primers F, R3, and R4, as indicated in panels C and D. The wild-type allele gives a product of 173 bp (primers F × R3), the flox allele gives a product of 273 bp (primers F × R3), the negative allele gives a product of 428 bp (primers F × R4), and R3 does not hybridize anymore. Lane 1 shows the Mfrn1flox/− genotype, lane 2 shows the Mfrn1+/− genotype, lanes 3 and 4 show the Mfrn1flox/+ genotype, and lane 5 shows the Mfrn1+/+ genotype.
Embryonic lethality of Mfrn1−/− mice
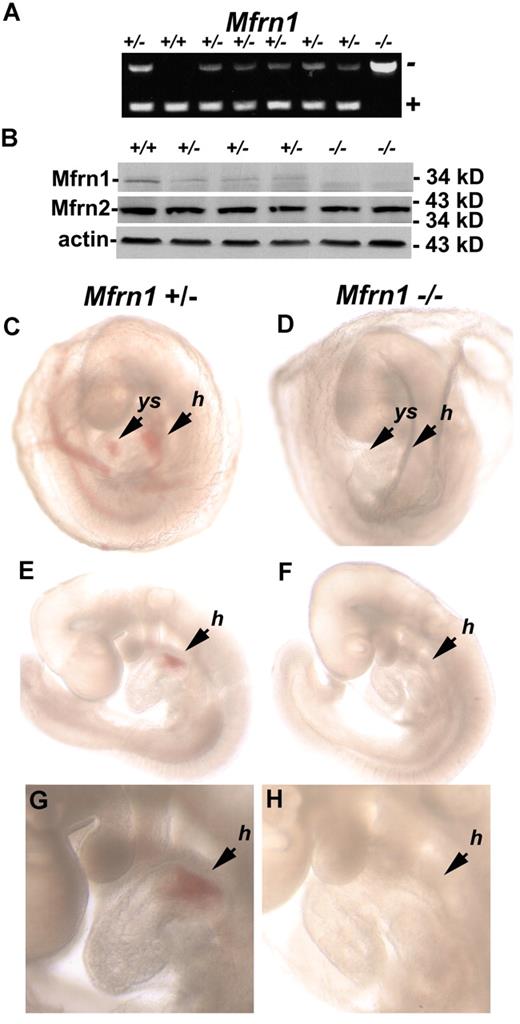
Crossing mice heterozygous for a Mfrn1 deletion (Mfrn1+/−) in all tissues did not yield any live Mfrn1−/− animals. Mendelian ratios of littermate genotypes indicated that Mfrn1−/− mice died prenatally (64 pups analyzed; Table 1). Analysis of embryos showed that embryonic lethality in Mfrn1−/− mice occurred between embryonic day 9.5 (E9.5) and E11.5. Inactivation of Mfrn1 in embryos was confirmed by PCR analysis (Figure 2A) and Western blot analysis (Figure 2B). The removal of exon 2 in the Mfrn1 locus leads to the expression of a shorter mRNA, but does not result in the accumulation of the Mfrn1 protein. Compared with wild-type embryos at E9.5 (Figure 2C,E,G), Mfrn1−/− embryos were noticeably paler (Figure 2D,F,H), with an absence of hemoglobinization in the yolk sac and heart. Mfrn1−/− embryos were not found after E11.5. These data demonstrate that it is the deletion of Mfrn1 and not the expression of the neomycin-resistance gene that leads to embryonic lethality
Embryonic lethality of due to deletion of Mfrn1
| Embryos/pups . | ≤ E9.5 (n = 37) . | ≥ E11.5 (n = 41) . | Newborn (n = 64) . |
|---|---|---|---|
| Mfrn1+/+ | 10 | 10 | 20 |
| Mfrn1+/− | 19 | 22 | 44 |
| Mfrn1−/− | 8 | 0 | 0 |
| Not determined (necrotic embryos) | 0 | 9 | 0 |
| Embryos/pups . | ≤ E9.5 (n = 37) . | ≥ E11.5 (n = 41) . | Newborn (n = 64) . |
|---|---|---|---|
| Mfrn1+/+ | 10 | 10 | 20 |
| Mfrn1+/− | 19 | 22 | 44 |
| Mfrn1−/− | 8 | 0 | 0 |
| Not determined (necrotic embryos) | 0 | 9 | 0 |
Mice (Mfrn1+/−) heterozygous for a Mfrn1 deletion were intercrossed and the progeny were genotyped.
Embryonic lethality of a homozygous Mfrn1 deletion. (A) PCR analysis of the offspring from the breeding of Mfrn1+/− with Mfrn1+/− mice. These littermates were genotyped at E9.5. In this particular litter, 1 animal was wild-type, 6 were heterozygous, and 1 was homozygous for the Mfrn1 deletion. PCR was done using primers F, R3, and R4, as indicated in the legend to Figure 1. (B) Western blot of whole embryos probed with antibodies against Mfrn1, Mfrn2, and tubulin using peroxidase-conjugated goat anti–rabbit IgG or goat anti–mouse IgG. (C-H) Stereomicroscopy of E9.5 embryos from Mfrn1+/− × Mfrn1+/− breeding. Panels C, E, and G are the Mfrn1+/− mouse; panels D, F, and H are the Mfrn1−/− mouse. Panels C and D show embryos kept in the yolk sac, panels E and F show embryos without their yolk sac, and panels G and H are enlarged images from panels E and F. The Mfrn1−/− mouse is deprived of hemoglobin, as shown by the lack of RBCs in vasculature of the yolk sac (ys) in panel D compared with panel C, and by the hemoglobinization of the heart and the common atrial chamber of the heart (h) in panels F and H compared with Mfrn1+/− mice panels E and G.
Embryonic lethality of a homozygous Mfrn1 deletion. (A) PCR analysis of the offspring from the breeding of Mfrn1+/− with Mfrn1+/− mice. These littermates were genotyped at E9.5. In this particular litter, 1 animal was wild-type, 6 were heterozygous, and 1 was homozygous for the Mfrn1 deletion. PCR was done using primers F, R3, and R4, as indicated in the legend to Figure 1. (B) Western blot of whole embryos probed with antibodies against Mfrn1, Mfrn2, and tubulin using peroxidase-conjugated goat anti–rabbit IgG or goat anti–mouse IgG. (C-H) Stereomicroscopy of E9.5 embryos from Mfrn1+/− × Mfrn1+/− breeding. Panels C, E, and G are the Mfrn1+/− mouse; panels D, F, and H are the Mfrn1−/− mouse. Panels C and D show embryos kept in the yolk sac, panels E and F show embryos without their yolk sac, and panels G and H are enlarged images from panels E and F. The Mfrn1−/− mouse is deprived of hemoglobin, as shown by the lack of RBCs in vasculature of the yolk sac (ys) in panel D compared with panel C, and by the hemoglobinization of the heart and the common atrial chamber of the heart (h) in panels F and H compared with Mfrn1+/− mice panels E and G.
To investigate the mechanism underlying embryonic lethality, we targeted the disruption of Mfrn1 in endothelial and hematopoietic cells using Tie2-Cre mice. Tie2-Cre is active in blood islands, aorta-gonad-mesonephros region, and endothelial cells starting at E7.5 and E8.5, and fetal liver hematopoietic cells and in circulating blood cells starting at E11.5.11 No live Tie2-Cre;Mfrn1flox/− pups were identified in more than 50 pups analyzed (data not shown). Because Mfrn1 is also expressed in the developing heart, we used a cardiac-specific Cre, αMHC-Cre, to determine whether Mfrn1 expression in that tissue was critical for early development. Cre expression is under the regulation of the αMHC promoter, which is expressed in cardiac myocytes starting at E9.5. αMHC-Cre, Mfrn1-deleted mice were viable and showed no phenotype up to 3 months of birth. A cross between αMHC-Cre and ROSA-LacZ mice showed that the αMHC-Cre was expressed and functional in cardiomyocytes (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). It is possible that loss of Mfrn1 in the Tie2-Cre;Mfrn1flox/− pups resulted in the dysfunction of multiple tissues, but the data are consistent in showing that it is the disruption of Mfrn1 in endothelial and/or hematopoietic cells at the early stage of development and not in cardiac myocytes that is responsible for embryonic lethality of Mfrn1−/− animals.
Mice with an induced deletion of Mfrn1 develop anemia
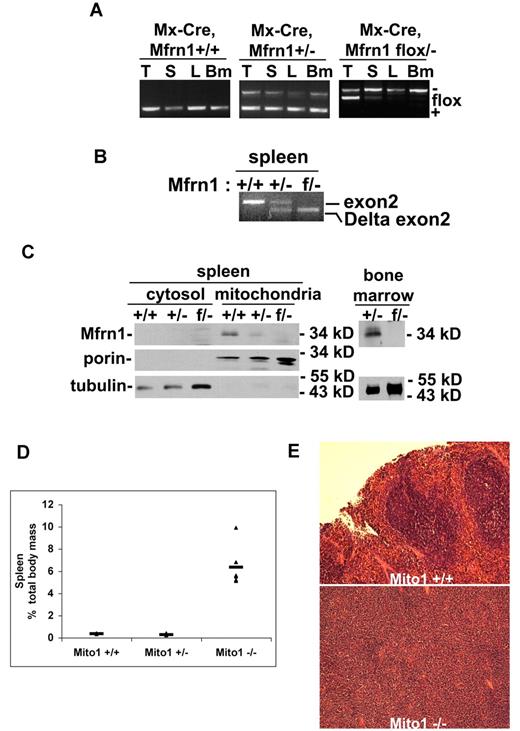
To assess the role of Mfrn1 in definitive (adult) RBC development, we used a hematopoietic Cre construct that was inducible after birth. We crossed the Mfrn1flox/− allele into mice carrying an inducible Mx-Cre, which is expressed in hematopoietic tissue. Four weeks after birth, we exposed Mfrn1flox/−;Mx-Cre mice to poly(I:C), which induces expression of Cre in most cell types, including hematopoietic precursors. Examination 4 weeks later showed that there was a significant loss of Mfrn1 in the spleen, liver, and bone marrow in Mfrn1flox/−;Mx-Cre animals exposed to the poly(I:C) inducer (Figure 3A-B). Animals not exposed to poly(I:C) showed no loss of the Mfrn1flox/− allele. Western analysis of mitochondria obtained from bone marrow or splenic cells showed a marked loss of Mfrn1 (Figure 3C). These results show an efficient deletion of Mfrn1 in erythropoietic tissues.
Deletion of Mfrn1 in adult mice leads to loss of Mfrn1 in hematopoietic tissues.Mfrn1flox/− mice were crossed to mice carrying Mx-Cre. At 3 weeks of age, mice (Mfrn1flox/−;Mx-Cre) were injected with poly(I:C) to induce Cre expression. The mice were analyzed 4-8 weeks later. (A) PCR analysis of Mfrn1 of genomic DNA from thymus (T), spleen (S), liver (L), and bone marrow (Bm) of mice with noted genotypes. (B) RT-PCR analysis of mRNA in the spleens of mice with the noted genotypes using primers specific to exon1 and exon4, which flank exon 2. A wild-type mRNA gave rise to a transcript amplification of 443 bp; a deletion of exon 2 at genomic level gave rise to a transcript amplification of 214 bp. (C) Western analysis of Mfrn1, porin, and tubulin in cytosol, mitochondria, and total bone marrow isolated from mice with the specified genotypes. (D) Bar graph of spleen weight/total weight of mice with the noted genotypes. (E). Hematoxylin and eosin–stained sections of spleen from Mfrn1+/+ mice and Mx-Cre–deleted Mfrn1−/− mice.
Deletion of Mfrn1 in adult mice leads to loss of Mfrn1 in hematopoietic tissues.Mfrn1flox/− mice were crossed to mice carrying Mx-Cre. At 3 weeks of age, mice (Mfrn1flox/−;Mx-Cre) were injected with poly(I:C) to induce Cre expression. The mice were analyzed 4-8 weeks later. (A) PCR analysis of Mfrn1 of genomic DNA from thymus (T), spleen (S), liver (L), and bone marrow (Bm) of mice with noted genotypes. (B) RT-PCR analysis of mRNA in the spleens of mice with the noted genotypes using primers specific to exon1 and exon4, which flank exon 2. A wild-type mRNA gave rise to a transcript amplification of 443 bp; a deletion of exon 2 at genomic level gave rise to a transcript amplification of 214 bp. (C) Western analysis of Mfrn1, porin, and tubulin in cytosol, mitochondria, and total bone marrow isolated from mice with the specified genotypes. (D) Bar graph of spleen weight/total weight of mice with the noted genotypes. (E). Hematoxylin and eosin–stained sections of spleen from Mfrn1+/+ mice and Mx-Cre–deleted Mfrn1−/− mice.
Analysis of bone marrow and spleen showed evidence of ineffective erythropoiesis with compensatory splenic hematopoiesis. Spleens were greatly enlarged (Figure 3D) and showed a marked change in architecture that is suggestive of increased erythropoiesis (Figure 3E). After induction by poly(I:C), there was a time-dependent change in RBC number in mice carrying the Mfrn1flox/−;Mx-Cre allele, but no significant changes in these hematologic parameters were observed in control Mx-Cre mice. Poly(I:C)–treated Mfrn1flox/−;Mx-Cre mice developed anemia, as shown by the decrease in hemoglobin concentration (Figure 4A), RBC number (Figure 4B), hematocrit (Figure 4C), and MCV (Figure 4D). FACS analysis of erythroid cells from poly(I:C)–treated Mfrn1flox/−;Mx-Cre mice showed a marked increase in erythroblasts, which represented 69% ± 4% (mean ± SEM; n = 6) of splenocytes compared with 1.9% ± 0.5% (n = 9) in poly(I:C)–treated control mice (Figure 4E-I). Splenic erythroid progenitor cells were increased by 5-fold, whereas T- and B-cell frequencies were markedly reduced (by 4-fold and 6-fold, respectively), possibly because of the relative increase in erythroblasts (data not shown). Splenic myeloid cell frequency, however, was not altered. Analysis of bone marrow showed normal numbers of cells with several notable differences in cellular distribution. The erythroblast compartment was increased by 2-fold in the deleted mice, whereas mature erythrocytes numbered 50% of control levels, which is consistent with compensatory erythropoiesis due to peripheral anemia (data not shown). Our data show that the terminal stages of erythropoiesis (TER-119+) are largely similar between control and mitoferrin 1–knockout mice, with some differences in the intensity of staining because TER-119 levels were lower and CD71 levels were higher in the knockout mice (Figure 4F). However, the early stages of erythropoiesis (TER-119−) were markedly distinct, with the knockout mice exhibiting a 10-fold increase in early erythroid progenitors (CD71+TER-119−). The data indicate that the distribution of progenitors within the TER-119− progenitor compartment with respect to CD71 and c-kit (Figure 4G) showed a 10-fold increase in primitive erythroid progenitors, which includes both c-kit+ and c-kit− progenitors. The frequency of nonviable cells was increased by an average of 2- to 3-fold in both spleen and bone marrow (Figure 4H). These results confirm that Mfrn1 is essential for hemoglobinization in developing adult erythrocytes. The frequency of B-lineage cells in bone marrow was reduced 4-fold, with no reduction in myeloid cell frequency (Figure 4I). Coupled with the decrease in splenic B-cell frequency, this result suggests that Mfrn1 may also play an important role in B-cell development.
Changes in hematologic values after deletion of Mfrn1 in MX-Cre mice. Mice with the noted genotypes were injected with poly(I:C) to induce Cre recombinase. At times subsequent to induction of Cre, blood was taken for analysis of hemoglobin (HGB; A), RBC number (B), hematocrit (C), and MCV (D). Flow cytometric analysis of hematopoiesis in Mfrn1-knockout mice. Groups of Mx-Cre (Control) or floxed Mfrn1/Mx-Cre mice were treated with poly(I:C) and killed 8 weeks later for analysis of hematopoietic tissues. (E) Spleen cell suspensions were treated with ammonium chloride to lyse mature erythrocytes and evaluated for expression of the erythroid marker TER-119, which indicates erythroblast cells. (F-I) Bone marrow cell suspensions were isolated and evaluated for hematopoietic markers as indicated. (F) Selective gating for the hematopoietic stem and progenitor cell compartment (viable cells lacking expression of the lineage markers B220 and CD11b). Mfrn1-deleted mice exhibited increased numbers of erythroid progenitor cells (CD71+TER-119−). (G) Selective gating to exclude B220, CD11b, and TER-119+ cells. It can be seen that Mfrn1-deleted mice exhibit an increase in erythroid progenitor cells (c-kit+CD71+) and proerythroblasts (c-kit−CD71+). We also observed increased numbers of nonviable CD71+ cells (DAPI+CD71+; H) and decreased numbers of B-lineage cells based on B220 expression (I) in Mfrn1-deleted mice. Panels are representative of 6 Mfrn1-deleted mice and 9 control animals.
Changes in hematologic values after deletion of Mfrn1 in MX-Cre mice. Mice with the noted genotypes were injected with poly(I:C) to induce Cre recombinase. At times subsequent to induction of Cre, blood was taken for analysis of hemoglobin (HGB; A), RBC number (B), hematocrit (C), and MCV (D). Flow cytometric analysis of hematopoiesis in Mfrn1-knockout mice. Groups of Mx-Cre (Control) or floxed Mfrn1/Mx-Cre mice were treated with poly(I:C) and killed 8 weeks later for analysis of hematopoietic tissues. (E) Spleen cell suspensions were treated with ammonium chloride to lyse mature erythrocytes and evaluated for expression of the erythroid marker TER-119, which indicates erythroblast cells. (F-I) Bone marrow cell suspensions were isolated and evaluated for hematopoietic markers as indicated. (F) Selective gating for the hematopoietic stem and progenitor cell compartment (viable cells lacking expression of the lineage markers B220 and CD11b). Mfrn1-deleted mice exhibited increased numbers of erythroid progenitor cells (CD71+TER-119−). (G) Selective gating to exclude B220, CD11b, and TER-119+ cells. It can be seen that Mfrn1-deleted mice exhibit an increase in erythroid progenitor cells (c-kit+CD71+) and proerythroblasts (c-kit−CD71+). We also observed increased numbers of nonviable CD71+ cells (DAPI+CD71+; H) and decreased numbers of B-lineage cells based on B220 expression (I) in Mfrn1-deleted mice. Panels are representative of 6 Mfrn1-deleted mice and 9 control animals.
Targeted disruption of the Mfrn1 gene in hepatocytes
To determine whether there is a role for Mfrn1 in tissues other than hematopoietic cells, we focused on hepatocytes because these cells show a high demand for heme. Mfrn1flox/− mice were crossed with mice containing an integrated Cre gene driven by the albumin promoter (Alb-Cre). To confirm that Alb-Cre was expressed and functional, we crossed Alb-Cre mice with Rosa-LacZ mice and examined the livers and spleens for the expression of β-galactosidase. No expression of β-galactosidase was seen in the spleens of Alb-Cre;Rosa-LacZ mice, whereas abundant β-galactosidase activity was seen in the hepatocytes of these mice (supplemental Figure 2A). We note that the deletion was not complete, perhaps reflecting the presence of nonhepatocyte cell types in the liver. We checked the efficacy of disruption by PCR analysis of genomic DNA from the livers of Mfrn1flox/− and Alb-Cre;Mfrn1flox/− mice (supplemental Figure 2B), and were able to demonstrate reduced levels of the floxed allele of Mfrn1 due to Alb-Cre. Examination of mRNA by RT-PCR using primers specific for Mfrn1 mRNA exon1 and exon 4 showed the appearance of the mRNA lacking exon 2 in the liver of an Alb-Cre;Mfrn1flox/− mouse compared with the spleen from the same animal (supplemental Figure 2C). The level of Mfrn1 protein in normal liver was too low to be detected by Western blots using the current antibodies. Loss of Mfrn1, however, did not affect the level of Mfrn2 mRNA or Mfrn2 protein (supplemental Figure 2D). The loss of Mfrn1 in hepatocytes had no effect on viability or on other measured parameters of iron and heme metabolism (data not shown).
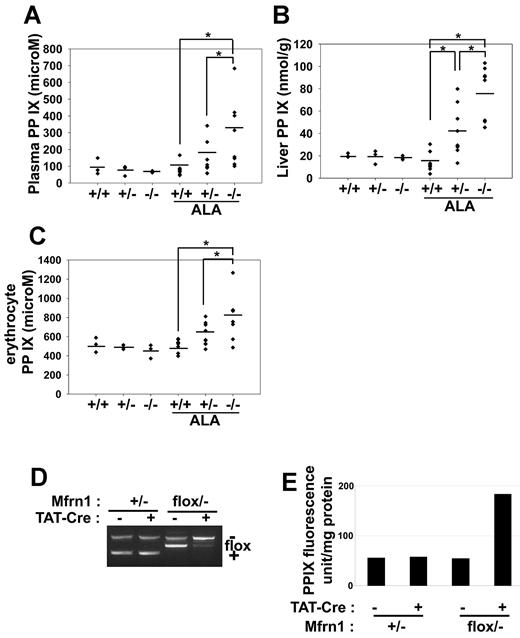
We next examined whether there was a role for Mfrn1 under conditions of increased heme synthesis. Mice were given ALA in their drinking water for 4 weeks. ALA bypasses the rate-limiting step in porphyrin biosynthesis and is expected to lead to increased porphyrin synthesis. Wild-type mice fed ALA showed little evidence of increased porphyrin intermediates in the urine, liver, or blood. In contrast, higher levels of protoporphyrin IX were seen in the plasma (Figure 5A), liver (Figure 5B), and RBCs (Figure 5C) of ALA-fed Alb-Cre;Mfrn1flox/− mice. Mice heterozygous for hepatocyte Mfrn1 (Alb-Cre;Mfrn1flox/+) also had higher levels of protoporphyrin than control mice but lower levels than homozygous deleted mice. Iron deficiency often leads to increased levels of Zn-protoporphyrin,12 but we did not observe increased Zn-protoporphyrin in tissue, RBCs, or plasma (data not shown). Further, we did not observe significant changes in the activity of the cytosolic iron-sulfur cluster–containing enzyme xanthine oxidase, the mitochondrial iron-sulfur–containing succinate dehydrogenase or ferrochelatase, respiratory complex III, or the heme-containing respiratory complex IV (supplemental Figures 3-4). The changes in the activities of these enzymes were minimal and were on the same order of magnitude as changes in the activity of the iron-free mitochondrial enzyme malate dehydrogenase. We did observe changes in the activities of mitochondrial and cytosolic aconitase. The fact that the activity of other mitochondrial and cytosolic iron-sulfur–containing enzymes were not altered suggests that, in general, the synthesis of iron-sulfur clusters was not affected by the loss of Mfrn1 in hepatocytes.
Deletion of Mfrn1 in hepatocytes leads to increased protoporphyrin IX in mice fed ALA. ALA was added to the drinking water of mice of the noted genotypes. Four weeks later, the ALA-fed mice and control mice were killed and the level of protoporphyrin IX (PPIX) measured in plasma (A), liver (B), and erythrocytes (C). (D) PCR analysis of Mfrn1 deletion in fibroblasts incubated with Tat-Cre. (E) Levels of PPIX in wild-type and Mfrn1-deleted fibroblasts. *P < .05.
Deletion of Mfrn1 in hepatocytes leads to increased protoporphyrin IX in mice fed ALA. ALA was added to the drinking water of mice of the noted genotypes. Four weeks later, the ALA-fed mice and control mice were killed and the level of protoporphyrin IX (PPIX) measured in plasma (A), liver (B), and erythrocytes (C). (D) PCR analysis of Mfrn1 deletion in fibroblasts incubated with Tat-Cre. (E) Levels of PPIX in wild-type and Mfrn1-deleted fibroblasts. *P < .05.
We confirmed that the increase in protoporphyrin IX was the result of decreased expression of Mfrn1. Cultured fibroblasts from Mfrn1flox/− mice were incubated with TAT-Cre, which induced loss of the floxed allele in greater than 90% of cultured cells (Figure 5D). The addition of ALA to wild-type or Mfrn1flox/− cells did not lead to increased protoporphyrin IX levels (Figure 5E). In contrast, incubation of Tat-Cre–treated Mfrn1flox/− cells with ALA led to a 4-fold increase in protoporphyrin IX levels.
Deletion of hepatocyte Mfrn1 in ALA-fed animals results in hepatotoxicity
We noted that deletion of hepatocyte Mfrn1 in ALA-fed animals affected the size of the liver. There was no difference in liver size between wild-type animals and hepatocyte Mfrn1-deleted animals in the absence of added ALA (Figure 6A). Further, ALA feeding had no effect on liver weight in wild-type animals compared with wild-type animals not fed ALA. In contrast, ALA-fed animals showed an increase in liver size that was graded with the loss of Mfrn1, because the homozygous deletion showed a greater effect than the heterozygous deletion. We also noted a small increase in spleen size in homozygous Mfrn1 hepatocyte–deleted animals fed ALA (Figure 6B).
Hepatotoxicity in livers of hepatocyte-deleted Mfrn1 animals fed ALA. ALA was added to the drinking water of mice of the noted genotypes (Mfrn1+/+, Mfrn1flox/+, and Alb-Cre;Mfrn1flox/−). Four weeks later, the ALA-fed mice and control mice were killed and liver weight/body weight (A) and spleen weight/body weight (B) were determined. *P < .05.
Hepatotoxicity in livers of hepatocyte-deleted Mfrn1 animals fed ALA. ALA was added to the drinking water of mice of the noted genotypes (Mfrn1+/+, Mfrn1flox/+, and Alb-Cre;Mfrn1flox/−). Four weeks later, the ALA-fed mice and control mice were killed and liver weight/body weight (A) and spleen weight/body weight (B) were determined. *P < .05.
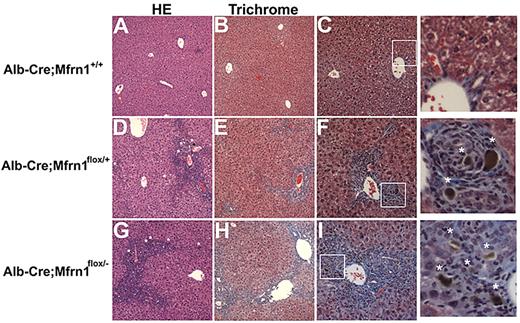
Histochemical analysis of the livers from ALA-fed, hepatocyte-deleted (Alb-Cre;Mfrn1flox/−) mice revealed pathologic changes that were not seen in either ALA-fed normal mice (Figure 7) or hepatocytes of Alb-Cre;Mfrn1flox/− mice not fed ALA (data not shown). Most notably was the presence of increased collagen deposition in the Alb-Cre;Mfrn1flox/+ heterozygotes and the Alb-Cre;Mfrn1flox/− homozygous mouse livers. Livers from heterozygous mice showed bile duct proliferation in the periportal area and chronic periportal inflammation. A brown pigment was also seen in the bile ducts of the ALA-fed Alb-Cre;Mfrn1flox/− mice that was not seen in normal animals fed ALA. This pigment was consistent with accumulated bile, suggesting cholestasis. Heterozygous mice showed a chronic cholestatic pattern and early fibrosis compared with wild-type mice. The homozygous mice showed a more severe pattern with a more reactive epithelium and lymphocytes in the lobule area. Fibrosis was also more developed, bridging periportal and centrolobular areas and starting to form nodules.
Histochemical analysis of wild-type and hepatocyte-deleted Mfrn1 livers. Livers from mice fed ALA were harvested at 6 weeks and stained with hematoxylin and eosin (A) or trichrome (B) in Alb-Cre;Mfrn1+/+. (C) Enlarged image from panel B. Alb-Cre;Mfrn1flox/+ were stained with hematoxylin and eosin (D) or trichrome (E). Panel F is an enlarged image from panel E. Alb-Cre;Mfrnlflox/− were stained with hematoxylin and eosin (G) or trichrome (H). Panel I is an enlarged image from panel G. Asterisks in the enlarged images of panels F and I denote areas of fibrosis.
Histochemical analysis of wild-type and hepatocyte-deleted Mfrn1 livers. Livers from mice fed ALA were harvested at 6 weeks and stained with hematoxylin and eosin (A) or trichrome (B) in Alb-Cre;Mfrn1+/+. (C) Enlarged image from panel B. Alb-Cre;Mfrn1flox/+ were stained with hematoxylin and eosin (D) or trichrome (E). Panel F is an enlarged image from panel E. Alb-Cre;Mfrnlflox/− were stained with hematoxylin and eosin (G) or trichrome (H). Panel I is an enlarged image from panel G. Asterisks in the enlarged images of panels F and I denote areas of fibrosis.
Discussion
Previous studies and those reported here show that Mfrn1 is essential for heme synthesis in developing erythroid cells.4,5 The loss of Mfrn1 in mouse embryos results in lethality and, as shown here, the loss of Mfrn1 in adults results in severe anemia. Our data show that it is the specific loss of Mfrn1 in erythroid cells that leads to anemia, because deletion of Mfrn1 in (definitive) hematopoietic stem cells in adult mice resulted in a severe anemia. Our data also indicate a decrement in the number of B cells, suggesting that Mfrn1 may play a role in B-cell lineage development. Studies are ongoing to test that hypothesis.
We then focused on the effect of deletion of Mfrn1 in hepatocytes because these cells have a high demand for heme due to the synthesis of heme containing P450. The original description of mitoferrins showed that Mfrn1 was expressed in hepatocytes, although (on the mRNA level) to a lesser extent than Mfrn2. Loss of hepatocyte Mfrn1 has no consequences either in terms of protoporphyrin accumulation or pathology in mice maintained on standard laboratory diets. In developing erythroid cells, the turnover rate of Mfrn1 is affected by the presence of Abcb10, which stabilizes Mfrn1 and increases its half-life. There is also evidence of Abcb10 transcripts in the liver, although it is not clear whether Abcb10 has an effect on hepatocyte Mfrn1 turnover.13 The deletion of Mfrn1 does not affect the level of Mfrn2 transcripts, which do not interact with Abcb10.6
Changes in pathology and porphyrin levels were only seen when porphyrin synthesis was increased in hepatocyte Mfrn1–deleted mice. Under these conditions, the hepatocyte Mfrn1–deleted mice showed a liver phenotype indistinguishable from erythrocyte protoporphyria (EPP), with increased protoporphyrin IX and a liver pathology of bridging cirrhosis with apparent cholestasis. EPP most commonly results from mutations in ferrochelatase, the enzyme that catalyzes the final step in the heme-biosynthetic pathway. Decreased ferrochelatase activity would result in decreased iron insertion into the protoporphyrin IX macrocycle, leading to the increased accumulation of protoporphyrin IX. Thus, conditions that result in a relative increase in porphyrin synthesis relative to the ability to insert iron into the porphyrin ring would result in increased protoporphyrin IX levels. Such conditions include the increased expression of ALA synthase due to homozygous deletions in mouse IRP214 or mutations in the carboxyl-terminal end of ALA synthase, which modulates enzyme activity.15 In mice with mutations in ferrochelatase16,17 or a deletion in IRP2,14 increased levels of protoporphyrin IX result predominately from increased protoporphyrin synthesis in erythrocytes. When normal bone marrow is transplanted into ferrochelatase mutant mice, the ferrochelatase defect is retained in liver cells. In all existing models of EPP, defects in either ferrochelatase activity or ALA synthase are the most predominant in the erythroid lineage, although ferrochelatase deficiencies are expressed in erythroid precursors and in the liver. Bone marrow transplantation “normalizes” erythroid ferrochelatase activity but not hepatocyte ferrochelatase activity. It is unclear whether the liver pathology seen in transplanted mouse EPP models reflects preexisting pathology or pathology due to the continued production of protoporphyrin IX by the defective liver enzyme.18
As shown here, decreased hepatocyte Mfrn1 levels lead to increased protoporphyrin IX when heme synthesis is expanded by addition of ALA. The finding that protoporphyrin IX levels increase indicates that Mfrn2-mediated iron delivery is not sufficient to accommodate the increase in porphyrin demand. This result confirms studies in cultured cells using RNA interference to lower the levels of Mfrn1 or Mfrn2.5 Reduction in either Mfrn1 or Mfrn2 had no effect on heme synthesis or porphyrin accumulation under normal conditions. In the presence of ALA, depletion of Mfrn1 or Mfrn2 resulted in increased protoporphyrin IX, indicating that mitochondrial iron delivery was insufficient to catalyze iron incorporation into the porphyrin macrocycle. The expression of Mfrn2 in Mfrn1-silenced cells or vice versa normalized protoporphyrin levels, indicating that the accumulation of protoporphyrin IX was due to iron limitation into mitochondria. These findings indicate that the activity or level of Mfrn2 cannot increase in response to heme demand or Mfrn1 deficiency.
Our most striking finding was the appearance of liver fibrosis and cirrhosis that were dependent on the copy level of Mfrn1, with heterozygotes and homozygotes showing pathology and normal mice not showing pathology. Mutations in ferrochelatase affect the enzyme in both hepatocyte and erythroid precursors, leading to protoporphyria and liver pathology. Reconstitution of normal bone marrow in mutant animals did not completely suppress liver pathology. Bone marrow transplantation, while robust (90%), was not complete, and the failure to suppress cirrhosis might reflect protoporphyrin IX generation by hepatocytes and/or by the residual host mutant RBCs. The results presented here show that defects in hepatocyte porphyrin production can itself lead to hepatic toxicity, suggesting that liver-specific defects can result in protoporphyria.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank our colleagues Dr Matt Hockin, Anne Moon, Dale Abel, and Nancy Andrews for their gracious sharing of reagents and mouse strains. ES cell manipulations and blastocyst injections were carried out by the University of Utah transgenic facility.
This work was supported by National Institutes of Health grants R01 DK070838 and P01 HL032262 to B.H.P. and by National Institutes of Health grant DK052380 to J.K.
National Institutes of Health
Authorship
Contribution: M.-B.T. designed and performed research and wrote the paper; D.W. and J.W. performed experiments; K.T. designed strategy, analyzed data, and assisted in manuscript preparation; G.J.S performed FACS analysis; J.P. performed porphyrin analysis; O.K. performed mitochondrial enzyme assays; B.H.P. analyzed data; D.M.W. analyzed data and wrote the paper; and J.K. designed research, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for M.-B.T. is Institut de Génétique et Développement de Rennes, Faculté de Médecine, Rennes, France.
Correspondence: Jerry Kaplan, University of Utah School of Medicine, Dept of Pathology, 50 North Medical Dr, 5C124, Salt Lake City, UT 84132-2501; e-mail: jerry.kaplan@path.utah.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal